Abstract
The human chromokinesin Kid/kinesin-10, a plus end-directed microtubule (MT)-based motor with both microtubule- and DNA-binding domains, is required for proper chromosome alignment at the metaphase plate. Here, we performed RNA interference experiments to deplete endogenous Kid from HeLa cells and confirmed defects in metaphase chromosome arm alignment in Kid-depleted cells. In addition, we noted a shortening of the spindle length, resulting in a pole-to-pole distance only 80% of wild type. The spindle microtubule-bundles with which Kid normally colocalize became less robust. Rescue of the two Kid deficiency phenotypes—imprecise chromosome alignment at metaphase and shortened spindles— exhibited distinct requirements. Mutants lacking either the DNA-binding domain or the MT motor ATPase failed to rescue the former defect, whereas rescue of the shortened spindle phenotype required neither activity. Kid also exhibits microtubule bundling activity in vitro, and rescue of the shortened spindle phenotype and the bundling activity displayed similar domain requirements, except that rescue required a coiled-coil domain not needed for bundling. These results suggest that distinct from its role in chromosome movement, Kid contributes to spindle morphogenesis by mediating spindle microtubules stabilization.
INTRODUCTION
The mitotic spindle is a highly dynamic molecular machine composed of tubulin, molecular motors, and other microtubule (MT)-associated protein complexes. At the onset of mitosis, the interphase microtubule network disassembles, and mitotic microtubules reassemble around condensed chromatin and the two centrosomes. During this process, multiple proteins associate with the mitotic spindle and play essential roles in spindle assembly and function (Sharp et al., 2000; Karsenti and Vernos, 2001; Scholey et al., 2003). Although MT dynamics and motor proteins are thought to be involved, the mechanisms by which spindle length is controlled remain elusive. Among models proposed are force balance models that highlight the role of MT polymerization dynamics (Inoue and Sato, 1967; Mitchison et al., 1986; Scholey et al., 2001), the action of motor proteins (Sharp et al., 2000; Nedelec, 2002; Cytrynbaum et al., 2003), or the “spindle matrix,” a hypothetical, non-MT, tensile element (Scholey et al., 2001). Other types of models can be grouped in that they seek to explain spindle length with the microtubule system, including dynamics regulators, motors, and cross-linkers, as the sole mechanochemical element (Gruss and Vernos, 2004).
The human chromokinesin Kid/Kinesin-10 is a plus end-directed MT-based motor (Shiroguchi et al., 2003; Yajima et al., 2003) that binds both MTs and chromosomes (Tokai et al., 1996). Kid has been reported to play an important role in chromosomal movement along MTs during prometaphase and metaphase. Xkid, a Xenopus homologue of Kid, is essential for chromosome alignment (Antonio et al., 2000; Funabiki and Murray, 2000), and human Kid is necessary for orientation of chromosome arms and chromosome oscillation (Levesque and Compton, 2001). Kid colocalizes with spindles and chromosomes during the period from prophase through metaphase, and recent studies revealed that Kid contains a second MT-binding site (Ohsugi et al., 2003; Shiroguchi et al., 2003) whose affinity for MT is controlled by Cdc2-mediated phosphorylation (Ohsugi et al., 2003). In addition, Kid together with nuclear mitotic apparatus (NuMA) protein directs spindle formation (Levesque et al., 2003). Another report showed that Ran, a GTPase in the Ras superfamily, modulates the affinity of Kid for MTs (Trieselmann et al., 2003). Ran is proposed to be a spatial regulator of spindle–MT assembly, which maintains spindle assembly factors in their active states near the chromatin (Kahana and Cleveland, 1999; Wiese et al., 2001; Gruss et al., 2002). The findings suggest the possibility that Kid is involved in spindle assembly. Although previous reports implicate Kid in chromosome movement along the spindle, the functions of Kid on spindle MTs remain unknown. In the present study, we depleted Kid from HeLa cells using RNA interference (RNAi) to investigate what functions Kid may play over the course of mitosis. Our results demonstrate that in addition to its role in metaphase chromosome movement, Kid is responsible for maintenance of the length of spindle MTs.
MATERIALS AND METHODS
RNA Interference
HeLa cells were cultured in DMEM with 10% fetal calf serum. RNAi experiments were carried out with small interfering RNA (siRNA) as described previously (Elbashir et al., 2002). Target sequences were 5′-AAGATTGGAGCTACTCGTCGT-3′ for Kid and 5′-AAGCGCGCTTTGTAGGATTCG-3′ for control Euglena gracilis chloroplast DNA. Synthetic siRNAs (JbioS, Tokyo, Japan) were transfected into cells with Oligofectamine (Invitrogen, Karlsruhe, Germany).
Immunocytochemistry
For immunofluorescence microscopy, cells were fixed in methanol and stained with primary antibodies (i.e., anti-Kid [Tokai et al., 1996], anti-α-tubulin [Sigma-Aldrich, St. Louis, MO], anti-γ-tubulin [Sigma-Aldrich], and kinetochore-specific CREST serum provided by Y. Takasaki [Tokyo Medical and Dental University, Tokyo, Japan]) and secondary antibodies coupled to Alexa488, Cy3, or fluorescein isothiocyanate (Invitrogen; Jackson ImmunoResearch Laboratories, West Grove, PA; or MP Biomedicals, Irvine, CA). DNA was stained with Hoechst33342 (Invitrogen). Immunofluorescence images were collected on a DeltaVision system (Applied Precision, Issaquah, WA) and subsequently analyzed by iterative constrained deconvolution.
Immunoblotting was performed as described previously (Tokai et al., 1996) with antibody against NuMA (NeoMarkers, Fremont, CA) and other antibodies described for indirect immunofluorescence.
For spindle intensity quantification, 48 h after the addition of the siRNA, cells were transferred onto ice for 10 min and fixed. Cells were stained as described above. Because fluorescence levels often varied among different coverslips, we selected areas that contained at least one completely depleted cell and a cell that retained normal Kid levels. Each mitotic cell was optically sectioned using the DeltaVision system. The three-dimensional data sets (z-series of 5 face optical sections in 1-μm increments) were projected onto a single image plane. The fluorescence intensity within the spindle area was measured by NIH Image 1.62, and the background intensity was subtracted. The resulting intensity value was defined as the spindle intensity. The average of spindle intensities in nondepleted cells was set to 1.
Laser Scanning Cytometry (LSC)
Cells were fixed in ethanol and stained with primary antibodies (i.e., anti-Kid [Tokai et al., 1996] and anti-α-tubulin [Sigma-Aldrich]) and secondary antibodies conjugated with Alexa488 or Cy3 (Invitrogen or Jackson ImmunoResearch Laboratories). DNA was stained with propidium iodide (PI). Cell fluorescence was measured by LSC (LSC2; CompuCyte, Cambridge, MA) as described previously (Luther and Kamentsky, 1996; Sasaki et al., 1996). For each cell, we measured DNA content and examined the Alexa488 fluorescence profile for Kid expression. At least 7000 cells were measured per sample. The experiments were repeated at least four times.
Plasmid Construction and Transfection
To prepare plasmids for expression of Kid-rescue (Kidr) mutants, nucleotide sequence 124–133 base pairs (GGA GCT ACT) of Kid were changed to GGT GCA ACG. Site-directed mutagenesis was performed by PCR (Ohsugi et al., 2003). Portions of the Kid cDNA corresponding to amino acids 1–515, 1–462, and 1–388 (for Kid-delDB, Kid-delC, and Kid-motor, respectively) were cloned into the mammalian expression vector pME18SMyc (Tokai et al., 1996) and pGEX-2T (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). HeLa cells were transfected with siRNAs and then synchronized by double thymidine block. pME18SMyc-Kidr vectors were transfected using LipofectAMINE Plus (Invitrogen) during the interval between the first and second thymidine blocks. Cells were released into fresh medium for 12 h and analyzed by indirect immunofluorescence.
Quantitation of Protein Expression
Control cells and cells rescued with the Myc-tagged Kid constructs were fixed and immunostained with anti-Myc antibody to detect exogenous protein and with anti-Kid antibody to detect all Kid protein. The efficiency of RNAi was confirmed to be >90% for each rescue experiment. Five control cells and five Myc-positive cells showing normal localization of Myc-Kid-WT protein were randomly selected and the amount of exogenous Myc-Kid-WT expressed in rescued cells was compared with that of endogenous Kid in control cells by quantifying the fluorescence intensity of anti-Kid and anti-Myc using NIH Image 1.62. The average fluorescence intensity of endogenous Kid protein was defined as 1 Kid expression unit. The amount of exogenous Kid in cells showing normal localization fell within the range of 0.5–1.1 Kid expression units.
Normal spindle localization of Kid was observed in cells expressing low levels of exogenous Kid-mutants. In contrast, exogenous Kid-mutants localized unusually near centrosomes in cells expressing high levels of exogenous protein. The average fluorescence intensity corresponding to Myc antibody in cells showing spindle localization of Kid was defined as 1. The fluorescence intensity corresponding to Myc antibody in cells showing abnormal localization fell into a range between 3 and 20. Therefore, for rescue experiments, we selected cells showing fluorescence intensity within a range of 0.6–1.4.
MT Cross-linking Assay
MT cross-linking assay were performed as reported previously (Shiroguchi et al., 2003). Tubulin (prepared by Y. Y. Toyoshima, University of Tokyo, Tokyo, Japan) labeled with rhodamine (Invitrogen) was polymerized as described previously. For the purification of glutathione S-transferase (GST) fusion proteins expressed in Escherichia coli [BL21Star(DE3)], a high-speed supernatant was adsorbed to glutathione-agarose (Sigma-Aldrich) and eluted. GST fusion protein of Kid mutant (75 nM) was mixed with MTs (150 nM) in the assay buffer, incubated for 2 min at 25°C, and then observed using a DeltaVision system (Applied Precision)
Statistical Analysis
Statistical analyses were achieved with paired t test by using StatView J (Abacus Concepts, Berkeley, CA). A p value <0.05 was considered significant.
RESULTS
Suppression of Kid by RNAi Leads to Misaligned Chromosome Arms and Malformed Nuclei
HeLa cells were transfected with siRNA-Kid, a small interfering RNA duplex targeting a region 94 base pairs downstream of the first nucleotide of the start codon of the human Kid mRNA. Western blot analysis of lysates from siRNA-Kid-transfected cells confirmed that, by 48 h after transfection, the amount of Kid was reduced to ∼3% of that in control cells (Figure 1A). Levels of other proteins, such as dynein, NuMA, and tubulin, were unaffected by siRNA-Kid.
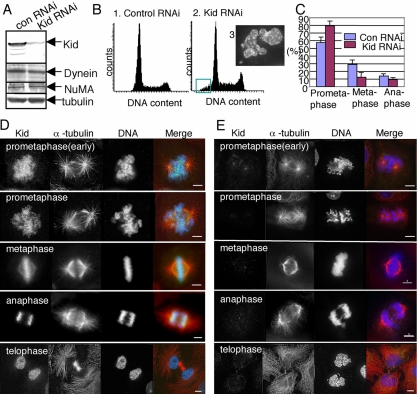
Figure 1.
Effects of Kid RNAi on HeLa cells. (A) Reduction of Kid expression after siRNA-Kid transfection. HeLa cell lysates prepared 48 h after transfection with either siRNA-Kid (Kid RNAi) or siRNA-control (con RNAi) were analyzed by Western blotting with antibodies against Kid, dynein, NuMA, and α-tubulin (internal control). (B) DNA contents histograms for control (1) and Kid-depleted (2) cells analyzed by LSC at 72 h after transfection. The green box in histogram 2 indicates cells with a DNA content <2n. Subpanel 3 shows a micrograph of typical cells that fall within the green box in histogram 2. (C) Bar graph. Quantification of mitotic states in control and Kid-depleted cells. One hundred fifty mitotic cells were scored from each of four independent experiments. Error bars represent the SD. (D and E) Immunofluorescence images of control (D) and Kid-depleted (E) cells. Forty-eight hours after transfection, cells were fixed and stained to reveal the distribution of Kid (green), α-tubulin (red), and DNA (blue). Bars, 5 μm.
Analysis with LSC revealed that the profile of the number of cells at different stages of the cell cycle did not differ appreciably between the Kid-depleted cells and control cells (Figure 1B, 1 and 2). Therefore, cell cycle progression per se was not grossly affected by Kid depletion. Nevertheless, the histogram suggests some enrichment of mitotic cells. We also noticed that >5% of transfected cells had a DNA content of <2n (Figure 1B, 2) 3 d after transfection of siRNA-Kid. Morphological observation of cells of interest by LSC revealed that most of the cells with less than 2n DNA content (Figure 1B, 2, boxed area) contained satellite or multiple nuclei, which were probably the consequence of lagging chromosomes (Figure 1B, 3). Nevertheless, we observed severe chromosome segregation or nucleus reformation abnormalities in only a minority of treated cells. Functional redundancy or residual Kid activity may explain the continued ability of most cells to divide normally.
We next quantified frequency of each mitotic phase of Kid-depleted cells (Figure 1C) and found that 80% of Kid-depleted mitotic cells showed a prometaphase-like morphology, whereas only 60% of control mitotic cells showed a similar morphology. The data suggest delay of the cell cycle during prometaphase. Then, we examined the spindle and chromosome morphology of the Kid-depleted mitotic cells in greater detail. In control prometaphase and metaphase cells, Kid was distributed along the mitotic chromosomes and spindles (Figure 1D). In contrast, in Kid-depleted cells, we detected little or no immunostaining of Kid (Figure 1E). Kid-depleted prometaphase cells had condensed chromosomes and two MT asters that were clearly separated from each other. In metaphase cells, the chromosome arms frequently failed to align properly at the metaphase plate. The data confirm a critical role of Kid in generating the polar ejection force.
On entry into anaphase, Kid is enriched at the spindle pole proximal side of the chromosomes in control cells (Figure 1D). In Kid-depleted cells, a lagging chromosome phenotype was observed at the onset of anaphase (Figure 1E). Moreover, although cytokinesis and nuclear reassembly had occurred, the resulting nuclei in the daughter cells showed irregular profiles (Figure 1E, bottom). Further studies are needed to understand the abnormalities in anaphase chromosome movement and daughter nucleus configuration in the absence of Kid.
Kid Is Required for Maintenance of Normal Spindle Size
The Kid-depleted cells were stained with anti-γ-tubulin antibody for centrosomes and cells with both centrosomes in one focal plane were subjected to the analysis. In Kid-depleted HeLa cells, although the bipolar spindles formed, the distance between the two centrosomes was reduced to ∼80% of that in control cells (Figure 2A). The short spindle phenotype was also observed in Kid-depleted U2OS cells (our unpublished data). We further tested the effects of Kid depletion on monopolar spindles induced by monastrol, an inhibitor of the mitotic kinesin Eg5 (Mayer et al., 1999). To do so the monastrol-treated cells were stained with anti-γ-tubulin antibody and CREST serum for centrosomes and kinetochores, respectively. The average distance between the centrosome and kinetochores for monopolar spindles in Kid-siRNA-transfected cells was also reduced to ∼80% of that in control cells (Figure 2, B and C).
Figure 2.
Suppression of Kid expression decreases spindle size. (A) Bar graph shows the average distance between two centrosomes in siRNA-control (con RNAi)-treated cells, siRNA-Kid (Kid RNAi)-treated cells, and siRNA-Kid-treated HeLa cells transfected with RNAi-resistant Kid-WT construct (Kid). Cells were fixed and stained with antibodies against Myc-tag and γ-tubulin, and the pole-to-pole distance was measured. Twenty metaphase cells were scored in each of five independent experiments. Error bars represent the SD. *p < 0.0005. (B) immunofluorescence images of monopolar HeLa cell. Kid-depleted (Kid RNAi) and control (con RNAi) HeLa cells were treated with 100 μM monastrol for 2 h, fixed, and processed for immunofluorescence staining with antibodies against γ-tubulin (green) and kinetochore-specific CREST (red). DNA was visualized with Hoechst33342 stain (blue in the right panels). (C) Histograms of the average distances between centrosomes and kinetochores monopolar spindles obtained from siRNA-Kid- (Kid RNAi) and siRNA-control (con RNAi)-transfected monopolar cells. The average value of 10 centrosome-to-kinetochore distances was calculated for each spindle. Each bar value, in turn, reflects the average of five spindle values. Error bars represent the SD. *p < 0.0005. (D) Immunofluorescence analysis of Kid-depleted HeLa cells demonstrating that NuMA localization is unaffected. Cells were fixed, and stained with antibodies against Kid (green) and NuMA (red). DNA (blue) was visualized with Hoechst stain.
To verify that the observed decrease in spindle size was due specifically to loss of Kid, we performed a rescue experiment. We constructed a Kidr cDNA containing three silent mutations within the siRNA target sequence to prevent destruction of exogenous mRNA. As shown in Figure 2A, the myc-tagged Kidr-WT protein rescued the short-spindle phenotype caused by Kid siRNA. The data indicate that Kid is involved in the formation of the mitotic spindles. Previously it has been reported that short bipolar spindles were formed after simultaneous perturbation of Kid and NuMA (Levesque et al., 2003). Immunoblot and immunofluorescent analyses showed that depletion of Kid did not affect levels of NuMA, dynein, or α-tubulin proteins (Figure 1A) nor the localization of NuMA in metaphase cells (Figure 2D). The results suggest that Kid contributes to spindle formation independently of NuMA.
For maintaining consistent pole-to-pole spacing, MT polymer dynamics is important (Waters et al., 1996). To examine whether altered MT dynamics contributes toward the failure to maintain spindle size in mitotic cells lacking Kid, we suppressed MT dynamics by addition of 100 nM nocodazole (Supplemental Figure 1). The pole-to-pole distances in Kid-depleted cells were reduced to ∼80% of that in control cells (Supplemental Figure 1B). The failure of the low dose of nocodazole to rescue the short spindle phenotype implies that MT dynamics is unlikely to be an important factor in explaining the short spindle phenotype of Kid-depleted cells.
We noticed that spindle microtubules were less robust after the depletion of Kid. This was obviously demonstrated when we chilled the cells for 10 min 48 h after siRNA transfection to depolymerize the nonbundled MTs (Figure 3A). Quantitative analysis showed that the microtubule density of these spindles in Kid-depleted cells was reduced by ∼18% (Figure 3B) compared with that in control cells. The observations suggest that Kid contributes to stable spindle formation.
Figure 3.
Less robust kinetochore fibers in Kid-depleted cells. (A) siRNA-Kid-treated cells were incubated on ice for 10 min to induce complete disassembly of all nonbundled microtubules and then fixed and processed for immunofluorescence using Kid (green in merge panel) and α-tubulin (left and red in merge panel) antibodies and Hoechst33342 for DNA staining (blue in merge panel). (B) Spindle fluorescence intensities in Kid-depleted or -nondepleted mitotic cells were measured using NIH Image software. The average of spindle fluorescence in nondepleted cells has been normalized to 1. The ratio of spindle fluorescence intensities in Kid-depleted cells to nondepleted cells is shown.
The Maintenance of Spindle Size Does Not Require Kid's DNA-binding Region and Motor Activity
To test the functional domains of Kid for maintenance of spindle size, we constructed expression vectors encoding a series of Kidr mutant proteins (Figure 4A). After immunofluorescent staining of Kidr-transfected cells with antibodies against α-tubulin and myc or Kid, we quantified the level of exogenous protein expression by fluorescent intensity (see Materials and Methods). The cells expressing the Kidr protein at a level comparable with endogenous Kid of non-siRNA transfected cells were selected for measurements of the pole-to-pole distance. Note that exogenous Kidr-wild-type (WT) protein at this level localized to chromosomes and spindles similar to endogenous Kid (Figure 4B). To test the need for DNA-binding activity, the Kidr-delDB fragment lacking the DNA-binding region was expressed in Kid-depleted cells. The Kidr-delDB mutant protein localized only to the spindle (Figure 4B). Kidr-delDB expression rescued spindle size as effectively as Kidr-WT (Figure 4C), indicating that Kid DNA-binding activity was not essential for spindle length maintenance. Previously, it was reported that perturbation of Kid–DNA interaction by microinjection of Kid antibodies into HeLa cells causes defects in chromosome arm orientation, but no spindle lengths abnormality was reported (Levesque and Compton, 2001). These facts support the idea that Kid maintains spindle size independently of its role in chromosome movement.
Figure 4.
DNA binding region and motor activity of Kid were not required to rescue the short spindle phenotype caused by Kid depletion. (A) Schematic representation of truncated and mutated Kid constructs. Kid has a kinesin-like motor domain (red box), a coiled-coil region (gray box), a helix-hairpin-helix domain (blue box), and the second microtubule-binding region (green box). Amino acid positions are indicated to the right. (B) Immunofluorescence analysis of Kid-depleted HeLa cells transfected with RNAi-refractory Kid mutant constructs. Cells were fixed and stained with antibodies against Myc-tag (green in merge) and α-tubulin (red in merge). DNA (blue in merge panel) was visualized with Hoechst33342 stain. (C) Intercentrosome distances in siRNA-Kid-treated HeLa cells transfected with RNAi-refractory Kid mutant constructs. Cells were fixed and stained with antibodies against Myc-tag and γ-tubulin and measured the pole-to-pole distance. Twenty metaphase cells were scored in each of five independent experiments. Error bars represent the SD. *p < 0.0005.
We then examined the necessity of Kid motor activity using the Kidr-rigor mutant. The Kidr-rigor mutant has a threonine-to-asparagine mutation in the conserved ATP loop of the motor domain such that it lacks ATP hydrolysis activity; therefore, it interacts with MTs constitutively (Ohsugi et al., 2003). Kidr-rigor localized to the spindle (Figure 4B) and also rescued spindle length (Figure 4C), indicating that the motor activity of Kid was also irrelevant to spindle length in vivo. Because the rigor mutant has higher affinity to MTs than wild type, Kidr-rigor protein may stabilize MTs in a manner that is different from Kidr-WT. To test this possibility, Kidr-rigor-motor that consists of only motor domain was examined. Kidr-rigor-motor also strictly localized to the spindle (Figure 4B) but could not rescue spindle length (Figure 4C), suggesting that the strong MT-binding character of Kidr-rigor was not enough to stabilize MTs. These results strongly support the idea that Kid's motor activity—and by extension, its activity in chromosome movement—are not essential for spindle size maintenance.
In contrast to spindle length, the misalignment of chromosome arms in Kid-depleted cells was not rescued by any tested mutant (Figure 4B). The average length of chromosome arms along the pole-to-pole axis was 4.4 ± 0.5 μm (n = 11) in Kid-depleted cells and 4.9 ± 0.6 μm (n 11) in Kidr-delDB-rescued cells, but 3.5 ± 0.8 μm (n = 11, p < 0.0005) in Kidr-WT-rescued cells. In particular, chromosomes in cells expressing the Kidr-rigor mutant were oriented with their arms conspicuously poleward, parallel to the long axis of the spindle (Figure 4B, Kidr-rigor, DNA, 6.6 ± 1.2 μm, n = 10, p < 0.0005). This effect may be explained by the properties of the Kidr-rigor protein: the Kidr-rigor protein contains an immotile motor domain but retains strong binding activity to both MTs and DNA, and it may immobilize chromosomes along the spindle.
Both the Coiled-Coil Region and the Second MT-binding Region Are Required to Rescue the Short Spindle Phenotype
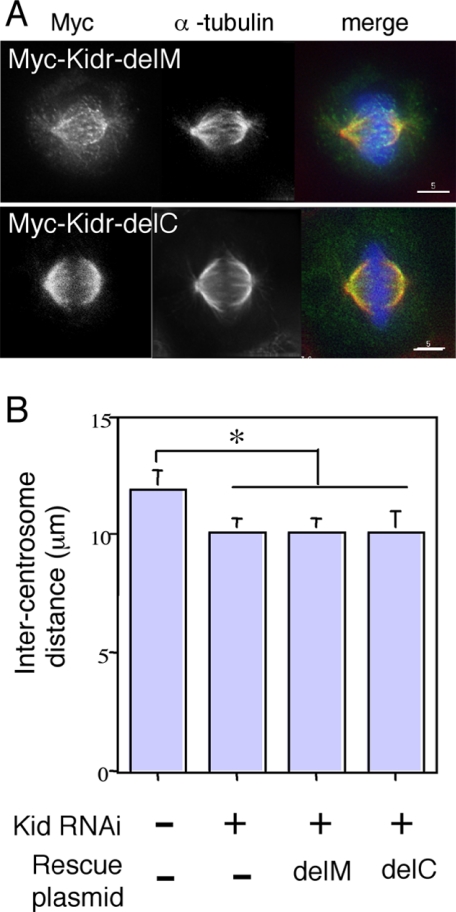
A previous study showed that the Kid-delDB fragment induces formation of MT bundles in vitro in an ATP-sensitive manner, but the Kid-motor fragment, which lacks both the second MT-binding region and the coiled-coil region present in Kid-delDB, does not (Shiroguchi et al., 2003). Kidr-delDB protein rescued spindle size but Kidr-motor protein did not (Figure 4). Thus, the ability to rescue spindle size seems to correlate with the ability of inducing MT bundles. However, a possibility remains that the coiled-coil region is sufficient for rescue of the short spindle phenotype. To examine the necessity of the second MT-binding region and the coiled-coil region of MT bundle formation, we assayed the in vitro MT bundle-inducing activity of Kid constructs lacking the second MT-binding region and DNA-binding domains (Kidr-delM in Figure 5A) or the coiled-coil region and DNA binding domains (Kidr-delC in Figure 5A). As shown in Figure 5B, Kid-delC protein induced formation of MT bundles to about the same extent as Kid-delDB, but Kid-delM did not. In the previous report by Shiroguchi et al. (2003), MT-bundling activity of Kid was abolished in the presence of ATP in vitro. Consistent with the results of the in vitro MT-bundling assay, we found that overexpressed Kidr-delDB and Kidr-delC proteins induced formation of MT bundles and colocalized with MT bundles in the nucleus (Figure 5C, top and second panels). In contrast, Kidr-motor protein and Kidr-delM protein colocalized with MTs in interphase cells but did not induce formation of MT bundles even when expressed at high levels (Figure 5C, third and bottom panels). Kidr-delDB protein rescued spindle size (Figure 4, B and C). To the contrary, both Kidr-delC and Kidr-delM expression failed to rescue spindle size (Figure 6B), despite localization of the mutant proteins to the spindle (Figure 6A). These data indicate that the MT bundle-inducing activity alone is insufficient to rescue the short spindle phenotype of Kid-depleted cells and that the coiled-coil region is insufficient for the rescue, either. Together, these results strongly suggest that both the coiled-coil region and the second MT-binding region of Kid are needed for the maintenance of spindle size.
Figure 5.
Domain requirements for Kid-induced MT bundling in vitro and in cells. (A) Schematic representation of truncated Kid constructs. Kid has a kinesin-like motor domain (red box), a coiled-coil region (gray box), a helix-hairpin-helix domain (blue box), and a second microtubule-binding region (green box). Amino acid positions are indicated to the right. (B) Images observed by fluorescent microscopy when Kid fragments were mixed with rhodamine-labeled MTs in vitro. (C) immunofluorescence images of Kid-depleted cells expressing Kid mutant fragments at high levels. The Kid-depleted HeLa cells were transfected with Myc-Kidr-delDB (top), Myc-Kidr-delC (second panels), Myc-Kidr-delM (third panels), or Myc-Kidr-motor (bottom) constructs and then fixed and stained for mutant Kid protein, α-tubulin, and DNA with antibodies against Myc-tag and α-tubulin, and Hoechst33342, respectively.
Figure 6.
Both the coiled-coil region and the second MT-binding region were required to rescue the short spindle phenotype caused by Kid depletion. (A) Immunofluorescence analysis of Kid-depleted HeLa cells transfected with RNAi-refractory Kid mutant constructs. Cells were fixed and stained with antibodies against Myc-tag (green in merge panel) and α-tubulin (red in merge panel). DNA (blue in merge panel) was visualized with Hoechst33342 stain. (B) Intercentrosome distances in siRNA-Kid-treated HeLa cells transfected with RNAi-refractory Kid mutant constructs. The pole-to-pole distance of the cells from experiments shown in A was measured. Twenty metaphase cells were scored in each of five independent experiments. Error bars represent the SD. *p < 0.0005.
DISCUSSION
In this study, we used RNAi coupled with rescue assays to analyze the function of the chromokinesin Kid/kinesin-10. The study showed that Kid contributes to the formation of the prometaphase/metaphase mitotic apparatus in at least two independent ways: pushing away the chromosome arms to generate the polar ejection force (Figure 7, left) and maintaining correct spindle size (Figure 7, right), the latter of which represents a novel role of Kid. These two functions may be regulated and coordinated through molecular modifications such as phosphorylation. We previously showed that Kid is phosphorylated by Cdc2 kinase and that the phosphorylation of Kid down-regulates its affinity for MTs (Ohsugi et al., 2003). Thus, it is conceivable that the phosphorylated and nonphosphorylated forms of Kid play separate roles; phosphorylated Kid might associate reversibly with MTs to shepherd chromosome arms, whereas nonphosphorylated Kid would interact with MTs to facilitate microtubule stability.
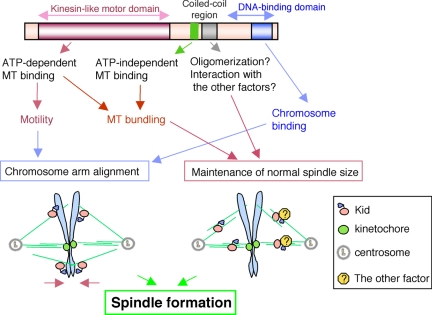
Figure 7.
Proposed model for Kid's roles during prometaphase/metaphase. Role of chromosomes-bound Kid (left): Kid localized on or around prometaphase/metaphase chromosome arms exert a force to transport the arms toward the plus end of the spindle MTs. Role of spindle-bound Kid (right): Kid may induce MT bundles to enhance the integrity of the spindle. Here, two possible mechanisms are shown. 1) MT cross-linking: Kid may directly cross-link parallel MTs such as kinetochore MTs or cross-link centrosome-nucleated MTs with chromosome-emanating MTs. 2) Factor recruitment: Kid may recruit other factors to stabilize the spindle MTs bundled by Kid.
Our findings seem to contradict an existing report that microinjection of CFPAC-1 cells with antibodies against the DNA-binding region of Kid caused defects in chromosome arm orientation, whereas spindle lengths were unaffected (Levesque et al., 2003). Furthermore, in Kid-depleted mitotic cells, the proportion of prometaphase cells was increased, suggesting delay of the cell cycle during prometaphase, whereas microinjection with Kid antibodies did not result in delay of anaphase onset (Levesque et al., 2003). The apparent discrepancy may be explained if the antibodies used by Levesque et al. (2003) against the DNA-binding region of Kid left other Kid activities unaffected. Obviously, in our RNAi-based experiments, Kid functions were inhibited more completely. These observations also suggest the possibility that Kid is not only required for proper spindle length but also is involved in prometaphase events such as spindle formation, independent of chromosome motility.
How might Kid be involved in maintenance of spindle size? Considering Kid's properties, at least three hypotheses may explain the short spindle phenotype caused by Kid siRNA. First, the loss of Kid-generated polar ejection force may shorten spindle length. Second, Kid may cross-link and slide overlapping antiparallel MTs within interpolar MT bundles. Third, Kid may be involved in stabilization of spindle MTs. By the rescue assay with various Kid-mutant constructs, we were able to define the functional relevance of Kid's different domains. The expression of Kid constructs Kidr-delDB or Kidr-rigor, lacking either the DNA-binding or motor activity, respectively, nevertheless rescued the short spindle phenotype. Thus, the function of Kid in organizing spindle MTs is independent of its activity in chromosome movement. Furthermore, the observation that the motor activity of Kid is not required for spindle length rescue rules out the hypothesis that interpolar MT sliding plays a role in this phenotype. Therefore, we were left with the third hypothesis that Kid is involved in stabilization of spindle MTs.
In Kid-depleted metaphase cells, suppression of MT dynamics by low levels of nocodazole could not rescue the short spindle phenotype. Therefore, it is likely that Kid stabilizes the spindle MTs in a manner distinct from controlling MT dynamics during metaphase. On the other hand, MT bundles were subtly less robust in Kid-depleted metaphase cells compared with control cells. In addition, immunoelectron microscopic analysis revealed Kid signals along bundled MTs as well as on chromosomes in prometaphase HeLa cells (Supplemental Figure 2, A–E). Based on these observations, we speculate that Kid present on MT bundles may contribute to the stabilization of spindle MTs and spindle morphogenesis.
How might Kid be involved in stabilization of spindle MTs? Our rescue experiments with a series of Kid deletion mutants showed that all the constructs that could rescue the short spindle phenotype posses an ability to induce MT bundles. Therefore, we propose that Kid stabilizes the spindle MTs, at least in part, by directly cross-linking parallel MTs through its motor and second MT-binding sites. Actually, Kidr-delM, which possesses the coiled-coil domain but lacks MT-bundling activity, failed to rescue the short spindle phenotype (Figure 6). However, Kidr-delC, which retains MT-bundling activity but lacks the coiled-coil domain, also failed to rescue the short spindle phenotype (Figure 6). These results argue that bundling activity alone is insufficient for spindle length rescue but is nevertheless required in conjunction with the coiled-coil region for spindle length maintenance.
The function of the coiled-coil region of Kid remains unknown. Kid oligomerization is one mechanism through which MT stability might be enhanced. Previously, Kid was reported to be a monomeric motor (Shiroguchi et al., 2003). However, we found that when GST-tagged Kid was coexpressed with truncation mutants of FLAG-tagged Kid in 293T cell, FLAG-Kid was coprecipitated with GST-Kid, but only when it contained the coiled-coil region, suggesting that Kid can oligomerize through the coiled-coil region (Supplemental Figure 3). Another monomeric motor, Kif1A/kinesin-3, is reported to be converted into a processive dimer at high motor concentrations (Tomishige et al., 2002). Thus, one possibility is that Kid may also form oligomers in a situation-specific manner, resulted in enhanced MT-bundling activity of Kid conferred by the motor and second MT binding site. Another possibility is that the coiled-coil region of Kid directly binds and recruits other molecules that facilitate or regulate the stabilization of spindle MTs bundled by Kid (Figure 7, right).
The mechanism of spindle length determination is very complex and involves multiple activities (Mitchison et al., 2005). Recently, a model was proposed, in which spindle length is set by a concentration gradient of the GTP form of small GTPase Ran (Gruss and Vernos, 2004). Activities of a subset of spindle assembly factors such as TPX2 were inhibited by its direct binding to importin α/β (Wiese et al., 2001; Gruss et al., 2002). Their interaction between TPX2 and importin α/β, in turn, is inhibited by binding of RanGTP to importin β (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001). Thus, in the course of spindle formation, RanGTP has the net effect of promoting spindle assembly factor activity close to the chromosomes by dissociating importin α/β complex from them (Kahana and Cleveland, 1999; Wiese et al., 2001; Gruss et al., 2002; Trieselmann and Wilde, 2002). It has been reported that importin α/β also binds to Kid and lowers Kid's affinity for MTs (Trieselmann et al., 2003). In addition, our findings, that Kid stabilizes the spindle at least in part through inducing MT bundles, fit together well with these biochemical observations of Trieselmann et al. (2003). In Figure 5, unusual nuclear MT bundles were observed in cells overexpressing Kid mutant fragments that possess the motor and second MT-binding region of Kid. Although the exact mechanism of this nuclear MT bundle formation is unknown, we speculate that when RanGTP promotes dissociation of importin α/β from Kid within the nucleus, Kid may exhibit high affinity for MTs or tubulin (Germani et al., 2000) and that overexpression could emphasize this effect to the point of MT bundle formation. These observations suggest that Kid could be one more spindle assembly factor subject to RanGTP-importin α/β regulation in setting the spindle size through stabilizing chromosome-associated MTs.
Supplementary Material
Acknowledgments
We thank Y. Takasaki for CREST serum; Y. Y. Toyoshima for purified tubulin; R. F. Whittier for helpful discussions; and K. Haraguchi, Y. Horiuchi, and N. Oshimori for technical help in this study. T. Y. was supported by a Grant for Advanced Cancer Research from the Ministry of Education, Science, Sports and Culture of Japan. M. O. was supported by grants-in-aid from the Japan Society for the Promotion of Science and from the Ministry of Education, Culture, Sports, Science and Technology of Japan as well as grant from the Yamanouchi Foundation for Research on Metabolic Disorders.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-03-0244) on September 21, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Antonio, C., Ferby, I., Wilhelm, H., Jones, M., Karsenti, E., Nebreda, A. R., and Vernos, I. (2000). Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102, 425-435. [DOI] [PubMed] [Google Scholar]
- Cooke, C. A., Schaar, B., Yen, T. J., and Earnshaw, W. C. (1997). Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106, 446-455. [DOI] [PubMed] [Google Scholar]
- Cytrynbaum, E. N., Scholey, J. M., and Mogilner, A. (2003). A force balance model of early spindle pole separation in Drosophila embryos. Biophys. J. 84, 757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Weber, K., and Tuschl, T. (2002). Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26, 199-213. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., and Murray, A. W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411-424. [DOI] [PubMed] [Google Scholar]
- Germani, A., Bruzzoni-Giovanelli, H., Fellous, A., Gisselbrecht, S., Varin-Blank, N., and Calvo, F. (2000). SIAH-1 interacts with alpha-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene 19, 5997-6006. [DOI] [PubMed] [Google Scholar]
- Gruss, O. J., Carazo-Salas, R. E., Schatz, C. A., Guarguaglini, G., Kast, J., Wilm, M., Le Bot, N., Vernos, I., Karsenti, E., and Mattaj, I. W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83-93. [DOI] [PubMed] [Google Scholar]
- Gruss, O. J., and Vernos, I. (2004). The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 166, 949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, O. J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Sillje, H., Karsenti, E., Mattaj, I. W., and Vernos, I. (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871-879. [DOI] [PubMed] [Google Scholar]
- Inoue, S., and Sato, H. (1967). Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 50 (suppl), 259-292. [PMC free article] [PubMed] [Google Scholar]
- Kahana, J. A., and Cleveland, D. W. (1999). Beyond nuclear transport. RanGTP as a determinant of spindle assembly. J. Cell Biol. 146, 1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle: a self-made machine. Science 294, 543-547. [DOI] [PubMed] [Google Scholar]
- Levesque, A. A., and Compton, D. A. (2001). The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154, 1135-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque, A. A., Howard, L., Gordon, M. B., and Compton, D. A. (2003). A functional relationship between NuMA and kid is involved in both spindle organization and chromosome alignment in vertebrate cells. Mol. Biol. Cell 14, 3541-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther, E., and Kamentsky, L. A. (1996). Resolution of mitotic cells using laser scanning cytometry. Cytometry 23, 272-278. [DOI] [PubMed] [Google Scholar]
- Mayer, T. U., Kapoor, T. M., Haggarty, S. J., King, R. W., Schreiber, S. L., and Mitchison, T. J. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971-974. [DOI] [PubMed] [Google Scholar]
- Mitchison, T., Evans, L., Schulze, E., and Kirschner, M. (1986). Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45, 515-527. [DOI] [PubMed] [Google Scholar]
- Mitchison, T. J., Maddox, P., Gaetz, J., Groen, A., Shirasu, M., Desai, A., Salmon, E. D., and Kapoor, T. M. (2005). Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell 16, 3064-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M. V., Maresca, T. J., Salmon, W. C., Waterman-Storer, C. M., Heald, R., and Weis, K. (2001). Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95-106. [DOI] [PubMed] [Google Scholar]
- Nedelec, F. (2002). Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 158, 1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi, M., Tokai-Nishizumi, N., Shiroguchi, K., Toyoshima, Y. Y., Inoue, J., and Yamamoto, T. (2003). Cdc2-mediated phosphorylation of Kid controls its distribution to spindle and chromosomes. EMBO J. 22, 2091-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, K., Kurose, A., Miura, Y., Sato, T., and Ikeda, E. (1996). DNA ploidy analysis by laser scanning cytometry (LSC) in colorectal cancers and comparison with flow cytometry. Cytometry 23, 106-109. [DOI] [PubMed] [Google Scholar]
- Scholey, J. M., Brust-Mascher, I., and Mogilner, A. (2003). Cell division. Nature 422, 746-752. [DOI] [PubMed] [Google Scholar]
- Scholey, J. M., Rogers, G. C., and Sharp, D. J. (2001). Mitosis, microtubules, and the matrix. J. Cell Biol. 154, 261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., Rogers, G. C., and Scholey, J. M. (2000). Microtubule motors in mitosis. Nature 407, 41-47. [DOI] [PubMed] [Google Scholar]
- Shiroguchi, K., Ohsugi, M., Edamatsu, M., Yamamoto, T., and Toyoshima, Y. Y. (2003). The second microtubule-binding site of monomeric kid enhances the microtubule affinity. J. Biol. Chem. 278, 22460-22465. [DOI] [PubMed] [Google Scholar]
- Suzuki, E., and Hirosawa, K. (1994). Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J. Electron Microsc. 43, 183-189. [PubMed] [Google Scholar]
- Tokai, N., Fujimoto-Nishiyama, A., Toyoshima, Y., Yonemura, S., Tsukita, S., Inoue, J., and Yamamota, T. (1996). Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 15, 457-467. [PMC free article] [PubMed] [Google Scholar]
- Tomishige, M., Klopfenstein, D. R., and Vale, R. D. (2002). Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297, 2263-2267. [DOI] [PubMed] [Google Scholar]
- Trieselmann, N., Armstrong, S., Rauw, J., and Wilde, A. (2003). Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 116, 4791-4798. [DOI] [PubMed] [Google Scholar]
- Trieselmann, N., and Wilde, A. (2002). Ran localizes around the microtubule spindle in vivo during mitosis in Drosophila embryos. Curr. Biol. 12, 1124-1129. [DOI] [PubMed] [Google Scholar]
- Waters, J. C., Mitchison, T. J., Rieder, C. L., and Salmon, E. D. (1996). The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 7, 1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, C., Wilde, A., Moore, M. S., Adam, S. A., Merdes, A., and Zheng, Y. (2001). Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291, 653-656. [DOI] [PubMed] [Google Scholar]
- Yajima, J., Edamatsu, M., Watai-Nishii, J., Tokai-Nishizumi, N., Yamamoto, T., and Toyoshima, Y. Y. (2003). The human chromokinesin Kid is a plus end-directed microtubule-based motor. EMBO J. 22, 1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.