Abstract
Plant steroid hormones, brassinosteroids (BRs), play important roles throughout plant growth and development. Plants defective in BR biosynthesis or perception display cell elongation defects and severe dwarfism. Two dwarf mutants named bin3 and bin5 with identical phenotypes to each other display some characteristics of BR mutants and are partially insensitive to exogenously applied BRs. In the dark, bin3 or bin5 seedlings are de-etiolated with short hypocotyls and open cotyledons. Light-grown mutant plants are dwarfs with short petioles, epinastic leaves, short inflorescence stems, and reduced apical dominance. We cloned BIN3 and BIN5 and show that BIN5 is one of three putative Arabidopsis SPO11 homologs (AtSPO11-3) that also shares significant homology to archaebacterial topoisomerase VI (TOP6) subunit A, whereas BIN3 represents a putative eukaryotic homolog of TOP6B. The pleiotropic dwarf phenotypes of bin5 establish that, unlike all of the other SPO11 homologs that are involved in meiosis, BIN5/AtSPO11-3 plays a major role during somatic development. Furthermore, microarray analysis of the expression of about 5500 genes in bin3 or bin5 mutants indicates that about 321 genes are down-regulated in both of the mutants, including 18 of 30 BR-induced genes. These results suggest that BIN3 and BIN5 may constitute an Arabidopsis topoisomerase VI that modulates expression of many genes, including those regulated by BRs.
Plant steroid hormones, called brassinosteroids (BRs), affect many growth and developmental processes such as stem elongation, leaf development, xylem differentiation, root growth inhibition, pollen tube growth, apical dominance, and senescence (1, 2). Mutants defective in BR synthesis or perception show pleiotropic dwarf phenotypes characterized by short hypocotyls, stems and petioles, dark green and epinastic leaves, and reduced apical dominance, senescence, and male sterility (3). Dark-grown BR mutant seedlings grow like light-grown plants with short hypocotyls and open cotyledons; these seedlings also misexpress light-regulated genes. BR-deficient mutants have allowed the identification of enzymes for the biosynthesis of brassinolide (BL), the most active BR (4, 5). A BR-insensitive locus BRI1 encodes a plasma membrane localized leucine-rich repeat (LRR) receptor serine/threonine kinase, a critical component of a BL receptor (6–9). Genetic studies have also identified several genes implicated in BR signaling downstream from BRI1. A second BR-insensitive mutant, bin2, displays bri1-like dwarf phenotypes, and the semidominance of the mutation suggests that BIN2 is a negative regulator of the BR pathway (10). BIN2 encodes a serine/threonine kinase with homology to GSK-3, a negative regulator in the Wnt signaling pathway of animal systems (11, 12). Two homologous genes, BES1 and BZR1, were identified in genetic screens for bri1 suppressors and resistant mutants to a BR biosynthesis inhibitor brassinazole, respectively (13, 14). Both BES1 and BZR1 accumulate in the nucleus in the presence of BL. Nuclear-accumulated BES1 appears to activate BR-target gene expression, whereas BZR1 seems to play a major role in the activation of a BR feedback inhibition pathway. In addition, BES1 is a substrate of the BIN2 kinase and BES1 protein levels are negatively regulated by BIN2 (14). These results establish a signaling cascade that transduces the BR signal from a cell surface receptor to nuclear target genes.
Archaebacterial topoisomerase VI (TOP6) is a heterotetramer composed of two subunits, TOP6A and TOP6B (15). The catalytic subunit TOP6A is a close homolog of the yeast SPO11 that was originally identified as an important component for meiotic recombination (16). Consistent with its homology to a topoisomerase catalytic subunit, SPO11 was found to associate with and is likely to catalyze DNA double-strand breaks (DSBs) that are required for initiation of meiotic recombination (17). Up to now, all characterized SPO11 homologs have been implicated in meiosis (16). Mutations of SPO11 genes in fungi (18), flies (mei-W68) (19), nematodes (20), and mice (21, 22) cause defects in meiotic recombination. Unlike other systems with only one SPO11 gene, Arabidopsis has three SPO11-like genes, AtSPO11-1, AtSPO11-2, and AtSPO11-3 (23–25). Disruption of AtSPO11-1 causes severe meiotic phenotypes associated with drastic reduction (about 10-fold) in meiotic recombination (23), suggesting that AtSPO11-1 is a SPO11 ortholog. Archaebacterial TOP6B is involved in ATP binding and hydrolysis (15). With the exception of plants, there are no close TOP6B homologs in most eukaryotic systems (26). Arabidopsis AtTOP6B was identified by its close homology to TOP6B and found to interact with AtSPO11-2 and AtSPO11-3, but not with AtSPO11-1 in yeast two-hybrid assays (25). These results imply that AtSPO11-2 and AtSPO11-3 may form a complex with AtTop6B in vivo to constitute a TOP6-like activity. Although the high and ubiquitous expression patterns of AtSPO11-3 and AtTOP6B suggest possible involvement in somatic development (25), the biological functions of AtSPO11-2, AtSPO11-3, and AtTOP6B remain uncharacterized.
In attempts to identify other components that either transmit or modify BR signaling, we isolated dwarf mutants with some characteristics of the BR-insensitive mutant bri1, and named them bin3, bin4, and bin5 (brassinosteroid insensitive 3, 4, and 5). Here, we report the cloning of BIN5 and BIN3 and show that these genes represent AtSPO11-3 and AtTOP6B, respectively. Gene expression studies indicate that many BR-regulated genes are down-regulated in bin3 and bin5 mutants, suggesting a role for BIN3 and BIN5 in regulating gene expression. This work establishes a previously uncharacterized function for a SPO11 homolog in Arabidopsis somatic development and provides genetic evidence that BIN5/AtSPO11-3 and BIN3/AtTOP6B may constitute a TOP6-like activity that is essential for plant growth and development.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana ecotype Columbia (Col-0) was the wild type. Plants were grown in long-day (16 h light/8 h dark) or short-day (8 h light/16 h dark) conditions at 22°C.
Isolation and Characterization of bin3, bin4, and bin5 Mutants.
To identify additional BR-insensitive mutants, ethyl methanesulfonate (EMS)-mutagenized M2 seeds (Lehle Seeds, Round Rock, TX), or a lab collection of T-DNA transformed plant lines were screened for dwarf plants with similar phenotypes to those of bri1 mutants (6). The putative mutants were sprayed with 1 μm BL (CIDTech, Cambridge, ON, Canada) and the dwarf plants that showed no or little responses to BL treatments were further analyzed. bin3-1 was identified from an EMS-mutagenized pool and bin3-2, bin4, and bin5 were identified from T-DNA transformed plant lines. Segregation studies indicated that bin3-2 was tagged but that neither bin4 nor bin5 was tagged by T-DNA. For hormone sensitivity assays, the seeds were germinated and grown in ½ MS medium containing various concentrations of BL or abscisic acid (ABA) for 10 days under light. The ABA root growth inhibition assays were done in vertical plates. At least 20 plants were used to measure hypocotyl or root length for each treatment.
bin5 and bin3 Cloning.
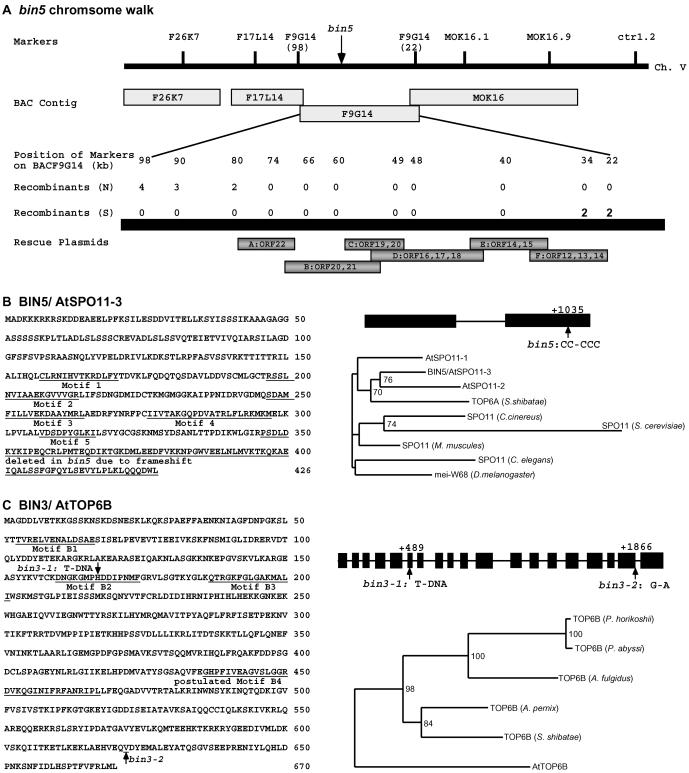
bin5 in Col-0 background was crossed to Landsberg-erecta (La-er) and F2 plants with bin5 phenotypes were used to map the mutation using molecular markers (27, 28). bin5 was mapped to the top arm of chromosome V linked to the marker ctr1.2 (Fig. 3A). Using about 2,000 recombinant chromosomes, the mutation was mapped to a region between markers derived from bacterial artificial chromosome (BAC) clones F26K7 and MOK16. Markers derived from BAC F17L14 and F9G14 further narrowed bin5 to a 46-kb fragment (from 34 to 80 kb) on BAC clone F9G14. BAC F9G14 was obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH) and BAC DNA was prepared using plasmid Midi Kit (Qiagen). Six DNA fragments (A–F) containing overlapping ORFs in the 46-kb region were generated by digestions of BAC DNA and cloned into the binary vector pZP211 (29). The resulting constructs were put into Agrobacterium strain GV3101 and transformed into bin5 mutant plants by infiltration (30). T1 seeds were screened on kanamycin plates to score for complementation of bin5 phenotypes.
Figure 3.
Identification of BIN5 and BIN3 genes. (A) bin5 chromosome walk. The bin5 mutation was mapped to a 46-kb region on BAC F9G14. Complementation constructs B and C, both containing ORF20, rescued the bin5 mutant phenotypes, indicating that ORF20 is mutated in bin5. (B) BIN5/AtSPO11-3 gene and bin5 mutation. BIN5 was identified as AtSPO11-3, one of three Arabidopsis homologs of archaebacterial topoisomerase VI subunit A (Top6A) (23, 25). BIN5 contains two exons and DNA sequencing of bin5 mutant DNA revealed a single base pair addition at nucleotide position +1035 with respect to the putative translation start site (ccg agt gat to ccc gag tga t). A phylogenetic distance tree shows that BIN5 is most closely related to AtSPO11-2 and archaebacterial TOP6A than to nematode, mouse, fly, fungus, and yeast SPO11. Numbers in branch nodes indicate the number of times that the group of sequences to the right of the node occurred of 100 bootstrap replicates. Numbers are only given when 70 or more occurrences were observed. (C) BIN3/AtTop6B gene and mutations. BIN3 was identified by plasmid rescue with a T-DNA-tagged allele, bin3-1. BIN3/AtTOP6B gene contains 18 introns and 19 exons that encode a predicted polypeptide of 670 aa (25). Phylogenetic distance tree shows relationship of BIN3/AtTOP6B, archaebacterial TOP6B, and prokaryotic homologs of Aeropyrum pernix, Archaeoglobus fulgidus, Pyrococcus horikoshii, and Pyrococcus abyssi.
Genomic DNA from young seedlings of bin3-2 was isolated according to protocols provided by Phytopure, Nucleon Biosciences (Glasgow, U.K.). Plasmid-rescue was carried out as described (31).
Phylogenetic Tree Construction.
Sequences were aligned using clustalw (32) with subsequent manual alignment; nonconserved amino and carboxy-terminal extensions were removed before tree reconstruction. Maximum likelihood phylogenetic distances were estimated using TREE-PUZZLE 5.0 (33) and 100 bootstrap replicates were performed using puzzleboot (M. Holder and A. Roger, http://hades.biochem.dal.ca/Rogerlab/Software/software.html). Tree topology was estimated using the fitch program from the phylip package (J. Felsenstein, Phylogeny Inference Package Version 3.5c, Department of Genetics, University of Washington, Seattle). Similar topologies were obtained by using tree-puzzle to estimate maximum likelihood trees and using protpars from phylip to estimate maximum parsimony trees.
Northern Blotting and Microarray Analysis.
RNA was isolated using TRIzol reagent according to the manufacturer's instructions (GIBCO/BRL). Northern blotting was carried out using standard procedures. For microarray experiments, 14-day-old light-grown seedlings were treated with 1 μM BL or mock treatment and used to prepare RNA for microarray analysis according to manufacturer's instructions (Affymetrix, Santa Clara, CA). The scanned images were analyzed using AFFYMETRIX MICROARRAY SUITE 5.0. The expression levels (raw data) of 30 previously identified BL-induced genes (14) were examined in wild-type, bin3, and bin5 mutants (Table 1). Mock treatments were used to identify genes down-regulated in the mutants. Genes that are down-regulated by at least 2-fold in both bin3 and bin5 compared with wild-type control were identified using GENESPRING 4.2 (Silicon Genetics, Redwood City, CA) and listed in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Table 1.
BR-induced genes in bin3 and bin5
| Gene no. | Gene | Col
|
bin3
|
bin5
|
bri1-116
|
||||
|---|---|---|---|---|---|---|---|---|---|
| −BL | +BL | −BL | +BL | −BL | +BL | −BL | +BL | ||
| At4g38850 | SAUR-AC1 | 442 | 2260 | 161 | 449 | 105 | 476 | 223 | 128 |
| At4g38860 | SAUR-AC1-like | 652 | 2808 | 99 | 834 | 264 | 975 | 285 | 208 |
| At3g15540 | IAA19 | 980 | 1958 | 301 | 535 | 372 | 472 | 360 | 438 |
| At2g14900 | GASA3 | 10567 | 13722 | 4754 | 6042 | 2698 | 5365 | 7635 | 7332 |
| At5g57560 | TCH4 | 19164 | 26329 | 14615 | 15691 | 12952 | 16996 | 5823 | 5046 |
| At2g26710 | BAS1 | 922 | 2527 | 589 | 1286 | 954 | 1150 | 627 | 669 |
| At2g31730 | Similar to LeER33 | 1939 | 2820 | 1311 | 1670 | 1549 | 1561 | 1023 | 1306 |
| At1g65310 | Putative xyloglucan endoglucanase | 1296 | 2431 | 302 | 394 | 419 | 287 | 274 | 397 |
| At1g04610 | YUCCA3 | 1372 | 1639 | 288 | 452 | 532 | 446 | 593 | 653 |
| At5g13870 | EXGT-A4 | 1025 | 1885 | 280 | 267 | 189 | 450 | 637 | 644 |
| At4g20780 | Putative Ca-dependent protein kinase | 5508 | 6829 | 1223 | 1225 | 1077 | 1265 | 2532 | 2130 |
| At2g40610 | Putative expansin | 3945 | 7978 | 615 | 2767 | 1011 | 2070 | 1178 | 802 |
| At4g28780 | Pro-rich APG like protein | 2263 | 4792 | 861 | 2559 | 531 | 1995 | 1373 | 859 |
| At1g21820 | Unknown | 2227 | 4069 | 1305 | 1617 | 1760 | 1982 | 1846 | 1851 |
| At2g32560 | Unknown | 2534 | 4380 | 1235 | 1817 | 1516 | 1519 | 1366 | 1769 |
| At2g36220 | Unknown | 1425 | 3803 | 1488 | 800 | 1034 | 1391 | 1311 | 1435 |
| At4g09890 | Unknown | 1497 | 2725 | 340 | 465 | 388 | 464 | 582 | 657 |
| At4g01950 | Unknown | 1327 | 2921 | 893 | 1802 | 946 | 1465 | 319 | 239 |
RNA from seedlings treated without or with BL was used for microarray experiments using Affymetrix arabidopsis gene chip. BR-induced genes have been described (14). The raw data are presented to indicate gene expression levels.
Results
Genetic Screen for New BR-Insensitive Mutants.
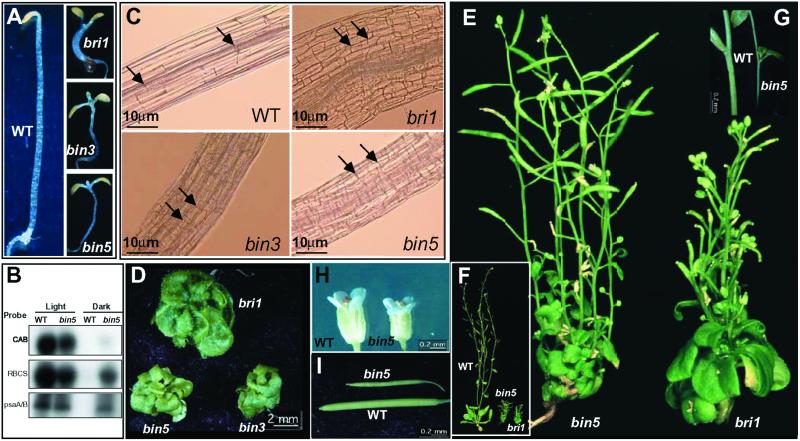
We identified three dwarf mutants—bin3, bin4, and bin5—that have identical phenotypes to each other and show reduced sensitivities to BL treatments. Two different alleles of bin3 were obtained, one from an ethyl methanesulfonate (EMS) pool (bin3-1) and another from T-DNA tagged transgenic lines (bin3-2). bin4 and bin5 were derived from T-DNA transformed plant lines, but neither line showed cosegregation of the bin phenotype with T-DNA (data not shown). Using mapping populations generated by crossing to La-er, bin3 was mapped to chromosome III while bin4 and bin5 were mapped to the middle (around 20 cM) and the top (around 7 cM) of chromosome V, respectively. bin3 and bin5 will be further described in this paper. Like bri1, bin3, and bin5 were de-etiolated in the dark with open cotyledons and short hypocotyls (Fig. 1A) and had inappropriate expression of light-regulated genes, such as CAB, RBCS, and psaA/B (Fig. 1B). The shorter hypocotyls were due to reduced cell elongation as revealed by the shorter cells in the mutants (Fig. 1C). On average, bin3 and bin5 hypocotyl cells were 3–5 times shorter than those of wild-type controls. Light-grown bin3 and bin5 plants were cabbage-like dwarfs with reduced leaf petioles and epinastic leaves (Fig. 1D). Mature plants were much reduced in stature when compared with wild type, with short and thin inflorescence stems, and reduced apical dominance and senescence (Fig. 1 E–G). These characteristics are reminiscent of strong bri1 alleles (Fig. 1). Despite these similarities, however, there were clear differences between bin3 or bin5 and bri1 phenotypes. In the dark, bin3 and bin5 had longer hypocotyls than bri1 (Fig. 1A); while in the light, the leaves of bin3 and bin5 were smaller than those of bri1 (Fig. 1D). Unlike bri1, which is male-sterile, bin3 and bin5 were moderately fertile although both their flowers and siliques were smaller than wild-type controls (Fig. 1 H and I). The overall mutant phenotypes indicate that bin3 and bin5 are defective in cell elongation and the similar but not identical phenotypes between these mutants and bri1 suggest that BIN3 and BIN5 may affect downstream components of BR signaling.
Figure 1.
BR-insensitive mutants bin3 and bin5 resemble bri1. (A) Dark grown bin3 and bin5 seedlings compared with wild-type and a bri1 mutant. Seedlings were grown in ½ MS medium plus 1% sucrose for 7 days. (B) Light-regulated genes are ectopically expressed in bin5 mutants. Ten micrograms of total RNA from wild-type and bin5 were used in Northern blots using CAB, RBCS, and psaA/B as probes (42). (C) bin3, bin5, and bri1 have shorter hypocotyl cells than wild type. Dark-grown hypocotyls shown in A were used to observe the cells under a microscope. The length of a typical cell from each of the genotypes is indicated by two arrows. (D) One-month-old bin3, bin5, and bri1 plants. (E) Mature bin5 and bri1 plants. (F) Mature bin5 and bri1 plants together with a wild-type control. (G–I). Inflorescence stems, flowers, and siliques of wild-type and bin5 plants.
bin5 Plants Are Partially Insensitive to BL and Hypersensitive to ABA.
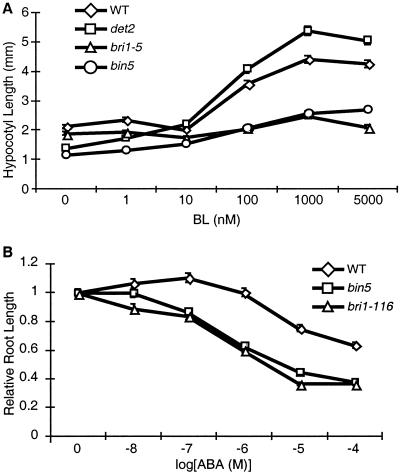
To test the involvement of BIN3 and BIN5 in BR signaling, the BL responses of the mutant plants were determined using a hypocotyl elongation assay in light-grown seedlings (Fig. 2A). While hypocotyl elongation in wild-type and BR-deficient det2 plants is stimulated by increasing concentrations of BL, this BL-induced hypocotyl growth was reduced in bin5, suggesting that BL-induced cell elongation is indeed impaired in the mutant. A weak allele of bri1 did not show any responses to BL, especially at lower concentrations (Fig. 2A). Similar BL responses were observed in bin3 (data not shown).
Figure 2.
bin5 is partially insensitive to BL and hypersensitive to ABA. (A) BL responses in bin5. Seeds of wild-type, det2, bin5, and bri1-5 were germinated and grown in ½ MS media plus indicated concentrations of BL for 10 days under white light, and hypocotyl lengths were measured. (B) bin5 is hypersensitive to ABA. Plants were grown in media containing the indicated concentrations of ABA on vertical plates and root lengths were measured.
bri1, bin2, and a BR-deficient mutant sax1 are reported to be hypersensitive to ABA in root-growth assays (10, 34, 35), suggesting an antagonistic interaction between BRs and ABA. The ABA responses in terms of root growth inhibition were measured in bin5, bri1, and wild-type plants (Fig. 2B). Like bri1, bin5 had a hypersensitive response to ABA (about two orders of magnitude), further supporting that BIN5 and BIN3 affect a BR signaling pathway.
BIN5 and BIN3 Encode Putative Arabidopsis Topoisomerase VI Subunits A and B.
BIN5 was cloned by chromosome walking (Fig. 3 A and B) and was found to be one of the Arabidopsis SPO11 homologous genes, named AtSPO11-3 (23, 25). The AtSPO11-3 gene contains two predicted exons and encodes a putative polypeptide of 426 aa with all of the conserved motifs proposed for the archaebacterial TOP6A (15). In the bin5 mutant a single base pair addition in the second exon resulted in a truncated BIN5 protein lacking the last 81 aa (Fig. 3B). Comparison of the SPO11 homologs indicated that BIN5/AtSPO11-3, AtSPO11-2 and archaebacterial TOP6A are more closely related to each other than to other SPO11 homologs known to be involved in meiosis (Fig. 3B).
T-DNA tagged bin3-1 was used to identify BIN3 by plasmid rescue (Fig. 3C). The T-DNA was found inserted in the 6th exon of a putative AtTOP6B gene that encodes a predicted protein sharing significant homology to the archaebacterial TOP6B (25). bin3-2, an ethyl methanesulfonate (EMS)-derived allele, was also sequenced and a single nucleotide change (G to A at + 1866) at the splice junction of the 18th intron was identified. The bin3-2 mutation is predicted to disrupt the splicing of the last intron, resulting in an aberrant cDNA predicted to encode a truncated protein without 49 aa from the last exon but with 20 unrelated amino acid residues encoded by the unspliced intron sequence. The putative BIN3 protein contained the conserved motif B1 (for ATP hydrolysis) and motifs B2 and B3 (for ATP binding) proposed for TOP6B (15), as well as a well conserved motif B4 with unknown function (25).
BR-Induced Genes Are Down-Regulated in bin3 and bin5 Mutants.
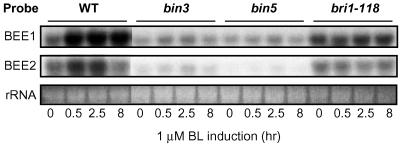
The similarities of the bin3 or bin5 phenotypes to those of bri1 led us to examine BR-regulated gene expression in these mutants to gain some clues about how BIN3 and BIN5 might affect BR signaling. Two Arabidopsis BR up-regulated genes, BEE1 and BEE2, were identified by a differential screening approach (D.F., T. Muramitsu, M. Furuya, and J.C., unpublished work). Basal expression of BEE1 and BEE2 were greatly reduced in bin3 and bin5 mutants and their induction by BL was also somewhat affected (Fig. 4), suggesting that BIN3 and BIN5 modulate BR-regulated gene expression. To further confirm this observation, we used microarray experiments to examine the expression of 30 known BR-induced genes (14) in bin3, bin5, and bri1 in the absence or presence of BL (Table 1). Like BEE1 and BEE2, 18 of these 30 BR-regulated genes were down-regulated in bin3 and bin5 (by at least 2-fold in most cases; Table 1). The other BR-regulated genes did not change dramatically in bin3 and bin5 mutants (data not shown). Four of the 18 genes that were down-regulated in the mutants encode putative xyglucan endotransglycosylase (XET), xyglucan endo 1,4-β-glucanase (EGase), and expansin (Table 1), all of which are implicated in cell elongation or expansion (4, 36–38). Interestingly, three of the BR-induced genes (SAUR-AC1, SAUR-AC1-like, and IAA19) are known to be regulated by auxin or have homologies to auxin-induced genes. It is worth noting that in addition to the BR regulated genes, another 303 genes were also down-regulated in both bin3 and bin5 mutants (see Table 2), suggesting that BIN3/BIN5 may affect the expression of a large number of genes.
Figure 4.
Down-regulation of BR-regulated genes, BEE1 and BEE2, in bin3 and bin5 mutants. Two-week-old light-grown seedlings of wild-type, bin3, bin5, and bri1 were treated with 1 μM BL for 0, 0.5, 2.5, and 8 h and 10 μg of total RNA was used in Northern blots to examine induction of BEE1 and BEE2 genes.
Discussion
In attempts to isolate additional BR-insensitive mutants, we identified two mutants, bin3 and bin5, that have similar phenotypes to those of bri1 and show cell-elongation defects throughout plant development. Surprisingly, BIN5 and BIN3 encode AtSPO11-3 and AtTOP6B that share sequence identity to archaebacterial topoisomerase VI subunits A and B, respectively. We showed that BR-induced genes including those implicated in cell elongation are down-regulated in bin5 and bin3, providing an explanation for the dwarf phenotypes of these mutants. This work establishes that AtSPO11-3 and AtTOP6B play crucial roles in plant growth and development, including cell-elongation processes mediated by BRs.
Our results show that a SPO11-like gene can play a major role in somatic development. SPO11 had been implicated in catalyzing double-strand breaks (DSBs) necessary for homologous recombination and all of the genetically characterized SPO11 homologs in eukaryotes are involved in meiotic recombination (16). One of the three Arabidopsis homologs, AtSPO11-1, like most other SPO11 genes, plays an important role during meiosis but does not play any roles in somatic cells (23). In contrast, the strong and pleiotropic bin5 mutant phenotypes establish that BIN5/AtSPO11-3 plays an essential role during somatic development. The facts that bin5 plants are fertile and no significant reduction in meiotic recombination was observed during bin5 chromosome walking (Fig. 3A) suggest that BIN5/AtSPO11-3 does not play a major role in meiosis.
Our results also establish a biological function for AtTOP6B, a close homolog of archaebacterial TOP6B. Archaebacterial TOP6 was biochemically purified and the biological functions of the corresponding genes are not known (15). The identical phenotypes of bin3 and bin5 and the identification of the same set of genes down-regulated in both mutants strongly suggest that BIN3/AtTOP6B and BIN5/AtSPO11-3 form a complex in plants constituting a TOP6-like activity that is essential for plant growth and development. Consistent with this, AtTOP6B interacts with AtSPO11-3 but not with AtSPO11-1 implicated in meiosis (25). The fact that no TOP6B homologs have been found in other eukaryotic systems whose genomes contain a single SPO11 gene involved in meiosis (26) suggests that ATSPO11-3 is evolutionally diverged from other SPO11 homologs, and AtSPO11-3 and AtTOP6B have evolved to perform a different biological function in somatic development. There is no other AtTOP6B-related gene in Arabidopsis genome.
Despite extensive efforts, we have been unable to detect consistent topoisomerase II activities by using purified BIN3 and BIN5 recombinant proteins expressed in Escherichia coli (unpublished results). Yeast SPO11 appears to function in conjunction with the Mre11-Rad50 complex to create DSBs (17), although no topoisomerase II or DNA cleavage activities have ever been directly demonstrated for any eukaryotic SPO11 homologs (16). In Arabidopsis, TOP6 activity may require additional component(s) to BIN3 and BIN5 for activity. In light of this, the BR-insensitive dwarf mutant bin4 that has identical phenotypes to bin3 and bin5 may define such a missing component.
To explore the mechanisms of BIN3/BIN5 gene function, we examined the chromosome ploidy levels from several cell types and found no differences between wild-type and mutant plants (J. Traas, H. Hofte, Y.Y., and J.C., unpublished results), suggesting that chromosome instability in somatic cells is an unlikely reason for the mutant phenotypes. Recent studies indicate that many factors involved in DNA replication, repair, and recombination are components of chromatin remodeling complexes and are involved in transcription regulation (39). It is tempting to speculate that BIN5/AtSPO11-3 and BIN3/AtTOP6B may constitute some kind of chromatin-remodeling complex with other factors that are required for the expression of a large set of genes including those regulated by BRs.
Consistent with this idea, microarray experiments showed that 321 Arabidopsis genes (of 5,500 genes analyzed) are down-regulated in both bin3 and bin5 mutant plants (see Table 2). Although many of the affected genes are probably due to secondary effects of the mutant phenotypes, at least some of them may be primary consequences of the mutations. Consistent with the similar mutant phenotypes between bin3, bin5, and bri1, many (18 of 30) BR-regulated genes are down-regulated in bin3 and bin5 mutants. The significance of the decreased BR-regulated gene expression in the mutants is further supported by the predicted function of the target genes. The 18 BR-induced genes that are down-regulated in bin3 and bin5 include putative cell wall-modifying proteins such as xyloglucan endotransglycosylase (XET), xyloglucan endo 1,4-β-glucanase (EGase), and expansin (Table 1; refs. 4, 36, and 37). XETs catalyze the cleavage and rejoining of the cell wall component xyloglucan and are involved in BL-induced cell elongation (38). Mutation of the KORRIGAN (KOR) gene encoding a putative EGase results in dwarf plants with defects in cell elongation and changes in cell wall composition (40). Our observations are consistent with the idea that BRs may regulate genes for cell wall-modifying enzymes, thereby regulating cell elongation. The down-regulation of these BR-regulated genes in bin3 and bin5 mutants provides a possible explanation for the similar dwarf phenotypes of bin3 and bin5 with those of bri1. It is surprising that several BR up-regulated genes have homology to auxin-regulated genes, including SAUR-AC1 that is known to be an early auxin-induced gene (41). It is known that BL and auxin act synergistically to regulate cell elongation (1). The induction of these genes by BL and auxin provides a potential mechanism for the interaction of these two plant hormones in regulating cell elongation.
In conclusion, the availability of the bin3 and bin5 mutants and cloning of the corresponding genes provide useful tools to characterize the enzyme activities of these unique genes in a eukaryotic system, to elucidate the molecular mechanisms of the gene functions in plant growth and development, and to understand their roles in modifying BR-regulated gene expression.
Supplementary Material
Acknowledgments
We thank Julin Maloof for constructing the phylogenetic trees. This work was supported by U.S. Department of Agriculture Grant 9935301-7903 (to J.C.) and the Howard Hughes Medical Institute. H.C. was supported by Crop Functional Genomics Center Grant CG1526, funded by the Republic of Korea. Y.Y. was supported in part by a National Research Service Award postdoctoral fellowship, D.F. by National Institutes of Health Training Grant T32 HD07495, Y.Z. by a fellowship from The Life Sciences Research Foundation, and S.M.-G. by a fellowship from the Human Frontier Science Program Organization.
Abbreviations
- ABA
abscisic acid
- BL
brassinolide
- BR
brassinosteroids
- TOP
topoisomerase
References
- 1.Mandava N B. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:23–52. [Google Scholar]
- 2.Clouse S, Sasse J. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Chory J. J Exp Botany. 1999;50:332–340. [Google Scholar]
- 4.Friedrichsen D, Chory J. BioEssays. 2001;23:1028–1036. doi: 10.1002/bies.1148. [DOI] [PubMed] [Google Scholar]
- 5.Mussig C, Altmann T. Plant Physiol Biochem. 1999;37:757–762. [Google Scholar]
- 6.Li J, Chory J. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z Y, Seto H, Fujioka S, Yoshida S, Chory J. Nature (London) 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 8.He Z, Wang Z Y, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 9.Friedrichsen D M, Joazeiro C A, Li J, Hunter T, Chory J. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Nam K H, Vafeados D, Chory J. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Pérez J M, Ponce M R, Micol J L. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Nam K H. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z Y, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 14.Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 15.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. Nature (London) 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 16.Keeney S. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 17.Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 18.Celerin M, Merino S T, Stone J E, Menzie A M, Zolan M E. EMBO J. 2000;19:2739–2750. doi: 10.1093/emboj/19.11.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKim K S, Hayashi-Hagihara A. Genes Dev. 1998;12:2932–2942. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dernburg A F, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve A M. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 21.Baudat F, Manova K, Yuen J P, Jasin M, Keeney S. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 22.Romanienko P J, Camerini-Otero R D. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 23.Grelon M, Vezon D, Gendrot G, Pelletier G. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartung F, Puchta H. Nucleic Acids Res. 2000;28:1548–1554. doi: 10.1093/nar/28.7.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartung F, Puchta H. Gene. 2001;271:81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- 26.Villeneuve A M, Hillers K J. Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 27.Neff M M, Neff J D, Chory J, Pepper A E. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 28.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 29.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 30.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 31.Weigel D, Ahn J H, Blazquez M A, Borevitz J O, Christensen S K, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil E J, Neff M M, et al. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 34.Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. Plant J. 1999;18:303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- 35.Clouse S D, Langford M, McMorris T C. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamport D T. Cell Mol Life Sci. 2001;58:1363–1385. doi: 10.1007/PL00000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darley C P, Forrester A M, McQueen-Mason S J. Plant Mol Biol. 2001;47:179–195. [PubMed] [Google Scholar]
- 38.Clouse S D. Plant J. 1996;10:1–8. doi: 10.1046/j.1365-313x.1996.10010001.x. [DOI] [PubMed] [Google Scholar]
- 39.Fyodorov D V, Kadonaga J T. Cell. 2001;106:523–525. doi: 10.1016/s0092-8674(01)00478-0. [DOI] [PubMed] [Google Scholar]
- 40.Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gil P, Liu Y, Orbovic V, Verkamp E, Poff K L, Green P J. Plant Physiol. 1994;104:777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.