Abstract
The maize (Zea mays) Viviparous 1 (Vp1) transcription factor has been shown previously to be a major regulator of seed development, simultaneously activating embryo maturation and repressing germination. Hexaploid bread wheat (Triticum aestivum) caryopses are characterized by relatively weak embryo dormancy and are susceptible to preharvest sprouting (PHS), a phenomenon that is phenotypically similar to the maize vp1 mutation. Analysis of Vp-1 transcript structure in wheat embryos during grain development showed that each homeologue produces cytoplasmic mRNAs of different sizes. The majority of transcripts are spliced incorrectly, contain insertions of intron sequences or deletions of coding region, and do not have the capacity to encode full-length proteins. Several VP-1-related lower molecular weight protein species were present in wheat embryo nuclei. Embryos of a closely related tetraploid species (Triticum turgidum) and ancestral diploids also contained misspliced Vp-1 transcripts that were structurally similar or identical to those found in modern hexaploid wheat, which suggests that compromised structure and expression of Vp-1 transcripts in modern wheat are inherited from ancestral species. Developing embryos from transgenic wheat grains expressing the Avena fatua Vp1 gene showed enhanced responsiveness to applied abscisic acid compared with the control. In addition, ripening ears of transgenic plants were less susceptible to PHS. Our results suggest that missplicing of wheat Vp-1 genes contributes to susceptibility to PHS in modern hexaploid wheat varieties and identifies a possible route to increase resistance to this environmentally triggered disorder.
The Viviparous1 (Vp1) gene is a major regulator of late embryo development in maize. Inactivation of this locus leads to disruption of embryo maturation, resulting in the promotion of germination of embryos while still attached to the cob (vivipary). The phenotypes of vp1 mutants show that this locus performs two distinct functions: to promote embryo maturation and embryo dormancy and simultaneously to repress germination (1, 2). Specific genes and enzyme activities have been identified that are activated or repressed by Vp1 in vivo. Expression of several enzymes (3) and maturation-related genes (4) is reduced in the mutants including those encoding seed-storage proteins and diverse LEA (late embryogenesis-abundant) proteins (1). On the other hand, α-amylase activity increases in the same mutants during aleurone maturation (3). These observations indicate a bifunctional role for Vp1 in regulating the balance between the activation of embryo-maturation genes and the repression of germinative hydrolases during kernel development and maturation.
Orthologues of Vp1 have been cloned from rice [Osvp1 (4)], wild oat [AfVp1 (5)], and several dicotyledonous species such as Arabidopsis [ABI3 (6)] and poplar [PtABI3 (7)]. Vp1 homeologues have been mapped in wheat to the long arms of the group 3 chromosomes at a location conserved relative to Vp1 in maize and Osvp1 in rice (8). Severe mutation of the Arabidopsis ABI3 gene causes a similar phenotype to that observed in maize vp1 embryos; the apical meristem is activated prematurely during embryo maturation, storage products do not accumulate, and the promoter of the germination-related CAB3 gene is activated before seed shedding (9). The observation that ABI3 functions in a similar way to Vp1 to control embryo maturation and repress germination indicates that this gene plays a central role in coordinating these processes in all flowering plants.
Biochemical analyses have shown that the Vp1 protein functions as a transcription factor, containing both transcriptional activation and DNA binding domains (1, 10). Comparison with orthologues from other species identified three basic domains (B1, B2, and B3) that are highly conserved. Both B2 and B3 show DNA binding activities, B2 nonspecifically enhances the DNA binding activity of other transcription factors, and B3 in isolation from the remainder of the protein shows highly cooperative sequence-specific DNA binding to the Sph cis element present in the promoter of the maize C1 gene.
Embryo dormancy has a profound influence on the bread-making quality of wheat and is influenced by interactions between the genotype and the environment of the developing grain (11). Water loss (desiccation) is an important signal involved in dormancy induction in wheat seeds (12), and both abscisic acid (ABA) synthesis (13) and ABA responsiveness are considered important for maintenance of embryo dormancy (14). Wheat can develop high levels of hydrolytic enzyme activity before harvest as a result of preharvest sprouting (PHS). This developmental disorder occurs when grains mature under cool, damp conditions. Little is known about how the environment influences the underlying molecular processes occurring during embryo maturation and which may be disrupted before visible PHS. PHS is similar to the maize vp1 phenotype in the sense that embryos germinate while still attached to the mother plant. In wild oat, expression of AfVp1 is controlled by interactions between the environment and the seed genotype such that there is a strong correlation between the AfVp1 mRNA content and the dormancy of the seed (5). Therefore changes in the environment can affect the level of Vp1 gene expression, which is associated with a transition from embryo development to germination.
In this paper, we analyzed the structure and expression of the three Vp-1 homeologues of wheat (TaVp1). Our analyses revealed that although each gene has the potential to encode a full-length functional protein, incorrect splicing of pre-mRNAs leads to a diverse RNA population, the majority of which encode aberrant translation products. Analysis of transcript structures in ancestral and closely related species indicates that missplicing of Vp-1 genes was present before the evolution and domestication of hexaploid wheat. Transgenic wheat seeds expressing the AfVp1 cDNA showed increased dormancy and resistance to PHS, suggesting that missplicing of native TaVp1 transcripts contributes to increased PHS.
Materials and Methods
Plant Material.
Seed stocks of hexaploid, tetraploid, and ancestral diploid wheats were obtained from the Institute of Plant Science Research Collection of Wheat and Related Species currently maintained at John Innes Centre (Norwich, UK). Both Soleil and Cadenza are red wheats.
RNA Isolation.
Poly(A)+ RNA was extracted from embryos as described (15). Reverse transcription (RT)–PCR was carried out by using Ready-To-Go RT-PCR beads (Amersham Pharmacia) with poly(A)+ RNA or polysomal RNA following manufacturer instructions. The oligonucleotide primers used were: forward (5′-ATCCAAACCGGCGGCTTCCCTCAAGAG-3′) and reverse (5′-CTTCTCTTTGCAACCACCGTCTTC-3′). Ten percent of each PCR was analyzed by Southern blotting after electrophoresis through 1% (wt/vol) agarose gels (16) by using the insert from Vp-A1 cDNA 10 as a probe, which showed that all cDNA products visible on agarose gel electrophoresis contained Vp-1 sequence (data not shown). Preliminary RT-PCR experiments were performed by using a range of RNA concentrations to ensure that the PCRs were carried out within a range that was quantitative, and RNase treatment of RNA samples before analysis resulted in no amplification products after RT-PCR (data not shown). The oligonucleotides used for analyses of progenitor/closely related sequences were based on wheat Vp-1 sequences; all oligonucleotides spanned the five introns, with the only difference being the starting point of the forward primers. Embryos from mature seeds all were imbibed for 6 h before RNA extraction.
Construction and Screening of cDNA and Genomic Libraries.
A wheat (cultivar Soleil) embryo-specific cDNA library was constructed with poly(A)+ RNA from mature, dormant seeds imbibed for 6 h. Screening was carried out according to manufacturer instructions. Isolated cDNA clones were sequenced by using the dRhodamine dye-terminator cycle-sequencing ready-reaction kit, and reaction products were analyzed on an ABI Prism 377 DNA sequencer (Perkin–Elmer Applied Biosystems, Warrington, UK). DNA sequence analysis was performed by using MACVECTOR software (Oxford Molecular Group, UK). Genomic DNA (cultivar Soleil) was purified as described (17). A genomic DNA library was constructed by cloning DNA partially digested with MboI into the XhoI site of Lambda Fix II (Stratagene). The primary library was divided into aliquots that were screened by PCR to identify those containing TaVp1 clones. Isolation of TaVp1 clones from positive aliquots was carried out by using standard procedures (16).
Genome Allocation of Wheat Vp-1 Genomic Clones.
Primers were designed to DNA-sequence from Vp-1 genomic clones at two positions downstream of the fifth intron. The primers used were: Vp-D1, forward (5′-AGGATCTAGCCAAGCACAAGAATGG-3′) and reverse (5′-CAACAGGTATACTTGAGCTAGCTAAG-3′); Vp-A1, forward (5′-AATATCTGATACGCGGCGTGAAGGTG-3′) and reverse (5′-CGGGTATACTTGAGCTAGCTAACTG-3′); Vp-B1, forward (5′-GGCGATCTCGCCAGGAGGCCAG-3′) and reverse (5′-CAGCGTCACATCTGACCAAGAAACC-3′).
Gene-specific amplification conditions were: 1 cycle of 94°C, 1 min; 5 cycles of 94°C, 45 sec, 70°C, 1 min, 72°C, 1 min; and 30 cycles of 94°C, 45 sec, 65°C, 1 min, and 72°C, 1 min.
Western Analysis of Wheat VP-1 Proteins.
Nuclear proteins were extracted from mature, dormant embryos imbibed for 6 h or young, expanding leaves as described (18). Proteins were precipitated from nuclear extracts in a 40% saturated ammonium sulfate solution to remove histones. Western analysis was carried out by using a polyclonal antibody produced against a synthetic peptide consisting of the generic B2 domain of Vp1 homologue proteins as described (16).
Production and Analysis of Transgenic Wheat Plants.
The AfVp1 cDNA was placed in the vector pUPLN (P.A.L., unpublished data), providing expression under the ubiquitin promoter (19). Wheat transformation was carried out as described (20) by using the marker plasmid (pCaI Neo) that confers resistance to the antibiotic geneticin (G418) under the control of the CaMV 35S promoter.
ABA Responsiveness of Excised Embryos.
Homozygous T4 seedlings of three independent transgenic wheat lines expressing wild oat VP1 (AfVP1) in the background cultivar, Cadenza, were grown in pots in a controlled-environment room under day/night temperatures and light intensities reflecting average UK spring/summer conditions. To test ABA responsiveness of embryos, grains from ears harvested at 5 weeks postanthesis were pooled, and embryos were dissected by making a cut parallel to the face of the scutellum. For each line and ABA concentration, three replicates of 10 embryos were placed on filter paper moistened with 1.0 ml of ABA solutions (0, 1.0, 3.0, 10, 30, and 100 μM) and incubated at 15°C in darkness for 7 days. The number of embryos where the coleorhiza had ruptured the pericarp was scored and removed at daily intervals. From these data a percentage germination index (%GI) (14), which gives greater weight to embryos that germinate sooner than those germinating later, was calculated for each ABA dose within a replicate for each line. An analysis of variance of the %GI values using a two-factor structure (line x ABA dose) was made by using an angular transformation of the data to improve variance homogeneity across the range.
“In-Ear” Grain Sprouting.
Five ripe ears from each line harvested at 9 and 14 weeks postanthesis were placed on paper towels on a free-draining flat surface in a mist propagator, and the numbers of nonsprouted and sprouted grains visible with the naked eye were scored after 7 days. Again an angular transformation of the data were used to improve variance homogeneity across the range before statistical analysis.
Results
Structures of Wheat Vp-1 Genes.
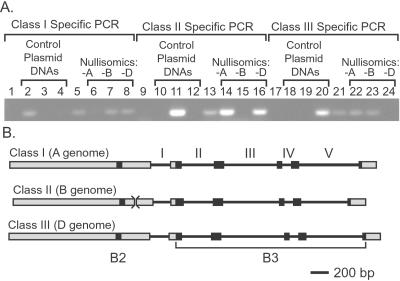
The Viviparous 1 family of transcription factors has been shown previously to be highly conserved in flowering plants (5, 6). We used the Avena fatua orthologue (AfVp1) as a probe to identify Vp-1 transcripts in mature wheat embryos (data not shown) and to clone cDNAs representing TaVp1 mRNAs. These cDNAs were used to identify three classes of TaVp1 genomic clones (Fig. 1) representing three homeologous, single-copy loci from the A, B, and D genomes of hexaploid wheat. Three sets of class-specific PCR primers, targeting sequences unique to each group in turn, were used to amplify from genomic DNA templates derived from the Chinese Spring (CS) nullisomic–tetrasomic aneuploid lines (Fig. 1A). Single-sized amplification products were obtained in each case, supporting a single-copy hypothesis. This analysis has allowed the unequivocal allocation of each gene class to a specific genome¶ (Fig. 1; class I Vp-A1, class II Vp-B1, class III Vp-D1). Analysis of homeologue sequences indicates that each has the capacity to encode a full-length protein. Comparison between sequences revealed a high level of similarity within coding regions, although the level of similarity is lower in predicted introns. Comparison of the ORFs of all the genes reveals two highly conserved basic regions (B2 and B3) at corresponding positions to those found in other Vp1 homologues (Fig. 1B).
Figure 1.
Allocation of wheat TaVp1 genomic clones to specific genomes. (A) Lanes 1–8 show class I genomic clone-specific PCR. 1, no template DNA; 2–4 class I, II, and III template DNAs, respectively; 5, Soleil genomic DNA; 6–8, CS euploid; 6, CS nullisomic 3A–tetrasomic 3D; 7, CSN3B–T3A; 8, CSN3D–T3A. Lanes 9–16 show class II genomic clone-specific PCR. 9, no template DNA; 10–12, as described for lanes 2–4; 13, as described for lane 5; 14–16, as described for lanes 6–8. Lanes 17–24 show class III genomic clone-specific PCR. 17, no template DNA; 18–20, as described for lanes 2–4; 21, as described for lane 5; 22–24, as described for lanes 6–8. (B) Schematic representation of Vp-1 homeologues (to scale). Exons (boxed) and introns (lines, numbered) are shown, as are the positions of the B2 and B3 domains within the coding regions (black boxes). The position of a 12-bp deletion in Vp-B1 is indicated (><).
The Majority of TaVp1 Cytoplasmic Transcripts Are Spliced Incorrectly.
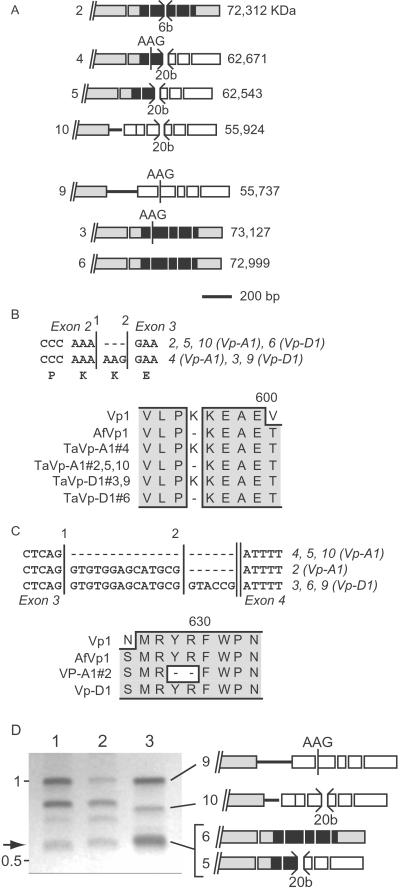
Preliminary experiments using Northern analysis showed that Vp-1 mRNA was expressed in imbibed mature embryos that were dormant (data not shown). To study the structure of these transcripts, we isolated and analyzed a number of Vp-1 cDNA clones from a cDNA library produced from mature imbibed dormant embryos. A total of seven Vp-1 cDNA clones were studied in detail (Fig. 2A). Comparison of cDNAs revealed a large variation in structural composition (Fig. 2A). Two cDNAs contained sequence derived from intron 1, either the complete intron, or the 5′ 85 bases. The cDNAs also showed alternative splicing at the beginning of exon 3, which gave rise to either the inclusion or exclusion of an AAG codon (introducing a lysine residue within the B3 domain; Fig. 2B). Deletions at the end of exon 3 were observed in several cDNAs, leading either to premature termination of the ORF (Fig. 2C, splice site 1) or the deletion of two codons within the B3 domain (splice site 2). Only three cDNA clones had the potential to encode full-length or near full-length VP-1 proteins. Other cDNAs encoded derivatives with frameshifts in the reading frame, resulting in introduced stop codons. In all cases introduction of a stop codon would terminate translation before the B3 domain. Analysis of the relative abundance of the different Vp-1 RNAs (Fig. 2D) indicates that correctly spliced transcripts represent a very small minority of the total cytoplasmic mature embryo Vp-1 RNA population, and therefore that the majority of Vp-1 mRNAs are unlikely to encode functional protein. The abundance of misspliced transcripts was relatively constant throughout grain development (data not shown).
Figure 2.
Structures of TaVp1 RNAs. (A) Structure of misspliced cDNAs derived from wheat Vp-1 homeologues [to scale; clone numbers are indicated (Left)]. Exons are indicated as boxes, and intron sequences are shown as horizontal lines. The positions of intron sequence, alternatively spliced AAG codons, and short deletions (b, bases) are shown. Where missplicing leads to incorporation of a stop codon in the ORF is indicated as open boxes following the stop codon, and the sizes of presumptive resulting proteins are indicated next to each transcript (Right). (B) Alternative splicing (either at position 1 or 2 at the junction of intron 2 and exon 3) leads to deletion of an AAG codon in several RNAs from both homeologues. Also shown is the predicted protein sequence compared with maize (Vp1) and A. fatua (AfVp1). (C) Premature splicing of exon 3 in Vp-A1 leads to termination of the ORF (position 1) or deletion of two residues (position 2) within the highly conserved B3 domain. Also shown is the predicted protein sequence compared with Vp1 and AfVp1. (D) Agarose gel electrophoresis analysis of RT-PCR amplification products. Comparison of products from polysomal (lane 1) and poly(A) RNA (lane 2) sources from mature embryos, with combined products from cDNA clones (lane 3), is shown (clones are indicated diagrammatically). The arrow indicates the position of correctly spliced transcript product. The positions of molecular weight markers are indicated (in kb).
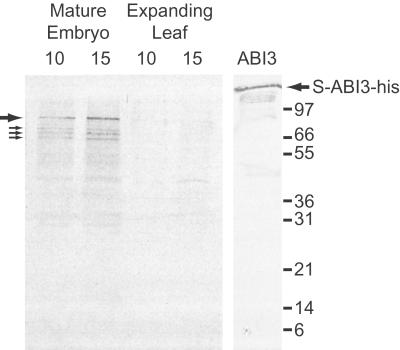
The composition of VP-1 proteins in wheat nuclei (Fig. 3) was analyzed by using a polyclonal antibody raised to the B2 domain (Fig. 1) that is conserved in all VP-1 orthologue proteins. Because all the alternative/missplicing events occur after the B2 domain, this antibody should detect all protein products derived from Vp-1 RNAs. The antibody specifically recognized a full-length derivative of Arabidopsis ABI3 produced in E. coli (Fig. 3). As anticipated, the antibody did not interact with antigens present in nuclear extracts derived from wheat leaf material but did recognize at least four proteins ranging in size from 83 to 64 kDa in nuclear extracts from mature wheat embryos. It is difficult to predict the position to which full-length VP-1 protein would migrate under SDS/PAGE (73 kDa based on the predicted ORF size), because the ABI3 protein (79.5 kDa) has been shown previously to migrate with a higher than expected molecular weight (ref. 21; Fig. 3, S-ABI3-his, 85 kDa).
Figure 3.
Western analysis of nuclear-localized VP-1 proteins. Western analysis was carried out with a B2-specific antibody by using nuclear proteins derived from mature wheat embryos or young, expanding leaves (with 10 or 15 μg of protein). Whole Escherichia coli extracts containing the ABI3 derivative protein, S-ABI3-his, were used to show the specificity of the antibody. The positions of proteins detected in embryo extracts are shown, the thick arrow indicates the major immunoreactive protein, and the thin arrows indicate three lower molecular weight proteins (83, 74, 70, and 64 kDa, respectively). Positions of molecular weight markers are indicated (in kDa).
Vp-1 Homeologue RNAs Are Misspliced in Diploid and Tetraploid Ancestral Wheats and Closely Related Species.
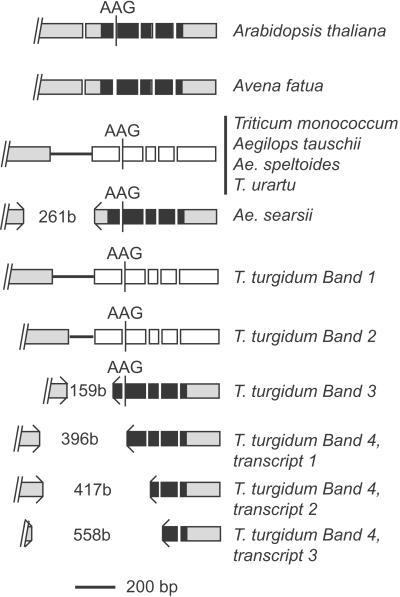
To determine whether missplicing of Vp-1 transcripts was specific to modern hexaploid wheat (genomes AABBDD) or present in ancestral and related species, we carried out RT-PCR experiments by using poly(A) RNA from embryos from the following species: Triticum monococcum (AmAm), Triticum urartu (AA), Aegilops speltoides (SS), Aegilops searsii (SsSs), Aegilops tauschii (DD), Triticum turgidum ssp. durum (AABB), the more distantly related A. fatua, and Arabidopsis (Fig. 4). The wheat relatives were chosen for their close genomic relationships to the hexaploid: A and Am to the donor of the A genome, S and Ss representing unknown Sitopsis group donor of the B genome, Ae. tauschii is thought to have been the D genome donor. These experiments used primers bounding all introns as described (Fig. 2). The RT-PCR products from A. fatua and Arabidopsis were of the expected size for correctly spliced transcripts. However, in all cases transcript sizes and numbers from Triticum and Aegilops species were not as expected for correct splicing of introns. A single Vp-1 cDNA band was produced in each of the diploid species analyzed, indicating that a single Vp-1 transcript is produced in these species. Tetraploid T. turgidum produced at least four cDNA bands visible on agarose gel electrophoresis, indicating that multiple Vp-1 transcripts are produced from homeologous genes. Intron 1 was not excised properly in several species, resulting in transcripts containing the complete intron (T. monococcum, T. urartu, Ae. tauschii, Ae. speltoides, and T. turgidum, band 1) or a truncated version containing only the first 85 bp of intron 1 (T. turgidum, band 2). These missplicing events result in loss of the ORF before the B3 domain. Transcripts derived from two species contained partial deletions of exon 1 and 2 sequences caused by missplicing within exons (Ae. searsii and T. turgidum, band 3 and band 4 types, respectively), in both cases resulting in deletions of coding regions, that leave the predicted protein in-frame. For Ae. searsii, missplicing results in a putative protein with a deletion of 87 aa, and for T. turgidum band 3, 53 aa, in the latter case with a deletion of the initial 8 aa of the B3 domain. For T. turgidum band 4, multiple RNA species were identified with deletions of exon material that included substantial portions of the B3 domain.
Figure 4.
Structure of Vp-1 RNAs in diploid and tetraploid wheat ancestors and related species. Exons are indicated as boxes, and intron sequences are indicated as horizontal lines. The positions of intron sequence, alternatively spliced AAG codons, and short deletions (b, bases) are shown (to scale). Where missplicing leads to incorporation of a stop codon in the ORF is indicated as open boxes following the stop codon. The positions of deletions are indicated (><).
Transgenic Wheat Seeds Expressing A. fatua Vp1 Have Increased Responsiveness to ABA and Resistance to PHS.
The observed missplicing of wheat Vp-1 homeologues might explain the propensity of this species to show PHS (22). To investigate whether it was possible to increase embryo dormancy and resistance to PHS in wheat, we produced transgenic wheat plants that expressed the A. fatua Vp1 homologue (AfVp1) under the control of the constitutive ubiquitin promoter (19). There were no obvious effects of ectopic expression of AfVp1 on plant height, flowering time, or tillering under our growth conditions. Similarly, there were no effects of tissue culture or transgene insertion on seed viability or long-term embryo dormancy in the dry seed. Developing embryos and ears from transgenic lines were analyzed to determine the effect of the expression of this gene on embryo responsiveness to ABA and resistance to PHS. At 5 weeks postanthesis, there was very strong evidence of overall ABA effects (F probability < 0.001) on germination index (angular transformed data) and overall differences between lines (F probability = 0.002). The test for statistical interaction, ABA × lines, was not significant (F probability = 0.356), implying that the shape of the dose-response curves for inhibition of embryo germination in response to ABA is similar for all lines. The overall differences between lines therefore can be summarized by the angular transformed means across all levels of ABA (Table 1). The mean germination index for the three transgenic lines expressing AfVp1 is significantly lower, and their embryos therefore were more responsive to exogenous ABA than the control. The influence of transgene expression on resistance to PHS in the ear was analyzed also. The proportion of sprouted grains in ears harvested at 9 and 14 weeks postanthesis, after misting for 1 week, showed highly significant increases with time, reflecting a gradual loss of embryo dormancy during grain after ripening (Table 2). Only one line showed a significant decrease in the proportion of sprouted grains in ears harvested at 9 weeks compared with control (angular transformed data). However, all three AfVp1 expression lines showed a significant decrease in the number of sprouted grains (P < 0.001 for lines 6 and 7, and P = 0.002 for line 11) in misted ears harvested 14 weeks postanthesis. These results confirm that expression of this transcription factor also decreases the potential for PHS in these lines.
Table 1.
Mean percentage germination indices (%GI) of wheat embryos at 5 weeks postanthesis incubated in ABA solutions at 15°C over 7 days
| Genotype | ABA, μM (back-transformed means, %GI)
|
Mean %GI (angular scale) | t* | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 3.0 | 10 | 30 | 100 | |||
| Cadenza | 65.8 | 45.7 | 34.2 | 24.7 | 9.5 | 0.2 | 30.43 | |
| Line 6 | 61.6 | 33.2 | 29.7 | 18.9 | 0.7 | 0.2 | 25.43 | <0.001 |
| Line 7 | 54.8 | 38.1 | 36.6 | 16.2 | 2.7 | 0.0 | 26.05 | 0.003 |
| Line 11 | 61.4 | 35.7 | 26.7 | 15.7 | 2.7 | 0.5 | 25.76 | 0.001 |
| Least significant difference (P = 0.05) = 2.77 | ||||||||
, t probability is for testing each line against the control genotype, Cadenza.
Table 2.
Mean percentage of sprouted grains in ears of wheat lines after 7 days in a mist propagator
| Genotype | Time postanthesis, weeks
|
|||
|---|---|---|---|---|
| 9 | 14 | 9 | 14 | |
| (back-transformed, %) | (angular scale) | |||
| Cadenza | 30.2 | 79.4 | 33.3 | 63.0 |
| Line 6 | 20.7 | 34.8 | 27.1 | 36.1 |
| Line 7 | 9.3 | 41.1 | 17.7 | 39.9 |
| Line 11 | 30.5 | 53.8 | 33.5 | 47.2 |
| Least significant difference (P = 0.05) = 9.5 | ||||
Discussion
Genome-specific PCR enabled unequivocal allocation of each of the Vp-1 genes to one of the three constituent genomes of hexaploid bread wheat (Fig. 1). The expression of these genes was analyzed in detail and revealed that transcripts derived from all three homeologues are highly misspliced (Fig. 2). Observed missplicing events include nonremoval of whole or partial introns, removal of exon material, and alternative splicing of a codon. Examples of alternative splicing of plant genes similar to those seen in Vp-1 transcripts have been described previously, including the retention of intron material, and splicing within exonic regions (23–25). In addition to naturally occurring splicing variation, analysis of splicing in mutated genes has indicated that exon definition is an important feature of control and also that there may be interactions between introns such that inefficient splicing of one intron affects splicing of associated introns (26). Analysis of Vp-1 transcript structure showed many combinations of intron insertions/exon deletions that do not fit easily with these models. Described examples of alternative pre-mRNA splicing in plants usually results in conservation of the ORF and therefore production of functional protein. For the majority of Vp-1 transcripts this was not the case, and missplicing resulted in termination of the ORF before the B3 domain. Extensive missplicing of transcripts has been observed also for the maize Intensifier1 (In1) transcription factor gene (27). Mutations at In1 cause an increase in flavonoid production in the aleurone, suggesting that this transcription factor can function, similar to Vp1, as a repressor of gene transcription. Similar to the results described here, analysis of In1 expression showed that the majority of transcripts were misspliced, indicating that very little functional transcript was made. Missplicing occurred within In1 transcripts before the helix–loop–helix DNA binding domain, suggesting that truncated proteins would not contain specific DNA binding activity. Analysis of cDNA sequences indicated at least three different size ranges for VP-1 protein derivatives, and Western analysis showed at least four different VP-1-related proteins present in embryo nuclei (Fig. 4). The highly conserved B3 domain has been shown in maize to bind DNA in a sequence-specific highly cooperative manner, activating transcription through the Sph cis element present in the C1 promoter (10). Deletion studies using the VP1 protein in a transient expression system have shown that removal of the B3 domain significantly reduces reporter gene expression driven from the Em promoter (28). Analyses of different alleles of ABI3 in Arabidopsis have shown that removal of the latter half of the protein (including the B3 domain) severely reduces function of the protein in repressing postgerminative development (6, 9), and recently Vp1 has been shown to complement this function in a transgenic Arabidopsis abi3 mutant (29). Other data have shown that maize vp1 alleles that lack the B3 domain are still quiescent under normal growth conditions (25). It would be interesting to test these alleles under altered growth conditions that induce PHS (cool and damp) to test the relative contribution of the B3 to suppressing PHS in maize.
The observed missplicing of Vp-1 transcripts in ancestral diploid Triticum and Aegilops species and more modern tetraploids (Fig. 4) suggests that Triticum aestivum has inherited defects in Vp-1 homeologue gene splicing from wild ancestral species. It seems unlikely that this phenomenon resulted from forced evolution during domestication, and that missplicing of this gene may be a common feature within the genera Triticum and Aegilops. Missplicing also occurs in the barley Vp-1 homologue (analysis of sequences from the Japanese Barley expressed sequence tag-sequencing program, data not shown). We did not observe missplicing of the A. fatua Vp1 transcript, although this species is relatively closely related (30). The majority of missplicing events in related diploid and progenitor species were similar to those observed in modern wheat, and some specific missplicing events observed in ancestral species were present also in the hexaploid. For example, transcripts containing the entire intron 1, or the first 85 bp, were observed in progenitors [T. monococcum/T. urartu, that are considered equivalents to the A genome of wheat, Triticum tauschii (D genome) and T. turgidum (AABB)]. Diploid species showed in each case a single Vp-1-related transcript, whereas the tetraploid T. turgidum showed multiple missplicing examples in addition to those shown by the diploid species; it therefore may be possible that increasing ploidy levels (and hence homeologue gene number) also may increase the level of missplicing of the Vp-1 genes. This hypothesis can be tested directly by analyzing missplicing events by using synthetic wheat (“AABBDD”) hybrids that have been constructed recently from T. turgidum and various ancestral diploids (31).
The occurrence of multiple misspliced Vp-1 transcripts in the cytoplasm of embryos suggests that Vp-1 gene function may be compromised in modern wheat. The resulting reduction in VP-1 protein activity may lead to an increased susceptibility to PHS in view of the identified function of VP1 in the regulation of sensitivity to ABA and repression of germination potential (32). Mutations at other loci, related to ABA synthesis, have been shown also to lead to viviparous phenotypes in maize (33) and to reduce dormancy in Arabidopsis (34), and thus it is possible that ABA synthesis/levels could also affect PHS induction in wheat. However, previous results show that desiccation arrests embryo development and acts as a signal to induce embryo dormancy without any increase in ABA content (12, 35), suggesting that ABA levels per se are not related directly to dormancy induction in wheat. Other evidence shows that ABA levels in embryos are similar in both sprouting sensitive and resistant cultivars and that sprouting behavior is more related to the extent of ABA responsiveness of embryos (36). Analysis of transgenic wheat embryos showed that dormancy levels measured as increased ABA responsiveness could be manipulated via expression of the Vp1 orthologue AfVp1, previously implicated in the regulation of the high levels of embryo dormancy in A. fatua (5). As predicted from vp1 and abi3 mutant studies, inhibition of germination of AfVp1 transgenic wheat embryos is more responsive to ABA than the control. As well as influencing ABA sensitivity of isolated embryos, AfVp1 also increased resistance of wheat ears to PHS. Taken together, these data suggest that missplicing of wheat Vp-1 transcripts may contribute to increased susceptibility to PHS. Although we have shown that transgenic wheat seeds containing the AfVp1 cDNA under the control of the ubiquitin promoter have a heightened responsiveness to exogenous ABA, further experiments will be required to determine the interaction between this transgene and endogenous ABA signaling. Other genetic factors also have been shown to influence dormancy and susceptibility to PHS in wheat, for example effects of testa pigment (R loci) (11), of the Phs locus controlling embryonic dormancy status (37) and of multiple dormancy quantitative trait loci (38). However, our analysis of the expression of Vp-1 homeologues in wheat demonstrates that increasing Vp-1 function in seeds increases ABA sensitivity and PHS resistance and therefore offers one avenue for the manipulation of embryo dormancy levels.
Acknowledgments
We are grateful to Dr. Gillian Arnold, Institute of Arable Crops Research (IACR) Statistics Department, for the statistical analysis. R.S.M., M.D.W., and P.C.B. were funded by grants from the Biotechnology and Biological Sciences Research Council of the United Kingdom (BBSRC, Agri-Food Directorate). IACR–Long Ashton and The John Innes Centre receive grant-aided support from the BBSRC.
Abbreviations
- ABA
abscisic acid
- PHS
preharvest sprouting
- RT
reverse transcription
- CS
Chinese Spring
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AJ400714 (Vp-D1), AJ400713 (Vp-B1), and AJ400712 (Vp-A1)].
The symbol for the set of three wheat homeologues (homologues of the maize Vp1 locus; TaVp1) is Vp-1. Individual loci on A, B, and D genomes are Vp-A1, Vp-B1, and Vp-D1. Following convention, products of loci are VP-A1, etc.
References
- 1.McCarty D R, Hattori T, Carson C B, Vasil V, Lazar M, Vasil I K. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 2.Hoecker U, Vasil I K, McCarty D R. Genes Dev. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- 3.Wilson G F, Rhodes A M, Dickinson D B. Plant Physiol. 1973;52:350–356. doi: 10.1104/pp.52.4.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattori T, Terada T, Hamasuna S T. Plant Mol Biol. 1994;24:805–810. doi: 10.1007/BF00029862. [DOI] [PubMed] [Google Scholar]
- 5.Jones H D, Peters N C B, Holdsworth M J. Plant J. 1997;12:911–920. doi: 10.1046/j.1365-313x.1997.12040911.x. [DOI] [PubMed] [Google Scholar]
- 6.Giraudat J, Hauge B M, Valon C, Smalle J, Parcy F, Goodman H M. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohde A, ArdilesDiaz W, VanMontagu M, Boerjan W. J Exp Bot. 1998;49:1059–1060. [Google Scholar]
- 8.Bailey P C, McKibbin R S, Lenton J R, Holdsworth M J, Flintham J E, Gale M D. Theor Appl Genet. 1999;98:281–284. [Google Scholar]
- 9.Nambara E, Keith K, McCourt P, Naito S. Development (Cambridge, UK) 1995;121:629–636. [Google Scholar]
- 10.Suzuki M, Kao C Y, McCarty D R. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flintham J E, Gale M D. Plant Varieties Seeds. 1988;1:87–97. [Google Scholar]
- 12.King R W. J Exp Bot. 1993;44:1059–1066. [Google Scholar]
- 13.Garello G, Le Page-Degivry M T. Seed Sci Res. 1999;9:219–226. [Google Scholar]
- 14.Walker-Simmons M K. Plant Physiol. 1987;84:61–66. doi: 10.1104/pp.84.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rushton P J, Macdonald H, Huttly A K, Lazarus C M, Hooley R. Plant Mol Biol. 1995;29:691–702. doi: 10.1007/BF00041160. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Sharp P J, Chao S, Desai S, Gale M D. Theor Appl Genet. 1989;78:342–348. doi: 10.1007/BF00265294. [DOI] [PubMed] [Google Scholar]
- 18.Grierson C, Du J S, Zabala M D, Beggs K, Smith C, Holdsworth M, Bevan M. Plant J. 1994;5:815–826. doi: 10.1046/j.1365-313x.1994.5060815.x. [DOI] [PubMed] [Google Scholar]
- 19.Christensen A H, Quail P H. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 20.Rasco-Gaunt S, Riley A, Barcelo P, Lazzeri P A. Plant Cell Rep. 1999;19:118–127. doi: 10.1007/s002990050721. [DOI] [PubMed] [Google Scholar]
- 21.Parcy F, Valon C, Kohara A, Misera S, Giraudat J. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holdsworth M, Kurup S, McKibbin R. Trends Plant Sci. 1999;4:275–280. [Google Scholar]
- 23.Thorbjornsen T, Villand P, Kleczkowski L A, Olsen O A. Biochem J. 1996;313:149–154. doi: 10.1042/bj3130149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKnight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 25.Werneke J M, Chatfield J M, Ogren W L. Plant Cell. 1989;1:815–825. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorkovic Z J, Kirk D A W, Lambermon M H L, Filipowicz W. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 27.Burr F A, Burr B, Scheffler B E, Blewitt M, Wienand U, Matz E C. Plant Cell. 1996;8:1249–1259. doi: 10.1105/tpc.8.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carson C B, Hattori T, Rosenkrans L, Vasil V, Vasil I K, Peterson P A, McCarty D R. Plant J. 1997;12:1231–1240. doi: 10.1046/j.1365-313x.1997.12061231.x. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki M, Kao C Y, Cocciolone S, McCarty D R. Plant J. 2001;28:409–418. doi: 10.1046/j.1365-313x.2001.01165.x. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg E A. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozkan H, Levy A A, Feldman M. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarty D R, Carson C B. Physiol Plant. 1991;81:267–272. [Google Scholar]
- 33.Tan B C, Schwartz S H, Zeevaart J A D, McCarty D R. Proc Natl Acad Sci USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koornneef M, Léon-Kloosterziel K M, Schwartz S H, Zeevaart J A D. Plant Physiol Biochem (Paris) 1998;36:83–89. [Google Scholar]
- 35.Black M, Corbineau F, Gee H, Come D. Plant Physiol. 1999;120:463–471. doi: 10.1104/pp.120.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker-Simmons M, Sesing J. J Plant Growth Regul. 1990;9:51–56. [Google Scholar]
- 37.Flintham J E. Seed Sci Res. 2000;10:43–50. [Google Scholar]
- 38.Kato K, Nakamura W, Tabiki T, Miura H, Sawada S. Theor Appl Genet. 2001;102:980–985. [Google Scholar]