Abstract
Bacterial spores are renowned for their longevity, ubiquity, and resistance to environmental insults, but virtually nothing is known regarding whether these metabolically dormant structures impact their surrounding chemical environments. In the present study, a number of spore-forming bacteria that produce dormant spores which enzymatically oxidize soluble Mn(II) to insoluble Mn(IV) oxides were isolated from coastal marine sediments. The highly charged and reactive surfaces of biogenic metal oxides dramatically influence the oxidation and sorption of both trace metals and organics in the environment. Prior to this study, the only known Mn(II)-oxidizing sporeformer was the marine Bacillus sp. strain SG-1, an extensively studied bacterium in which Mn(II) oxidation is believed to be catalyzed by a multicopper oxidase, MnxG. Phylogenetic analysis based on 16S rRNA and mnxG sequences obtained from 15 different Mn(II)-oxidizing sporeformers (including SG-1) revealed extensive diversity within the genus Bacillus, with organisms falling into several distinct clusters and lineages. In addition, active Mn(II)-oxidizing proteins of various sizes, as observed in sodium dodecyl sulfate-polyacrylamide electrophoresis gels, were recovered from the outer layers of purified dormant spores of the isolates. These are the first active Mn(II)-oxidizing enzymes identified in spores or gram-positive bacteria. Although extremely resistant to denaturation, the activities of these enzymes were inhibited by azide and o-phenanthroline, consistent with the involvement of multicopper oxidases. Overall, these studies suggest that the commonly held view that bacterial spores are merely inactive structures in the environment should be revised.
The uncanny resistance of bacterial spores to physical and chemical insults (e.g., heat, desiccation, radiation, oxidants, and proteases) (35) enables them to persist in the environment for as long as thousands or millions of years without losing the capacity for germination and outgrowth (9, 21, 27, 39, 50). In the dormant resting state, totally devoid of metabolic activity, spores are generally assumed to have a negligible impact on the chemistry of the environments that they inhabit. There are, however, examples of how spores may influence the biology (biota) of a given habitat, such as the spore-associated insecticidal crystal proteins produced by Bacillus thuringiensis and Bacillus sphaericus (38, 42). Although a variety of enzymes are known to be associated with both the morphogenesis and germination of bacterial spores (16), the extent to which spore-associated enzymatic activities may actually influence their surrounding chemical environment is virtually unknown.
The only described example of bacterial spores that can significantly impact the distributions of heavy metals in the environment is the Bacillus sp. strain SG-1, isolated over 20 years ago from a shallow marine sediment off Scripps Pier, La Jolla, Calif. (33). Dormant spores of this organism enzymatically catalyze the oxidation of soluble manganese(II) to highly insoluble Mn(III, IV) oxide precipitates on the spore surface, thereby becoming encased in a metal oxide shell (40). The highly charged and reactive surfaces of Mn oxides are known to dramatically influence chemical distributions in the environment by oxidizing a wide array of organic and inorganic compounds as well as scavenging numerous heavy metals (e.g., Cu, Co, Cd, Zn, Ni, and Pb) and radionuclides out of solution (24, 32). SG-1 spores can increase the rate of Mn(II) oxidation, a thermodynamically favorable but kinetically slow reaction at neutral pH, by 4 to 5 orders of magnitude relative to abiotic rates (23). In addition, biogenic Mn oxides have been shown to have significantly greater surface areas and trace metal adsorption capacities than abiotically produced Mn oxides (34).
SG-1 has been studied as a model Mn(II)-oxidizing organism for many years (19, 47), due in part to the remarkable stability of the Mn(II)-oxidizing activity over a wide range of environmental conditions, including temperature (3 to 70°C), metal concentrations (less than nanomolar to more than millimolar), and ionic strength (freshwater to seawater). Molecular genetic studies of Mn(II) oxidation by SG-1 have revealed the involvement of a specific gene product, MnxG, which shares sequence similarity with multicopper oxidases (49). Members of this diverse family of proteins, which includes ascorbate oxidase, laccase, and ceruloplasmin, utilize copper ions of three spectroscopically distinct types as cofactors in the oxidation of a variety of substrates (41, 44). The recent identification of multicopper oxidase-like genes involved in Mn(II) oxidation in two other phylogenetically distinct proteobacteria, Pseudomonas putida GB-1 (8) and Leptothrix discophora SS-1 (11), suggests that these bacterial Mn(II) oxidases may represent a new functional group of multicopper oxidases. Recent spectroscopic studies have demonstrated that Mn(IV) minerals are the primary product of Mn(II) oxidation by SG-1 spores (4), formed most likely through sequential one-electron transfers with a transient Mn(III) intermediate, consistent with the involvement of a multicopper oxidase.
Spore-forming Bacillus species are generally considered to be environmentally ubiquitous, yet very few studies have focused on their significance in aquatic or sedimentary environments (5, 43). Our laboratory has demonstrated that, although spore-forming bacteria constitute a relatively small fraction (<1%) of the total colony-forming bacteria in coastal (San Diego, Calif.) surface sediments, a considerable portion (17 to 33%) of these organisms produce Mn(II)-oxidizing spores (31). In addition, the heat-resistant fraction of the microbial populations in these sediments accounted for a large fraction of the total Mn(II)-oxidizing activity (31), suggesting that spores may be major catalysts of this process in situ. In the present study, we report for the first time that Mn(II)-oxidizing sporeformers represent a phylogenetically diverse group of organisms within the genus Bacillus, based on both 16S rRNA and MnxG (multicopper oxidase) sequences obtained from marine sediment isolates. In addition, the extremely resilient Mn(II)-oxidizing spore enzymes of these organisms have the potential to significantly impact the biogeochemical cycling of elements in aquatic sedimentary environments.
MATERIALS AND METHODS
Sample collection and strain isolation.
Surface (top 1 to 3 cm) sediments were collected from three different environments: San Diego Bay, Mission Bay, and Point Loma (San Diego, Calif.). Samples were collected at the waterline from the shores of San Diego Bay and Mission Bay during low tide. Point Loma sediments were collected ≈10 miles off the San Diego coast using a box core deployed off the R.V. New Horizon. All sediment samples were transferred to sterile 50-ml centrifuge tubes and transported to the laboratory on ice. Sediment samples were subsequently diluted in sterile seawater, incubated at 80°C for 10 min, and spread on Mn(II)-containing K plates (48). K medium contains 2.0 mg of peptone (Difco) per ml and 0.5 mg of yeast extract (Difco) per ml in sterile 75% seawater with both 20 mM HEPES (pH 7.6) and 100 μM MnCl2 added after autoclaving. Mn(II)-oxidizing strains were isolated based on the ability to produce brown Mn oxide-encrusted colonies on plates. The presence of Mn oxides was confirmed using the colorimetric dye leucoberbelin blue (28). A number of additional Bacillus species (Table 1) were also tested for Mn(II) oxidation on K medium made with either seawater or deionized water, depending on the origin of the strain. The salt tolerances of isolates were determined on SW-10 medium (36) containing 10% (wt/vol) total salts, supplemented in some cases with an additional 5 to 10% NaCl.
TABLE 1.
Mn(II)-oxidizing and non-Mn(II)-oxidizing Bacillus strains used in this study
| Organism | Strain | Origin of Mn(II)-oxidizing isolates | Mn(II) oxidationa | Sourceb |
|---|---|---|---|---|
| Bacillus sp. | SG-1 | Scripps Pier sediments, La Jolla, Calif. | + | 33 |
| MB-1 | Mission Bay sediments, San Diego, Calif. | + | This study | |
| MB-3 | Mission Bay sediments, San Diego, Calif. | + | This study | |
| MB-5 | Mission Bay sediments, San Diego, Calif. | + | This study | |
| MB-7 | Mission Bay sediments, San Diego, Calif. | + | This study | |
| MB-9 | Mission Bay sediments, San Diego, Calif. | + | This study | |
| MB-12 | Mission Bay sediments, San Diego, Calif. | + | This study | |
| PL-7 | Point Loma sediments, San Diego, Calif. | + | This study | |
| PL-12 | Point Loma sediments, San Diego, Calif. | + | This study | |
| PL-16 | Point Loma sediments, San Diego, Calif. | + | This study | |
| PL-21 | Point Loma sediments, San Diego, Calif. | + | This study | |
| PL-26 | Point Loma sediments, San Diego, Calif. | + | This study | |
| PL-30 | Point Loma sediments, San Diego, Calif. | + | This study | |
| SD-18 | San Diego Bay sediments, Calif. | + | This study | |
| B. cereus | ATCC 10876 | − | ATCC | |
| B. circulans | ATCC 4513 | − | ATCC | |
| B. firmus | ATCC 14575 | − | ATCC | |
| B. licheniformis | ATCC 14580 | − | ATCC | |
| B. megaterium | ATCC 14581 | − | ATCC | |
| B. pumilus | ATCC 72 | ∗∗ | ATCC | |
| B. subtilis | PY79 | − | A. Driks | |
| B. thuringiensis | ATCC 35866 | − | ATCC | |
| H. litoralis | ATCC 700076 | − | ATCC | |
| S. marismortui | ATCC 700626 | − | ATCC |
Mn(II) oxidation was detected by the formation of brown Mn oxides (confirmed by reaction with leucoberbelin blue) on sporulated colonies after 10 days of incubation on K plates: −, no detectable Mn oxide formation; +, positive; ∗∗, weakly oxidized Mn(II) on plates but no Mn(II)-oxidizing activity was detected from purified spores or in gels.
ATCC, American Type Culture Collection, Manassas, Va.
DNA extraction, PCR, cloning, and sequencing.
DNA was extracted from cultures using the DNeasy DNA extraction kit (Qiagen). For amplification of 16S rRNA genes, the primers 27F and 1492R (30) were used in a standard 30-cycle PCR with Taq polymerase and an annealing temperature of 50°C. For amplification of mnxG homologues from various Bacillus isolates, PCR primers were designed based on the determinants of two of the copper-binding regions within the SG-1 sequence (49). The sequences are as follows: mnxGIF, 5′-ACGCATGTCTTTCACTATCATGTTCAT-3′, and mnxGIR, 5′-AAATAAGTGGTCATGGAAGAACCATGC-3′. The PCR program for mnxG amplification was 30 cycles of 94°C (30 s), 45°C (30 s), and 60°C (1 min), followed by one cycle of 72°C (15 min). The PCR products were cloned into the vector pCR2.1 using a TOPO-TA cloning kit (Invitrogen). Plasmid DNA was purified using the Qiagen miniprep kit, and both strands of the cloned PCR products were sequenced using an ABI 373A automated sequencer.
Phylogenetic analysis.
16S rRNA sequences were aligned manually using Sequencher and compared to alignments generated using CLUSTAL W and the Ribosomal Database Project Sequence Aligner program, and both gaps and ambiguously aligned regions were removed. Phylogenetic trees were generated by neighbor joining, using Kimura two-parameter corrected distances, or by maximum parsimony within the PAUP (version 4.0b3) software package. Derived MnxG amino acid sequences were aligned using CLUSTAL W, and phylogenetic trees were constructed using neighbor joining and parsimony methods within PAUP. Bootstrap analysis was used to estimate the reliability of phylogenetic reconstructions (1,000 replicates). The GenBank, EMBL, and DDBJ accession numbers for the 16S rRNA sequences used for comparison were as follows: Bacillus benzoevorans (X60611), Bacillus cereus (Z84581), Bacillus circulans (D78312), Bacillus cohnii (X76437), Bacillus firmus (D16268), Bacillus halmapalus (X76447), Bacillus lentus (D16272), Bacillus licheniformis (X68416), Bacillus megaterium (D16273), Bacillus methanolicus (X64465), Bacillus pseudomegaterium (X77791), Bacillus pumilus (AB020208), Bacillus simplex (D78478), Bacillus sporothermodurans (U49080), Bacillus subtilis (X60646), B. thuringiensis (D16281), Halobacillus litoralis (X94558), Salibacillus marismortui (AJ009793), and Virgibacillus pantothenticus (D16275). The 16S rRNA sequences from the following environmental clones and isolates were also used: “Bacillus jeotgali” strains YKJ-10 (AF22106) and YKJ-11 (AF221061), “Bacillus permians” strain 2-9-3 (AF166093), environmental clones BPC060 (AF15081), BPC094 (AF15082), and LMG-19416 (AJ276808), strain AS-38 (AJ391199), strain HTE831 (AB010863), strain IrT-RS2 (AJ295684), strain NRRLB-14850 (AF156316), and strain SB45 (AJ229238).
Isolation of Mn(II)-oxidizing acitivity from spores.
Spores were purified from 1-liter K cultures by the method of Rosson and Nealson (40). The Mn(II)-oxidizing activity was isolated by passing purified spores through a French pressure cell six times at 20,000 lb/in2. The stripped spores were then removed by centrifugation at 14,000 × g, and the supernatant, containing the outermost spore layer(s) and most of the activity, was recovered. The supernatants were assayed for Mn(II)-oxidizing activity by incubating them in 10 mM HEPES (pH 7.6) containing 200 μM MnCl2 and observing the formation of brown Mn oxides. The effect of azide (0.1 to 1 mM) on Mn(II)-oxidizing activity was also assayed relative to untreated controls.
SDS-PAGE analysis.
Supernatants from French-press-treated spores of various isolates were mixed with 2× Laemmli buffer, and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in standard 10% gels followed by staining with Coomassie blue for total protein (29). To assay for in-gel Mn(II) oxidation activity, the gels were first incubated in 0.5% Triton X-100-10% glycerol for 30 min to remove SDS and then incubated in 10 mM HEPES buffer (pH 7.6) containing 200 μM MnCl2 (20). Mn(II) oxidation was visualized by the formation of brown Mn oxide bands in the gels after several hours of incubation. To determine the sensitivity of the Mn(II)-oxidizing activity to copper chelators, gels were incubated in HEPES buffer (pH 7.6) containing o-phenanthroline (50 μM) for 15 min prior to the addition of 200 μM Mn(II).
Nucleotide sequence accession numbers.
The 16S rRNA sequences of the 15 Mn(II)-oxidizing Bacillus strains (including SG-1) determined in the present study have been deposited in GenBank under accession numbers AF326359 to AF326373. The partial mnxG gene sequences have been deposited under accession numbers AF326384 to AF326397.
RESULTS AND DISCUSSION
16S rRNA phylogeny of Mn(II)-oxidizing sporeformers.
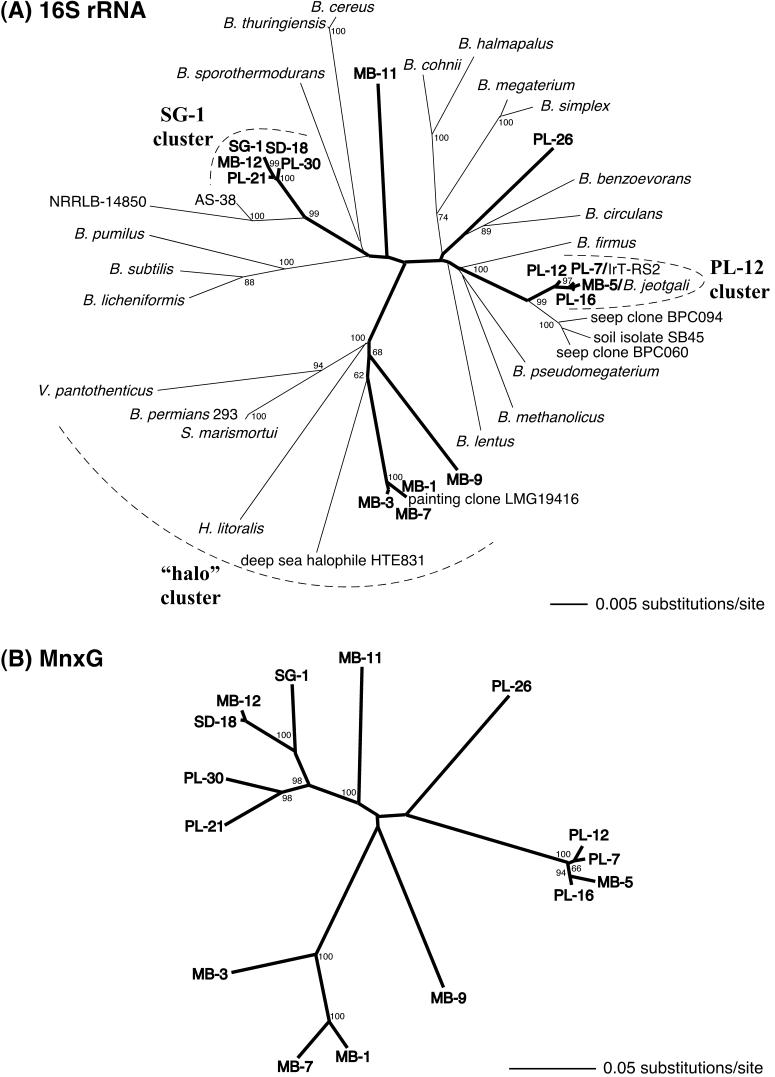
Although it has been recognized for over 2 decades that metabolically dormant spores of the marine Bacillus sp. strain SG-1 can enzymatically oxidize soluble Mn(II) to Mn oxides (33), how widespread this environmentally important capability is within the genus Bacillus has not previously been explored. In this study, a number of Mn(II)-oxidizing sporeformers were isolated from sediments collected within Mission Bay (MB strains) and San Diego Bay (SD strains) and off Point Loma (PL strains), near San Diego, Calif. Based on both phenotypic and phylogenetic characteristics, 14 different strains were chosen for further characterization, along with SG-1. Phylogenetic analysis of 16S rRNA genes obtained from these isolates revealed that the capacity to produce Mn(II)-oxidizing spores occurs in phylogenetically diverse organisms within the genus Bacillus (Fig. 1A). Many of the Mn(II)-oxidizing strains fell into distinct phylogenetic clusters with other Mn(II)-oxidizing isolates. Two of the three major clusters consist of closely related organisms isolated from different locations at different times. In fact, the five organisms within the SG-1 cluster were isolated from four different environments during five different years, suggesting that this group of organisms may be particularly ubiquitous in marine sedimentary environments. In addition, the closest relatives of the SG-1 cluster are from marine Bacillus strains (NRRLB14850 and AS-38) isolated from the Gulf of Mexico (43) and the Adriatic Sea, respectively, suggesting that together these organisms may constitute a “marine” branch of the Bacillus evolutionary tree (Fig. 1A). Strains MB-11 and PL-26 appear to represent lineages distinct from the three major clusters and are only distantly related (<97%) to other sequences in the databases.
FIG. 1.
Unrooted neighbor-joining trees based on 16S rRNA sequences (A) and MnxG amino acid sequences (B) obtained from 15 Mn(II)-oxidizing sporeformers (boldface). Additional sequences in panel A include diverse representatives within the genus Bacillus as well as the most closely related database sequences. The Bacillus species that have been tested for Mn(II) oxidation are given in Table 1. Percentages of bootstrap support (>60%) from 1,000 replicates are indicated at the branch points. The boldface branches highlight the topological similarities between the two trees. The strain designations of the Mn(II) oxidizers are based on the location of isolation: MB, Mission Bay; PL, off Point Loma; and SD, San Diego Bay.
Four of the isolates (MB-1, -3, -7, and -9) clearly clustered (bootstrap value, 100%) with a group of organisms that are all known to be either moderately halophilic (growing optimally in media containing 3 to 15% salt) or halotolerant (tolerating but not requiring high salt concentrations for growth). Phenotypically, many of the organisms within this cluster have been reported to be sufficiently different from the genus Bacillus to warrant classification into new genera, including Halobacillus, Salibacillus, Virgibacillus, and Gracibacillus (25, 45, 51). Database searches (BLAST and the Ribosomal Database Project) revealed that several of the sequences most closely related to MB-1, -3, and -7 were from “B. permians” strain 2-9-3, Salibacillus (formerly Bacillus) marismortui, and deep sea strain HTE831. Strain 2-9-3 was recently isolated from a brine inclusion within a reportedly 250-million-year-old salt crystal obtained from the Permian Salado near Carlsbad, N.Mex. (50). This organism can tolerate 20% NaCl and is very closely related (>99.5%) to S. marismortui, a moderate halophile isolated from a bottle of Dead Sea water collected in 1936 by Ben Volcani (2, 3). Strain HTE831 was recently isolated (46) from deep-sea (1,050 m) mud off the southern part of Japan and, consistent with its phylogenetic affiliation with this cluster, grows well on marine agar plates containing 2 to 23.4% NaCl. Similarly, the Mn(II)-oxidizing sporeformers within this cluster exhibited significant growth on media containing 10% salt (MB-9) and 20% salt (MB-1, -3, and -7), supporting their phylogenetic affiliation within the “halo” cluster. A rather intriguing finding was that the 16S rRNA sequence of an environmental clone (LMG-19416) obtained from a biodeteriorated Austrian wall painting (22) was 100% identical to the 16S rRNA sequence of MB-1 (1,501 bp considered). It is tempting to speculate that the production of Mn(II)-oxidizing spores by LMG-19416 may have contributed to the biodeterioration of these paintings.
A third group of four Mn(II)-oxidizing sporeformers formed a tight phylogenetic cluster (the PL-12 cluster), which also included several remarkably closely related environmental isolates: B. jeotgali strains YKJ-10 and YKJ-11 (99.9% identical to MB-5; 1,488 bp considered), isolated from the Korean traditional fermented seafood jeotgal (52), and strain IrT-RS2 (99.9% identical to MB-5 and PL-7; 1,404 bp considered), isolated from a uranium mine tailings pile near Dresden, Germany. The PL-12 cluster also clearly grouped with a clade consisting of two hydrocarbon seep clones (BPC060 and BPC094) and a numerically abundant isolate (SB45) obtained from rice paddy-associated anoxic bulk soil (10).
Based on the phylogenetic affiliations of the Mn(II)-oxidizing sporeformers described above, it is possible that Bacillus strains capable of producing Mn(II)-oxidizing spores are present in a wide variety of environments, including hypersaline environments, coastal and deep sea sediments, hydrocarbon seeps, uranium mine tailings, seafood, and dry solid surfaces (e.g., wall paintings), as well as soils, where spore-forming Bacillus species are often particularly abundant (7, 17). Evidence that this phenomenon is not limited to marine sedimentary environments comes from the fact that our laboratory has also isolated a number of Mn(II)-oxidizing sporeformers from Pinal Creek, an acid mine drainage-impacted, metal-contaminated stream near Globe, Ariz., in which Mn(II) is present at extremely high (0.5 to 1 mM) concentrations. In fact, several of the Pinal Creek organisms are closely related to organisms within the PL-12 cluster (>99.4% identity; 1,424 bp considered) (B. G. Clement and B. M. Tebo, unpublished results), indicating that closely related Mn(II)-oxidizing Bacillus strains may, in some cases, be found in both terrestrial and marine environments. It is possible that some of our marine sediment isolates may simply be halotolerant organisms rather than true marine (i.e., seawater-requiring) bacteria. It is worth noting that the capacity to grow in the presence of high salt concentrations was not restricted solely to organisms within the halo cluster. In fact, except for MB-11, all of the Mn(II)-oxidizing sporeformers described in this study were capable of growth on media containing 10% salt, while organisms within the PL-12 cluster were capable of growth even in the presence of 15% salt. This finding is consistent with the fact that two other members of the PL-12 phylogenetic cluster, B. jeotgali strains YKJ-10 and YKJ-11, were also shown to grow in the presence of up to 14% salt (52).
MnxG phylogeny.
The first genes shown to be involved in Mn(II) oxidation, the mnx genes, were previously identified in SG-1 using transposon mutagenesis (48, 49). The most downstream gene in this gene cluster, mnxG, encodes a multicopper oxidase believed to be directly involved in Mn(II) oxidation. Biochemical evidence for the involvement of a multicopper oxidase in this process comes from the fact that the Mn(II)-oxidizing activity of SG-1 spores has been shown to be enhanced by low concentrations of copper (49) and inhibited by azide (40), a potent metalloprotein inhibitor which bridges the type 2 and type 3 copper atoms of multicopper oxidases (12). In order to determine whether the Mn(II) oxidation-associated multicopper oxidase gene, mnxG, might also be involved in Mn(II) oxidation in these diverse isolates, the organisms were screened using PCR primers designed based on the determinants of two of the copper-binding regions of MnxG (HVFHYHVH and FFHDHL) expected to be highly conserved due to their functional roles. An ≈900-bp region of mnxG was successfully amplified from all 14 isolates. Multiple sequence alignments based on the derived amino acid sequences of this region of MnxG revealed that the protein is highly conserved in these phylogenetically diverse Bacillus strains, with identities (relative to SG-1) ranging from 75 to 93% and similarities ranging from 86 to 98%.
Phylogenetic trees based on the MnxG amino acid alignments (Fig. 1B) revealed the presence of three well-defined clusters, as well as several more distantly related sequences, suggesting that similar copper-dependent molecular mechanisms may exist in a wide variety of diverse Bacillus strains. The overall topologies of the 16S rRNA- and MnxG-based trees were remarkably similar, indicating that it is unlikely that the mnxG gene has been horizontally transferred throughout the genus but that it may instead be an evolutionarily and functionally important gene within these organisms. Despite the similarity between the two trees, as with many functional genes (e.g., rpoD and gyrB), the phylogeny based on the mnxG gene product appears to provide even greater resolution in distinguishing closely related organisms than the 16S rRNA phylogeny. For example, compared to the 16S rRNA tree, the organisms within the SG-1 cluster appear to be more distantly related in the MnxG tree.
To explore how widespread the ability to produce Mn(II)-oxidizing spores was within the genus Bacillus, a number of well-known strains were also tested for Mn(II) oxidation (Table 1). Of all these strains, only B. pumilus oxidized Mn(II) to any extent on plates, albeit this oxidation was relatively weak. In addition, mnxG was not successfully PCR amplified from any of these strains, although the possibility that mnxG homologues are present in some of these strains but differ in the primer sites cannot be totally ruled out. Analysis of the complete B. subtilis genome confirmed that the organism does not possess an mnxG homologue. However, B. subtilis does possess a gene encoding a spore coat protein, CotA (15), that appears to be a multicopper oxidase, based on sequence similarity as well as activity (26). This protein is roughly half the size of MnxG (65 versus 138 kDa) and appears to be a copper-dependent laccase which is responsible for the brown pigment (which is not due to Mn oxides) associated with B. subtilis spores.
Biochemical analysis of Mn(II)-oxidizing spore proteins.
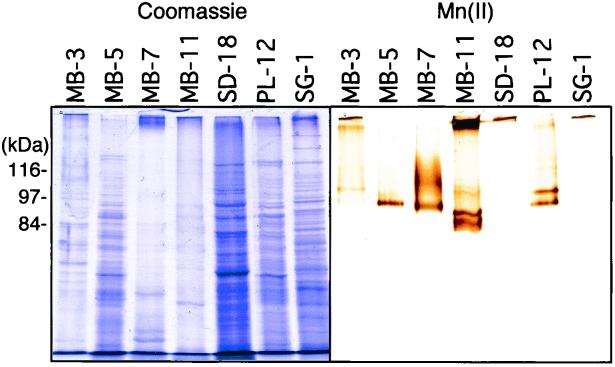
In order to explore the biochemical mechanism of Mn(II) oxidation in these diverse Bacillus isolates, SDS-PAGE analysis was used to compare the proteins which directly catalyze this reaction (Fig. 2). It has been previously demonstrated that use of a French press is an effective method for physically removing the Mn(II)-oxidizing outermost layer from SG-1 spores while still retaining activity (18). This method was applied to purified spores from all of the isolates, and in each case, a significant amount of Mn(II)-oxidizing activity was removed from the spores. As has been shown previously for SG-1 (40), the activity of the Mn(II)-oxidizing proteins from the isolates was completely inhibited by 1 mM azide (data not shown), consistent with the involvement of a metalloprotein (e.g., multicopper oxidase) (12, 44). SDS-PAGE analysis of the spore surface proteins revealed the presence of active Mn(II)-oxidizing protein bands of various sizes in all of the isolates (Fig. 2), the first such proteins identified in spores or gram-positive bacteria. There were, however, considerable differences in the overall protein profiles and sizes of Mn(II)-oxidizing proteins among the various isolates. For six of the isolates (PL-26 [not shown] and the SG-1 cluster), Mn(II)-oxidizing activity was only recovered in what appears to be a high-molecular-mass complex which barely enters the resolving gel, while for the remaining nine isolates, single Mn(II)-oxidizing bands were present in gels, ranging in mass from ≈90 to 120 kDa. MB-11 was unique in that significant activity was present both in a high-molecular-mass complex and in a doublet of ≈90 kDa, possibly indicating structural differences in the spore surface layer (either an exosporium or spore coat) of this organism relative to the other isolates. Overall, the relative sizes of the Mn(II)-oxidizing proteins from these Bacillus isolates correlated well with the phylogenetic groupings, suggesting a link between phylogeny and spore physiology.
FIG. 2.
SDS-PAGE gels of outermost spore layer extracts from phylogenetically diverse Mn(II)-oxidizing sporeformers. Following electrophoresis, replicate gels were incubated in either (i) Coomassie blue as a general protein stain (left), (ii) HEPES buffer containing 200 μM Mn(II) (after washing to remove SDS [see Materials and Methods]) to stain for Mn(II) oxidation activity (right), or (iii) HEPES buffer as for incubation ii but with pretreatment of the gel in HEPES buffer containing the copper chelator o-phenanthroline (50 μM) for 15 min prior to addition of Mn(II) to inhibit copper oxidases (resulting in a completely blank gel [data not shown]). A sufficient quantity of protein was loaded in each lane to yield visible Mn(II)-oxidizing proteins in gels incubated in Mn(II), as evidenced by the formation of brown Mn oxide bands.
The size differences of the various Mn(II)-oxidizing proteins could be due to different-sized genes, posttranslational modifications (e.g., proteolysis or glycosylation), or the requirement for additional proteins (e.g., in a complex) for activity. In L. discophora SS-1, a 110-kDa Mn(II)-oxidizing protein has been consistently recovered in SDS-PAGE gels (1, 6), yet the underlying multicopper oxidase gene, mofA, encodes a predicted 174-kDa protein (11), suggesting partial cleavage or proteolysis of the full-length polypepetide. In P. putida GB-1, Mn(II)-oxidizing activity has been recovered only in the form of high-molecular-mass complexes (250 and 180 kDa) in native gels (37), suggesting the requirement for several proteins, including the multicopper oxidase CumA, for activity (8, 13).
To determine whether the Mn(II)-oxidizing activities of the spore surface proteins were copper dependent (as in SG-1), gels were incubated in Mn(II) buffer with the addition of the copper chelator o-phenanthroline at a concentration (50 μM) well below that of Mn(II) (200 μM). The fact that the activities of all of the proteins were inhibited by this treatment (data not shown), combined with the azide inhibition of the spore surface extracts, is consistent with the involvement of Cu-dependent oxidases (e.g., MnxG) in these phylogenetically diverse marine Bacillus isolates. As might be expected of proteins located on the surfaces of bacterial structures designed to persist under extreme environmental conditions, the Mn(II)-oxidizing spore enzymes described in this study are extremely resilient. In fact, spores can be exposed to heat (70 to 80°C), multiple freeze-thaw cycles, fixatives (e.g., UV and glutaraldehyde), SDS, lysozyme, and reductants (to remove Mn oxides) with no significant loss of activity (14, 40). These properties suggest that these spores are stable enzymatic catalysts for the oxidative precipitation of metals in the environment. This is supported by the fact that, within the coastal sediments from which many of the Mn(II)-oxidizing sporeformers were isolated, the in situ Mn(II)-oxidizing activity is due primarily to heat-resistant organisms (e.g., spores) and is inhibited by azide (i.e., multicopper oxidase inhibitor) (31). Overall, the results of this study suggest that the commonly held view that bacterial spores are totally inactive structures in the environment should be revised.
Acknowledgments
We thank Margo Haygood and John Spear for helpful comments on the manuscript.
This research was funded by the National Science Foundation (MCB98-08915), the Collaborative UC/Los Alamos Research Program, and Superfund Basic Research Program (NIEHS) grant ES10337 from the National Institutes of Health. C.A.F. was supported in part by a STAR Graduate Fellowship from the U.S. Environmental Protection Agency and in part by a traineeship from the University of California Toxic Substances Research and Teaching Program.
REFERENCES
- 1.Adams, L. F., and W. C. Ghiorse. 1987. Characterization of extracellular Mn-oxidizing activity and isolation of a Mn-oxidizing protein from Leptothrix discophora SS-1. J. Bacteriol. 169:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arahal, D. R., M. C. Marquez, B. E. Volcani, K. H. Schleifer, and A. Ventosa. 1999. Bacillus marismortui sp. nov., a new moderately halophilic species from the Dead Sea. Int. J. Syst. Bacteriol. 49:521-530. [DOI] [PubMed] [Google Scholar]
- 3.Arahal, D. R., M. C. Marquez, B. E. Volcani, K. H. Schleifer, and A. Ventosa. 2000. Reclassification of Bacillus marismortui as Salibacillus marismortui comb. nov. Int. J. Syst. E vol. Microbiol. 50:1501-1503. [DOI] [PubMed] [Google Scholar]
- 4.Bargar, J. R., B. M. Tebo, and J. E. Villinsky. 2000. In situ characterization of Mn(II) oxidation by spores of the marine Bacillus sp. strain SG-1. Geochim. Cosmochim. Acta 64:2775-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonde, G. J. 1981. Bacillus from marine habitats: allocation to phena established by numerical techniques, p. 181-215. In R. C. W. Berkeley and M. Goodfellow (ed.), The aerobic endospore-forming bacteria: classification and identification. Academic Press, New York, N.Y.
- 6.Boogerd, F. C., and J. P. M. de Vrind. 1987. Manganese oxidation by Leptothrix discophora SS-1. J. Bacteriol. 169:489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouwers, G. J., J. P. M. de Vrind, P. L. A. M. Corstjens, P. Cornelis, C. Baysse, and E. W. de Vrind-de Jong. 1999. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 65:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cano, R. J., and M. K. Borucki. 1995. Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science 268:1060-1064. [DOI] [PubMed] [Google Scholar]
- 10.Chin, K. J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corstjens, P. L. A. M., J. P. M. de Vrind, T. Goosen, and E. W. de Vrind-de Jong. 1997. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol. J. 14:91-108. [Google Scholar]
- 12.da Silva, J. J. R. F., and R. J. P. Williams. 1991. The biological chemistry of the elements. Clarendon Press, Oxford, United Kingdom.
- 13.de Vrind, J. P. M., G. J. Brouwers, P. L. A. M. Corstjens, J. den Dulk, and E. W. de Vrind-de Jong. 1998. The cytochrome c maturation operon is involved in manganese oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 64:3556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vrind, J. P. M., E. W. de Vrind-de Jong, J.-W. H. de Voogt, P. Westbroek, F. C. Boogerd, and R. A. Rosson. 1986. Manganese oxidation by spores and spore coats of a marine Bacillus species. Appl. Environ. Microbiol. 52:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donovan, W., L. B. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 16.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felske, A., A. Wolterink, R. Van Lis, and A. D. Akkermans. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis, C. A. 2000. Diversity and molecular mechanisms of manganese(II)-oxidizing bacteria. Ph.D. thesis. University of California, San Diego.
- 19.Francis, C. A., and B. M. Tebo. 1999. Marine Bacillus spores as catalysts for oxidative precipitation and sorption of metals. J. Mol. Microbiol. Biotechnol. 1:71-78. [PubMed] [Google Scholar]
- 20.Francis, C. A., E.-M. Co, and B. M. Tebo. 2001. Enzymatic manganese(II) oxidation by a marine α-proteobacterium. Appl. Environ. Microbiol. 67:4024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gest, H., and J. Mandelstam. 1987. Longevity of microorganisms in natural environments. Microbiol. Sci. 4:69-71. [PubMed] [Google Scholar]
- 22.Gurtner, C., J. Heyrman, G. Pinar, W. Lubitz, J. Swings, and S. Roelleke. 2000. Comparative analyses of the bacterial diversity on two different biodeteriorated wall paintings by DGGE and 16S rDNA sequence analysis. Int. Biodeterior. Biodegradation 46:229-239. [Google Scholar]
- 23.Hastings, D., and S. Emerson. 1986. Oxidation of manganese by spores of a marine bacillus: kinetic and thermodynamic considerations. Geochim. Cosmochim. Acta 50:1819-1824. [Google Scholar]
- 24.Hem, J. D. 1978. Redox processes at surfaces of manganese oxide and their effects on aqueous metal ions. Chem. Geol. 21:199-218. [Google Scholar]
- 25.Heyndrickx, M., L. Lebbe, K. Kersters, P. De Vos, G. Forsyth, and N. A. Logan. 1998. Virgibacillus: a new genus to accommodate Bacillus pantothenticus (Proom and Knight 1950). Emended description of Virgibacillus pantothenticus. Int. J. Syst. Bacteriol. 48:99-106. [DOI] [PubMed] [Google Scholar]
- 26.Hullo, M. F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, M. J., S. L. Reader, and L. M. Swiercznski. 1994. Preservation records of micro-organisms: evidence of the tenacity of life. Microbiology 140:2513-2519. [DOI] [PubMed] [Google Scholar]
- 28.Krumbein, W. E., and H. J. Altman. 1973. A new method for detection and enumeration of manganese-oxidizing and -reducing microorganisms. Helgol. Wiss. Meeresunters. 25:347-356. [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, England.
- 31.Lee, Y. 1994. Microbial oxidation of cobalt: characterization and its significance in marine environments. Ph.D. thesis. University of California, San Diego.
- 32.Murray, J. W. 1975. The interaction of metal ions at the manganese dioxide-solution interface. Geochim. Cosmochim. Acta 39:505-519. [Google Scholar]
- 33.Nealson, K. H., and J. Ford. 1980. Surface enhancement of bacterial manganese oxidation: implications for aquatic environments. Geomicrobiol. J. 2:21-37. [Google Scholar]
- 34.Nelson, Y. M., L. W. Lion, W. C. Ghiorse, and M. L. Shuler. 1999. Production of biogenic Mn oxides by Leptothrix discophora SS-1 in a chemically defined medium and evaluation of their adsorption characteristics. Appl. Environ. Microbiol. 65:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nieto, J. J., R. Fernandez-Castillo, M. C. Marquez, A. Ventosa, E. Quesada, and F. Ruiz-Berraquero. 1989. Survey of metal tolerance in moderately halophilic eubacteria. Appl. Environ. Microbiol. 55:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okazaki, M., T. Sugita, M. Shimizu, Y. Ohode, K. Iwamoto, E. W. de Vrind-de Jong, J. P. M. de Vrind, and P. L. A. M. Corstjens. 1997. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl. Environ. Microbiol. 63:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter, A. G., E. W. Davidson, and J. W. Liu. 1993. Mosquitocidal toxins of bacilli and their genetic manipulation for effective biological control of mosquitoes. Microbiol. Rev. 57:838-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potts, M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosson, R. A., and K. H. Nealson. 1982. Manganese binding and oxidation by spores of a marine bacillus. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryden, L. G., and L. T. Hunt. 1993. Evolution and protein complexity: the blue copper-containing oxidases and related proteins. J. Mol. Evol. 36:41-66. [DOI] [PubMed] [Google Scholar]
- 42.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siefert, J. L., M. Larios-Sanz, L. K. Nakamura, R. A. Slepecky, J. H. Paul, E. R. Moore, G. E. Fox, and P. Jurtshuk, Jr. 2000. Phylogeny of marine Bacillus isolates from the Gulf of Mexico. Curr. Microbiol 41:84-88. [DOI] [PubMed] [Google Scholar]
- 44.Solomon, E. I., U. M. Sundaram, and T. E. Machonkin. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 96:2563-2605. [DOI] [PubMed] [Google Scholar]
- 45.Spring, S., W. Ludwig, M. C. Marquez, A. Ventosa, and K.-H. Schleifer. 1996. Halobacillus gen. nov., with descriptions of Halobacillus litoralis sp. nov. and Halobacillus trueperi sp. nov., and transfer of Sporosarcina halophila to Halobacillus halophilus comb. nov. Int. J. Syst. Bacteriol. 46:492-496. [Google Scholar]
- 46.Takami, H., K. Kobata, T. Nagahama, H. Kobayashi, A. Inoue, and K. Horikoshi. 1999. Biodiversity of deep-sea sites located near the south part of Japan. Extremophiles 3:97-102. [DOI] [PubMed] [Google Scholar]
- 47.Tebo, B. M., W. C. Ghiorse, L. G. van Waasbergen, P. L. Siering, and R. Caspi. 1997. Bacterially-mediated mineral formation: insights into manganese(II) oxidation from molecular genetic and biochemical studies. Rev. Mineral. 35:225-266. [Google Scholar]
- 48.van Waasbergen, L. G., J. A. Hoch, and B. M. Tebo. 1993. Genetic analysis of the marine manganese-oxidizing Bacillus sp. strain SG-1: protoplast transformation, Tn917 mutagenesis, and identification of chromosomal loci involved in manganese oxidation. J. Bacteriol. 175:7594-7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Waasbergen, L. G., M. Hildebrand, and B. M. Tebo. 1996. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J. Bacteriol. 12:3517-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vreeland, R. H., W. D. Rosenzweig, and D. W. Powers. 2000. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 407:897-900. [DOI] [PubMed] [Google Scholar]
- 51.Waino, M., B. J. Tindall, P. Schumann, and K. Ingvorsen. 1999. Gracilibacillus gen. nov., with description of Gracilibacillus halotolerans gen. nov., sp. nov.; transfer of Bacillus dipsosauri to Gracilibacillus dipsosauri comb. nov., and Bacillus salexigens to the genus Salibacillus gen. nov., as Salibacillus salexigens comb. nov. Int. J. Syst. Bacteriol. 49:821-831. [DOI] [PubMed] [Google Scholar]
- 52.Yoon, J. H., S. S. Kang, K. C. Lee, Y. H. Kho, S. H. Choi, K. H. Kang, and Y. H. Park. 2001. Bacillus jeotgali sp. nov., isolated from jeotgal, Korean traditional fermented seafood. Int. J. Syst. E vol. Microbiol. 51:1087-1092. [DOI] [PubMed] [Google Scholar]