Abstract
We describe the construction of TnFuZ, a genetic tool for the discovery and mutagenesis of proteins exported from gram-positive bacteria. This tool combines a transposable element (Tn4001) of broad host range in gram-positive bacteria and an alkaline phosphatase gene (phoZ) derived from a gram-positive bacterium that has been modified by removal of the region encoding its export signal. Mutagenesis of Streptococcus pyogenes with TnFuZ (“FuZ” stands for fusions to phoZ) identified genes encoding secreted proteins whose expression was enhanced during growth in an aerobic environment. Thus, TnFuZ should be valuable for analysis of protein secretion, gene regulation, and virulence in gram-positive bacteria.
Certain bacterial enzymes have activities that depend on their subcellular localization. For example, the alkaline phosphatase encoded by phoA of Escherichia coli is enzymatically active only when it has been transported across the cellular membrane into the periplasmic space. This property has been exploited to engineer PhoA as a molecular sensor of subcellular location (for a review, see reference 22). In a typical application, the region of phoA encoding its own promoter, translation initiation site, and signal peptide is removed and it is then fused with the gene that encodes the protein of interest. Should the fusion partner contribute an export signal, the bacterial cell expressing the hybrid protein will demonstrate alkaline phosphatase activity that can easily be detected through the use of several different assays (20). This strategy has been widely used in the analysis of the topologies of transmembrane proteins, for the identification and analysis of protein export signals, and for the identification of proteins that are targeted for export (19, 20). For the last application, the development of transposon-based methods for the construction of fusions to phoA (21) has allowed analyses to be conducted on a genome-wide scale with a wide variety of gram-negative bacterial species (for a review, see reference 13). Unfortunately, a similar technology has not been available for gram-positive bacteria.
As a group, gram-positive bacteria include species that are important for many industrial processes, for the production of food, and for the production of antibiotics as well as species that are model organisms for the study of development. Many important human and animal pathogens are gram positive, including several that are resistant to multiple antibiotics. While protein export in gram-positive bacteria shares a basic similarity with that in gram-negative bacteria, there are many notable differences and the pathways of export are not well characterized (35). Unlike most gram-negative bacteria, gram-positive bacteria typically secrete numerous proteins into the external environment surrounding the cell. This characteristic of protein secretion is particularly true for the pathogenic gram-positive species, and it is likely that many of these secreted proteins promote virulence (12, 16, 31). Thus, a transposon-based method for identifying mutations in genes that encode proteins targeted for export would be of broad application in the study of protein secretion by and the pathogenicity of gram-positive bacteria. The work reported here describes the construction of TnFuZ, a transposon-based genetic tool for the discovery and study of proteins exported from gram-positive bacteria.
Construction of TnFuZ
An essential component of TnphoA, the prototype element developed for gram-negative bacteria, is the modified phoA reporter (21). Unfortunately, E. coli-derived PhoA is poorly active when it is expressed in gram-positive hosts (27), which may reflect differences in the export pathway from that in gram-negative bacteria and which may result in inefficient dimerization and/or formation of a disulfide bond required for its activity (27). Other secretion reporters, including DNase (29), have been developed for gram-positive bacteria, but the fact that some gram-positive bacteria naturally secrete multiple DNases (37) has limited use of this enzyme. Recently, an alkaline phosphatase (PhoZ) derived from the gram-positive bacterium Enterococcus faecalis (15) has been developed as a reporter that is highly active in gram-positive bacteria (10, 15). The alkaline phosphatase activity of PhoZ, like that of PhoA, is dependent on its export from the cytoplasm (15), and the activity of a derivative of PhoZ lacking its leader peptide can be restored through fusion with a heterologous exported polypeptide (10, 15). The fusion partner can be large, and the resulting chimera can be stable and demonstrate high specific activity (10, 15).
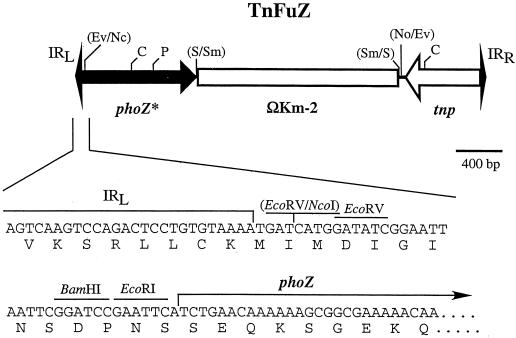
The second essential component of TnphoA is Tn5, a transposon that both inserts randomly and can accept a reporter gene immediately adjacent to one of the short inverted repeats that defines the end of the element (21). However, Tn5 does not transpose efficiently in gram-positive hosts. A suitable alternative is transposon Tn4001, a Tn5-like element derived from gram-positive bacteria that transposes with a high degree of randomness in gram-positive bacteria and mycoplasmas (11, 18). A modified derivative of Tn4001 which consists of a single copy of the insertion sequence IS256 modified to accept an antibiotic resistance gene immediately adjacent to its left-end short (28-bp) terminal inverted repeat has been constructed (18). In the present study, we used this site to introduce a modified phoZ as follows. Primers 5PhoEcoRI (GCGGGTTGTA CGAATTCATC TGAACAAAAA AGCGGCGAAA AAC) and PhoS-SalI (CGTTCTGCTT TGTCGACATT TTGTTATTTA CCAATACC) were used to amplify a 1,387-bp fragment of chromosomal DNA from Enterococcus faecalis ATCC 11700 (32). This fragment consisted of the region between nucleotides +52 and +1415 of the phoZ sequence (GenBank accession no. AF154100). The modified phoZ (phoZ∗) lacked the first 18 amino acids of its signal sequence, and fusion with a heterologous secreted protein at this junction has been sufficient for export of a chimeric protein with alkaline phosphatase activity (10). Sites for EcoRI and SalI were introduced into the primer sequences (underlined above), and the insertion of the amplified phoZ∗ fragment between the EcoRI and SalI sites of pET22(b)+ (Invitrogen) followed by the insertion of a 2.2-kb SmaI fragment containing ΩKm-2 (28) into a blunted SalI site generated pCMG7. A blunted 3.6-kb NcoI-NotI fragment containing both phoZ∗ and ΩKm-2 was inserted into the EcoRV site that was introduced into IS256 contained on pMGC57 (18) to generate the novel TnFuZ element (Fig. 1).
FIG. 1.
Construction of TnFuZ. The structure of TnFuZ is shown in the top line. The element contains the left and right inverted repeats (IRL and IRR, respectively), the transposase (tnp) of IS256, and the ahpA3 kanamycin resistance determinant contained on ΩKm-2. The gene encoding the Enterococcus faecalis alkaline phosphatase (phoZ) was altered by removal of the region which encoded its signal sequence, and the modified gene (phoZ∗) was introduced into the element as shown. The bottom of the figure shows the DNA sequence and open reading frame which extends across IRL and joins the open reading frame of phoZ∗. Abbreviations: C, ClaI; Ev, EcoRV; Nc, NcoI; P, PstI; S, SalI; Sm, SmaI. A slash indicates a junction of two restriction fragments joined during construction of the element, and restriction sites enclosed by parentheses are inactive.
Production of insertion libraries
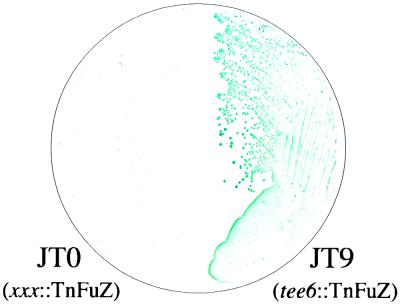
The group A streptococcus Streptococcus pyogenes is the causative agent of a wide variety of suppurative diseases that affect the skin (impetigo, necrotizing fasciitis, erysipelas) and pharynx (pharyngitis). In addition, S. pyogenes can cause the nonsuppurative sequelae of rheumatic fever and glomerulonephritis. The ability to cause a range of diseases in many different host tissues is likely facilitated by the ability of the organism to secrete upwards of 40 different polypeptides (16). Unfortunately, only a few of these gene products have been directly linked to pathogenesis and the regulation of these putative virulence determinants is still poorly understood, making S. pyogenes an ideal model for evaluation of the utility of TnFuZ. The ColE1-based plasmid used for the construction of TnFuZ does not replicate in gram-positive hosts (18). Thus, transformation of S. pyogenes with the TnFuZ-containing plasmid (pCMG8) by electroporation (6) with selection for the kanamycin resistance determinant of ΩKm-2 yielded between 102 and 104 transformants μg of DNA−1, depending on the specific S. pyogenes strain used. The incorporation of an alkaline phosphatase substrate directly into medium did not prove to be an efficient method for detection of alkaline phosphatase activity in colonies, likely because lactic acid bacteria like S. pyogenes acidify their surroundings. However, the use of an alternative method in which the pH of colonies was neutralized after they were lifted from the plates on filters (27) allowed the detection of blue colonies that ranged in color from intensely blue to light blue with the substrate XP (5-bromo-4-chloro-3-indolylphosphate) (Fig. 2). The yield of colonies with detectable alkaline phosphatase activity was between 1 and 4% of the total number of colonies analyzed. Untransformed S. pyogenes strains produced no detectable color (Fig. 2), which was consistent with the observation that there is no gene identified as encoding an alkaline phosphatase in the S. pyogenes strain whose genome sequence has been determined (9).
FIG. 2.
Detection of colony-associated PhoZ activity. Colonies from overnight culture on solid Todd-Hewitt yeast extract medium were transferred to a nitrocellulose filter and exposed to the chromogenic alkaline phosphatase substrate XP as described in the text. A representative S. pyogenes TnFuZ insertion strain that expressed TnPhoZ activity (JT9) (Table 1) appears bright blue, while a TnFuZ insertion strain (JT0) that did not express activity remains colorless.
Characterization of fusion strains
Since virulence factors are frequently subject to coordinate regulation, screening for TnphoA insertions that are regulated by the same environmental signals that control expression of known virulence factors has proved to be a powerful strategy for identification of novel virulence factors in gram-negative bacteria (22). Oxygen is one environmental signal that is known to be involved in the regulation of virulence determinants in S. pyogenes (36). To evaluate whether TnFuZ could be used in an environmental screen for potential virulence factors, a pool of 650 TnFuZ insertions from two independent transformations of S. pyogenes JRS4 (34) were screened to identify 17 colonies with detectable alkaline phosphatase activity. It was possible to pick individual blue colonies directly from the assay filters for restreaking on replica plates which were then cultured in aerobic or anaerobic environments as described previously (36). As expected, since the original screen was conducted on colonies grown aerobically, all 17 isolates demonstrated some level of alkaline phosphatase activity following aerobic culture (Table 1). Of these, nine showed reduced activity following anaerobic growth, and two of these nine strains (JT8 and JT13) demonstrated substantial decreases in activity (Table 1). In addition, analysis of alkaline phosphatase activity in cell-free culture supernatants (by the method described in reference 10) indicated that 11 of the 17 strains had an activity that was freely secreted from the streptococcal cell (Table 1).
TABLE 1.
Alkaline phosphatase activities of various strains
| Straina | Colony-associated activityb
|
Cell-free-supernatant activityc | |
|---|---|---|---|
| +O2 | −O2 | ||
| JRS4 | − | − | − |
| JT1 | ++ | + | ++ |
| JT2 | +++ | +++ | +++ |
| JT3 | ± | − | + |
| JT4 | ++ | + | − |
| JT5 | ++ | + | + |
| JT6 | ++ | ++ | + |
| JT7 | ++ | ++ | + |
| JT8 | +++ | + | + |
| JT9 | +++ | +++ | +++ |
| JT10 | +++ | +++ | +++ |
| JT11 | +++ | +++ | + |
| JT12 | +++ | +++ | − |
| JT13 | ++ | − | − |
| JT14 | ++ | + | − |
| JT15 | + | − | − |
| JT16 | +++ | +++ | ++ |
| JT17 | +++ | ++ | ++ |
Indicated strains were derived from JRS4 by insertion of TnFuZ.
Cultures on solid medium were incubated under aerobic (+O2) or anaerobic (−O2) conditions. Alkaline phosphatase activities were determined by assessment of colony color following staining with the XP substrate and are reported as follows: −, white; ±, very light blue;+, light blue; ++, medium blue; and +++, dark blue.
Strains were cultured in liquid medium. Alkaline phosphatase activities in cell-free supernatants were determined using the para-nitrophenylphosphate substrate as described previously (11) and are reported as optical densities at 405 nm as follows: −, 0.000 to 0.024; +, 0.0025 to 0.050; ++, 0.051 to 0.150; and +++, >0.150.
Sequence characterization of selected fusion strains
The observations that some fusion strains had alkaline phosphatase activity that was exclusively cell associated but that others had an activity that was also released from the cell suggested that TnFuZ had inserted into genes encoding both membrane proteins and proteins that are targeted for secretion past the cellular membrane. To examine the types of genes identified, chromosomal DNAs from selected fusion strains were purified as described previously (6) and directly used as templates in a DNA sequencing reaction with the primer EnPhoR1 (TGCCT TCGCT TCAGC AACCT CTGTT TG), fluorescently labeled dideoxynucleotides (Big Dye terminators; PE Applied Biosystems), and a hyperstable DNA polymerase (ThermoFidelase) according to the recommendations of the manufacturer (Fidelity Systems Inc.). Chromosomes from a total of 10 fusion strains, including representatives from both the oxygen- and non-oxygen-regulated groups, were analyzed, and in each case TnFuZ had inserted into the correct open reading frame in the correct orientation to encode a hybrid fusion protein. Of the 10 strains, 8 had a putative export signal that included a predicted signal sequence or transmembrane domain (Table 2).
TABLE 2.
Sequence analysis of selected mutants
| Straina | Locusb | Gene(s)c | Similar gene(s) (organism)d | % Amino acid identity (% homology) | Putative export signale | Function | Entrez accession no. |
|---|---|---|---|---|---|---|---|
| JT4* | Spy1618 | cysK (Bs) | 61 (74) | TM (3) | Cysteine biosynthesis | P37887 | |
| JT5* | Spy0092 | adcR (Spn) | 53 (76) | NI | Transcription repressor | CAA96184 | |
| JT7 | Spy2188 | trmU (Bs) | 63 (76) | NI | tRNA modifier | 035020 | |
| JT8* | Spy2192 | sai2/isaA (Sa) | 54 (60) | SS | Secreted antigen | BAA95959/AF144681-1 | |
| JT9 | tee-6 | SS | Surface antigen | P18481 | |||
| JT11 | Spy1882 | lppC | SSII | Acid phosphatase | CAA73175 | ||
| JT12 | Spy0763 | murA (Bs) | 58 (73) | TM (2) | Peptidoglycan synthesis | P70965 | |
| JT13* | Spy0242 | fasB/gasK1 | TM (7) | Histidine protein kinase | |||
| JT15* | Spy2211 | tmp5 (Lc) | 44 (60) | TM (7) | Unknown | AAC14608 | |
| JT16 | Spy2033 | tptA (Sc) | 59 (72) | SSII | ABC transporter | AAB97959 |
An asterisk indicates that alkaline phosphatase activity was decreased in anaerobically grown cultures. See Table 1.
Corresponding open reading frame in the sequenced genome of S. pyogenes SF370 (10) (GenBank accession no. AE004092).
Previously characterized for S. pyogenes or related species.
Determined by comparison to sequences in EMBL and GenBank databases using TBLASTN (1) and the National Center for Biotechnology Information network services (www.ncbi.nlm.nih.gov/BLAST/). Bs, B. subtilis; Spn, Streptococcus pneumoniae; Lc, L. lactis; Sa, Staphylococcus aureus; Sc, Streptococcus cristatus.
SS, signal sequence; SSII, lipoprotein signal sequence; TM, transmembrane domain; NI, none identified. For prediction of signal sequences by SignalP (25) and transmembrane domains by TopPred 2 (8), we used network services provided by Meta-PredictProtein (dodo.cpmc.columbia.edu/pp/predictprotein.html). Numbers of predicted transmembrane domains are indicated within parentheses.
An important facet of the use of TnFuZ is that it generates mutations through insertion into its target genes. Comparison of the sequences obtained for the set of fusion strains described above to data in the streptococcal genome database (9) was done to obtain the sequences of the entire targeted open reading frames, which were then compared to sequences in the Entrez nucleotide sequence database using TBLASTN (1). This analysis revealed that while a relatively modest number of colonies were analyzed to identify fusion strains, the screen developed a rich collection of mutants (Table 2). Strains with mutations in genes encoding surface and cell wall protein antigens were obtained, as well as strains with mutations in genes encoding various enzymatic activities, transporters, and regulators of transcription (Table 2).
The collection identified only three genes that have been previously characterized in S. pyogenes or a closely related streptococcal species (Table 2). Of the surface antigens identified, T protein (encoded by tee-6) (33) has been characterized as a cell wall protein anchored by the LPXTG sortase pathway (23). This pathway covalently links proteins via an LPXTG motif located toward the carboxy terminus of the protein to the cell wall peptidoglycan. The function of T protein is unknown, but it is recognized by the immune system during infection. However, there may be as many as 13 proteins in the S. pyogenes genome that contain the LPXTG anchor signal, and several of these have been implicated in pathogenesis (9).
In mutant JT8, TnFuZ had inserted into a gene that encodes a protein with homology to the IscA secreted antigen of Staphylococcus aureus (17). The function of IscA is unknown. However, as is the case for several other putative S. pyogenes virulence factors, the expression of the iscA::phoZ fusion was enhanced in an aerobic environment (Table 1). Other mutants led to the identification of fusions with enhanced aerobic expression, including a gene with homology to cysK (JT4) (Table 2) and a transcription repressor of a Zn2+/Mn2+ transporter (JT5) (Table 2). In Bacillus subtilis, cysK is induced by superoxide stress (2), and in many organisms Mn2+ is important for resistance to oxidative stress (3, 30). A second aerobic-culture-induced transcription regulator was a histidine protein kinase of a two-component regulator (JT13) (Table 2) whose expression is enhanced when cultures of S. pyogenes enter stationary phase (14). Recent data indicate that this regulatory system is important for promoting the ability of S. pyogenes to survive oxidative stress (N. Ruiz, K. Y. King, and M. Caparon, submitted for publication). The final mutant that contained a fusion with enhanced aerobic expression (JT15) (Table 2) had a TnFuZ insertion in a gene identified as encoding a transmembrane protein of Lactococcus lactis of unknown function (29).
Conclusions
The interactions of S. pyogenes and other lactic acid bacteria with aerobic environments are poorly understood (7). This group of bacteria does not make heme and, as a consequence, lacks many enzymes that are important for the ability of other bacterial species to survive and grow under aerobic conditions. Nevertheless, many lactic acid species flourish in aerobic environments (7) and many virulence genes of pathogenic lactic acid species are regulated in response to alterations in atmosphere (4, 5, 8, 36). In the present study, mutagenesis with TnFuZ identified several genes for secreted proteins whose expression was influenced by aerobic growth conditions. Thus, TnFuZ is a valuable addition to the developing technology for genetic manipulation of gram-positive bacteria.
It was interesting that, of all the fusion proteins produced, none represented any of the known exotoxins of S. pyogenes. This may be because at least one toxin appears to have a specialized export mechanism (24). In other cases, it may be that the colony method for screening was not optimal for identification of fusion proteins that do not remain associated with the cell surface or it may be a result of the fact that the number of colonies screened was relatively small. However, the simplicity of various alkaline phosphatase assays and the availability of substrates of diverse chemistries will allow development of screens optimized for detection of proteins that are completely released from the cell. For example, a large-scale screen could be conducted to analyze cell-free culture supernatants harvested from cultures arrayed in a microplate format or the low background obtained in the colony filter assay could be exploited through the use of a highly sensitive substrate.
Most of the characterized fusion proteins had identifiable export signals. As for the two genes which did not encode a putative export signal, it is possible that the location of the fusion junction generated a fortuitous export signal (38) or that the expression of the fusion protein produced cell lysis. Preliminary characterization suggests that neither of these scenarios was the case. However, it should be noted that pathways of protein secretion in gram-positive bacteria are not well understood. In S. pyogenes alone, there are several examples of proteins that are found exterior to the cell membrane and that lack a defined export signal (25, 26). The availability of TnFuZ will likely facilitate investigation of alternative pathways of protein secretion in gram-positive bacteria. The broad host range of the transposon used to construct TnFuZ (Tn4001) suggests that this element will find wide application in the analysis of protein secretion by and the virulence of gram-positive bacteria and mycoplasmas.
Acknowledgments
This work was supported by Public Health Service grant AI38273.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., J. Bernhardt, R. Schmid, H. Mach, U. Volker, and M. Hecker. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451-1463. [DOI] [PubMed] [Google Scholar]
- 3.Archibald, F. S., and I. Fridovich. 1981. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 145:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard, J. P., and M. W. Stinson. 1996. The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect. Immun. 64:3853-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of the pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 7.Condon, S. 1987. Responses of the lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 46:269-280. [Google Scholar]
- 8.Echenique, J. R., and M. C. Trombe. 2001. Competence modulation by the NADH oxidase of Streptococcus pneumoniae involves signal transduction. J. Bacteriol. 183:768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 12.Iandolo, J. J. 1989. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu. Rev. Microbiol. 43:375-402. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman, M. R., and R. K. Taylor. 1994. Identification of bacterial cell-surface virulence determinants with TnphoA. Methods Enzymol. 235:426-428. [DOI] [PubMed] [Google Scholar]
- 14.Kreikemeyer, B., M. D. P. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. H., A. Nittayajarn, R. P. Ross, C. B. Rothschild, D. Parsonage, A. Claiborne, and C. E. Rubens. 1999. Characterization of Enterococcus faecalis alkaline phosphatase and use in identifying Streptococcus agalactiae secreted proteins. J. Bacteriol. 181:5790-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei, B., S. Mackie, S. Lukomski, and J. M. Musser. 2000. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect. Immun. 68:6807-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz, U., K. Ohlsen, H. Karch, M. Hecker, A. Thiede, and J. Hacker. 2000. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 29:145-153. [DOI] [PubMed] [Google Scholar]
- 18.Lyon, W., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galatosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 20.Manoil, C. 2000. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol. 326:35-47. [DOI] [PubMed] [Google Scholar]
- 21.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manoil, C., J. J. Mekalanos, and J. Beckwith. 1990. Alkaline phosphatase fusions: sensors of subcellular location. J. Bacteriol. 172:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 24.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. S. De Azavedo. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancholi, V., and V. A. Fischetti. 1998. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 26.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce, B. J., Y. B. Yin, and H. R. Masure. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9:1037-1050. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Privalle, C. T., and I. Fridovich. 1992. Transcriptional and maturational effects of manganese and iron on the biosynthesis of manganese-superoxide dismutase in Escherichia coli. J. Biol. Chem. 267:9140-9145. [PubMed] [Google Scholar]
- 31.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 32.Ross, R. P., and A. Claiborne. 1992. Molecular cloning and analysis of the gene encoding the NADH oxidase from Streptococcus faecalis 10C1. J. Mol. Biol. 221:857-871. [DOI] [PubMed] [Google Scholar]
- 33.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characteristics of the trypsin-resistant T6 surface protein of group A streptococci. J. Bacteriol. 172:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, J. R., P. C. Guenther, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.VanHeyningen, T., G. Fogg, D. Yates, E. Hanski, and M. Caparon. 1993. Adherence and fibronectin-binding are environmentally regulated in the group A streptococcus. Mol. Microbiol. 9:1213-1222. [DOI] [PubMed] [Google Scholar]
- 37.Wannamaker, L. W., B. Hayes, and W. Yasmineh. 1967. Streptococcal nucleases II. Characterization of DNase D. J. Exp. Med. 126:497-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Y., and J. K. Broome-Smith. 1989. Identification of amino acid sequences that can function as translocators of β-lactamase in Escherichia coli. Mol. Microbiol. 3:1361-1369. [DOI] [PubMed] [Google Scholar]