Abstract
A microorganism whose growth is linked to the dechlorination of polychlorinated biphenyls (PCBs) with doubly flanked chlorines was identified. Identification was made by reductive analysis of community 16S ribosomal DNA (rDNA) sequences from a culture enriched in the presence of 2,3,4,5-tetrachlorobiphenyl (2,3,4,5-CB), which was dechlorinated at the para position. Denaturing gradient gel electrophoresis (DGGE) analysis of total 16S rDNA extracted from the culture led to identification of three operational taxonomic units (OTUs 1, 2, and 3). OTU 1 was always detected when 2,3,4,5-CB or other congeners with doubly flanked chlorines were present and dechlorinated. Only OTUs 2 and 3 were detected in the absence of PCBs and when other PCBs (i.e., PCBs lacking doubly flanked chlorines) were not dechlorinated. Partial sequences of OTUs 2 and 3 exhibited 98.2% similarity to the sequence of “Desulfovibrio caledoniensis” (accession no. DCU53465). A sulfate-reducing vibrio isolated from the culture generated OTUs 2 and 3. This organism could not dechlorinate 2,3,4,5-CB. From these results we concluded that OTU 1 represents the dechlorinating bacterium growing in a coculture with a Desulfovibrio sp. The 16S rDNA sequence of OTU 1 is most similar to the 16S rDNA sequence of bacterium o-17 (89% similarity), an ortho-PCB-dechlorinating bacterium. The PCB dechlorinator, designated bacterium DF-1, reductively dechlorinates congeners with doubly flanked chlorines when it is supplied with formate or H2-CO2 (80:20).
Polychlorinated biphenyls (PCBs) remain an environmental concern because of their chemical stability and potential toxicity to humans and wildlife. Although production of PCBs has been banned in much of the industrial world, these compounds are still used to some extent in electrical equipment and can be found in the environment many years after they are released. For these reasons microbial degradation and dechlorination of PCBs have been investigated in great detail (reviewed in references 1, 5, 24, and 25). Aerobic PCB-degrading microorganisms degrade the biphenyl ring, but usually they do not transform the more extensively chlorinated PCB congeners that dominate many commercial PCB mixtures. In contrast, microbial dechlorination of PCBs under anaerobic conditions results in the dechlorination of PCB congeners that are more extensively chlorinated and more resistant to aerobic attack. Although there have been several reports concerning the microbes that catalyze the aerobic biodegradation of PCBs, dechlorinating anaerobes have eluded isolation and identification. This has impeded further research into the mechanisms of anaerobic PCB dechlorination.
Recently, two PCB-dechlorinating enrichment cultures derived from estuarine sediment were established in a defined and sediment-free medium. One of these cultures is capable of ortho PCB dechlorination (8), and the other (the DF culture) is restricted to dechlorination of PCB congeners with doubly flanked chlorines (27). The available evidence suggests that different microorganisms with distinct dehalogenase and congener specificities are responsible for the various processes attributed to PCB dechlorination (reviewed in references 5 and 25). The DF culture is a nonmethanogenic microbial community that can meta dechlorinate 2,3,4-chlorobiphenyl (2,3,4-CB) and 2,3,4,6-CB and para dechlorinate 3,4,5-CB and 2,3,4,5-CB, but it does not dechlorinate congeners lacking doubly flanked chlorines, including 3-CB, 4-CB, 2,3-CB, 2,4-CB, 2,5-CB, 3,4-CB, 3,5-CB, 2,3,5-CB, 2,3,6-CB, 2,4,6-CB, 2,3,5,6-CB, and 2,4,5-2,4,5-CB (27). The species in the DF culture that catalyze reductive PCB dechlorination have not been identified previously. Identification of any PCB-dechlorinating species has been precluded by the lack of pure cultures. Although many microorganisms have resisted isolation and growth in pure culture, molecular identification techniques have been used to identify individual microbial species in natural or enriched communities (4, 10, 11, 13, 15, 22). Reductive analysis of 16S ribosomal DNA (rDNA) by denaturing gradient gel electrophoresis (DGGE) combined with selective enrichment and comparative sequence analysis was used in this study to identify for the first time a bacterium that dechlorinates PCB congeners with doubly flanked chlorines.
MATERIALS AND METHODS
Culture procedures.
A defined estuarine medium (E-Cl medium) was prepared under anaerobic conditions as described by Berkaw et al. (6), except that Na2S · H2O was not added. The final pH of the medium was 7.0, and the medium was sterilized by autoclaving it at 121°C for 30 min. The dechlorinating enrichment culture used in this study, the DF culture, was enriched as described by Wu et al. (27). Sodium formate (10 mM) was added to the medium as a potential carbon and energy source. Following inoculation of the medium, 2,3,4,5-CB (1 mg in 10 μl of acetone, resulting in a final PCB concentration of 350 μM) was added to 10 ml of E-Cl medium. Control cultures containing no PCB were prepared by adding acetone (10 μl) without PCB to E-Cl medium (10 ml) inoculated with the DF culture. All cultures were incubated at 30°C in the dark, and all tests with the DF culture were done in duplicate. Unless stated otherwise, transfers of the DF culture were made by using 1% (vol/vol) inocula.
Sequence analysis of excised fragments of 16S rDNA obtained from DGGE gels (see below) showed that an organism related to a Desulfovibrio sp. was present in the DF culture (see below). In order to isolate this organism, the DF culture was transferred to E-Cl medium containing 10 mM Na2SO4 and 10 mM sodium lactate (Desulfovibrio isolation medium [DIM]). After two sequential extinction dilution series (with dilution up to 10−8) of the DF culture in DIM, the second serially diluted culture (0.1 ml of the 10−8 dilution) was used to inoculate 1 ml of molten top agar medium (DIM plus 0.5% [wt/vol] agar), which was poured over a solidified agar medium (DIM plus 2% [wt/vol] agar). The plates were prepared in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.) with an N2-H2-CO2 (90:5:5) atmosphere. The inoculated plates were incubated in closed canning jars at room temperature in the dark in the anaerobic chamber. Isolated colonies with light yellow pigmentation were inoculated into liquid DIM. Culture purity was confirmed by reisolation on agar as described above, and then the isolates were analyzed by DGGE and for PCB-dechlorinating activity. A similar agar overlay procedure was used in an attempt to isolate other organisms from the DF culture. In this case 10 mM sodium formate and 350 μM 2,3,4,5-CB were added to the agar medium.
Specificity of PCB-dechlorinating activity.
E-Cl medium (10 ml) was inoculated with 0.1 ml of an actively dechlorinating DF culture. Each culture was supplemented with 10 mM sodium formate and one of the following congeners (final concentration, 100 μM): 2,3,4-CB, 2,3,5-CB, 2,4,6-CB, 3,4,5-CB, 2,3,4,5-CB, or 2,3,5,6-CB.
PCB analysis.
PCBs were extracted from the cultures with ethyl acetate, and the solvent extracts were passed through a Florisil-copper column (6). The PCBs were analyzed by using a Hewlett-Packard 5890 series II gas chromatograph equipped with an RTX-1 capillary column (30 m by 0.25 mm [inside diameter] by 0.25 μm; Restek Corp., Bellefonte, Pa.) and a 63Ni electron capture detector as described previously (6). PCBs were identified by matching their gas chromatography retention times with those of authentic standards (purity, 99%; AccuStandard) and were quantified with a 16-point calibration curve for each congener (6).
Extraction of genomic DNA and PCR amplification.
Samples (1.5 ml) of DF cultures were withdrawn under anaerobic conditions by using the Hungate technique (7). Genomic DNA was extracted by using an InstaGene matrix (Bio-Rad, Hercules, Calif.) according to the manufacturer's instructions or by bead beating the cells (22). Bacterial 16S rDNA was amplified as described by Cutter et al. (9). DGGE analysis produced the same profile whether the DNA was extracted by the InstaGene procedure or by bead beating. To check for the presence of archaeal DNA, genomic DNA was extracted by bead beating and the 16S rDNA was amplified with primer sets specific for archaea (9).
DGGE.
Analysis of 16S rDNA PCR products by DGGE was performed as described by Cutter et al. (9), with the following modifications. The PCR products were applied to 7% (wt/vol) polyacrylamide gels that contained a 45 to 75% denaturing gradient of formamide and urea and were electrophoresed at 60°C for 17 h in 1× TAE buffer at a constant voltage of 50 V. To excise DNA fragments, the gels were stained with ethidium bromide (0.5 μg ml−1) and visualized with a UV transilluminator. Excised DNA fragments were amplified by using the PCR parameters described by Cutter et al. (9). Amplification and gel analysis of the excised DNA were repeated until only one band with the same migration distance as the original band was detected by DGGE.
Sequence analysis.
Following DGGE gel purification, DNA fragments at unique migration positions were sequenced by using the method of Ferris et al. (12) and an ABI 373 automated sequencer (Applied Biosystems, Foster City, Calif.). To obtain larger fragments of 16S rDNA for phylogenetic analysis, universal primers 29-47 forward (5"-GAG TTT GAT CCT GGC TCA G-3") and 1541-1525 reverse (5"-AGA AAG GAG GTG ATC AGC C-3") were used to amplify rDNA from cultures (23). Sequences of DNA fragments were submitted to the National Center for Biotechnology Information basic local alignment search tool (BLAST) (3) and the Ribosomal Database Project (18) to determine similarity with other 16S rDNA molecules. Partial 16S rDNA sequences were manually compiled and aligned, and evolutionary trees were generated, as described by Cutter et al. (9).
Nucleotide sequence accession numbers.
Sequences of the partial 16S rDNA of the predominant DNA fragments have been deposited in the GenBank database under accession numbers AF389905 (OTU 1), AF389906 (OTU 2), AF389907 (OTU 3), and AF393781 (bacterium DF-1). The accession numbers for organisms compared in this study are as follows: uncultured bacterium SJA-15, AJ009453; uncultured bacterium SJA-61, AJ009469; uncultured bacterium SJA-170, AJ009500; uncultured bacterium SHA-105, AJ249113; uncultured bacterium SJA-116, AJ009487; unidentified eubacterium vadinBA26, U81649; Dehalococcoides ethenogenes, AF004928; D. ethenogenes FL2, AF357918; bacterium CBDB1, AF230641; bacterium o-17 (9), AF058005; uncultured eubacterium t0.8.f, AF005746; uncultured eubacterium H1.4.f, AF005748; uncultured eubacterium H3.93, AF005750; and “Desulfovibrio caledoniensis,” DCU53465.
RESULTS
DGGE profile of the DF culture and effect of 2,3,4,5-CB on sustainability of PCB dechlorination.
The ability of the DF culture to maintain PCB-dechlorinating activity in the absence of PCBs was tested. An actively dechlorinating culture (10 μl) was transferred into 10 ml of E-Cl medium with and without 350 μM 2,3,4,5-CB. The 0.1% (vol/vol) inoculum ensured that the amount of residual PCB transferred into the medium lacking PCBs would be negligible. After 46 days, 60 mol% of the 2,3,4,5-CB was dechlorinated to 2,3,5-CB in the culture supplemented with 2,3,4,5-CB. At this time, the cultures maintained without 2,3,4,5-CB were transferred (0.1%, vol/vol) back into E-Cl medium with 2,3,4,5-CB, and the cultures were incubated for 45 days. No dechlorination was observed in the latter cultures. These results indicate that the DF culture must be grown with 2,3,4,5-CB in order to maintain the ability to dechlorinate this congener.
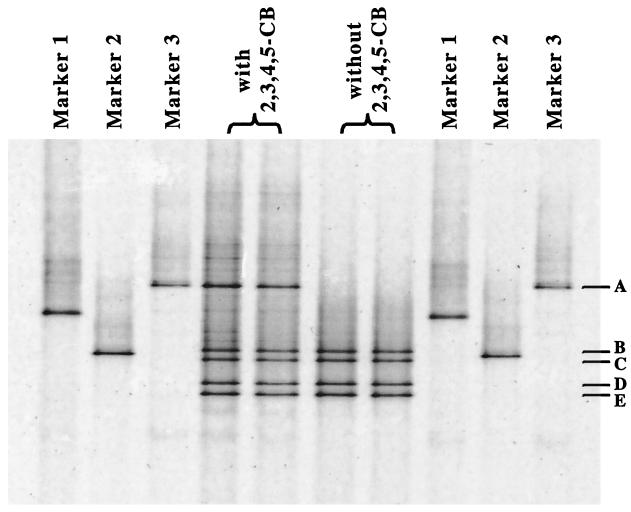
The community diversity of the DF culture when it was dechlorinating 2,3,4,5-CB to 2,3,5-CB (60% conversion after 46 days) was examined by PCR-DGGE. Five DNA fragment bands (bands A through E) were obtained by DGGE (Fig. 1). Each of the DNA fragments (DGGE bands) was excised, reamplified, and reexamined by DGGE until an isolated band was available for sequence analysis. Three of the bands (bands A, D, and E) were given operational taxonomic unit designations (OTUs 1, 2, and 3, respectively). Following comparative sequence analysis of the partial 16S rDNA (260 bp), OTU 1 was found to be most similar (93.6% similarity) to the uncultured eubacterium H1.4.f. Excision and amplification of bands B and C after three sequential DGGE analyses revealed the same band pattern (bands B through E) as the band pattern obtained with the DF culture. This purification procedure was repeated with a narrower denaturing gradient (55 to 60%) to ensure that the DNA fragments were pure. This resulted in clearly separated bands, but regardless of the band separation and the number of excisions and amplifications the four bands (bands B through E) were always obtained following amplification of purified DNA from bands B and C. Comparative sequence analysis of bands D and E (221 bp) showed that there was only one base pair difference (99.6% similarity) between the two fragments. Based on these results, we concluded that bands B and C are heteroduplex DNA of bands D and E. Therefore, only bands D and E were given operational taxonomic unit designations (OTUs 2 and 3, respectively). Comparative sequence analysis of OTUs 2 and 3 showed that they are most similar (98.2% similarity for both operational taxonomic units) to “D. caledoniensis.”
FIG. 1.
DGGE resolution of PCR-amplified 16S rDNA fragments from the DF culture incubated with and without 350 μM 2,3,4,5-CB in E-Cl medium. Marker 1, amplified DNA from bacterium o-17 (9); Marker 2, amplified DNA from Burkholderia cepacia; Marker 3, excised and purified 16S rDNA from band A of DGGE gel. The duplicate lanes contained individual samples from duplicate cultures.
In contrast to the results obtained with the DF culture incubated with 2,3,4,5-CB, OTU 1 was not detected in cultures incubated without 2,3,4,5-CB (Fig. 1). Cultures incubated without PCB and then transferred (1%, vol/vol) back to E-Cl medium with 2,3,4,5-CB did not dechlorinate the PCB. DGGE analysis of these cultures revealed only OTUs 2 and 3 (bands B through E) after 45 days of incubation (data not shown). Since no dechlorination of 2,3,4,5-CB occurred in the subcultures reintroduced into PCB-containing medium, the results indicated that the number of microorganisms responsible for dechlorination of 2,3,4,5-CB was reduced following incubation without 2,3,4,5-CB. Overall, the results support the conclusion that OTU 1 was the microorganism that dechlorinated the PCB in these cultures.
Microscopic observation (phase-contrast microscopy, with the results confirmed by electron microscopy) of the DF culture revealed the presence of vibrio-shaped cells and very small (diameter, <1 μm) irregularly shaped cocci (data not shown). A Desulfovibrio sp. was isolated from the DF culture in medium that was selective for sulfate-reducing bacteria (see Materials and Methods). Single colonies were obtained after two sequential isolations by streaking on lactate-sulfate agar plates. Only one type of cells, motile vibrios whose morphology was identical to the morphology of the vibrios in the DF coculture, was observed microscopically. This isolate could not dechlorinate 2,3,4,5-CB. All attempts to isolate the coccus-shaped microorganism were unsuccessful. These attempts included serial dilution with and without ampicillin. Attempts to grow the coccus-shaped organism in an agar overlay medium did result in successful passage of the PCB-dechlorinating community to the plates and then back to liquid E-Cl medium. However, the colonies, which were <1 mm in diameter, off-white, and irregular, remained a mixture of the same organisms, and colonies could not be successfully grown a second time on the agar medium.
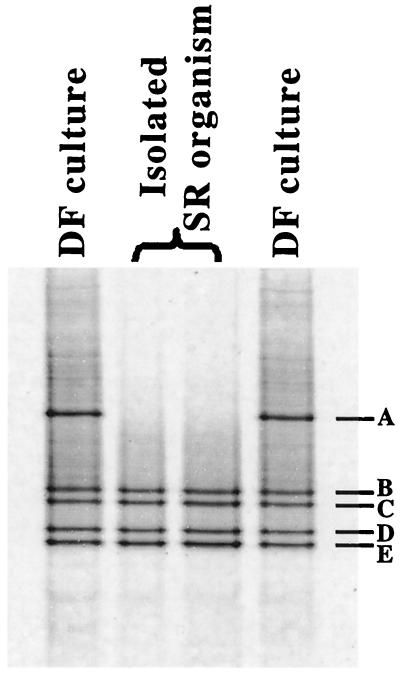
DGGE analysis of the sulfate-reducing vibrio revealed OTUs 2 and 3 (bands B through E) (Fig. 2). Since the purity of the culture was confirmed by reisolation of the Desulfovibrio sp. and the sequences of OTUs 2 and 3 are so similar (the partial sequences of OTUs 2 and 3 differ by only 1 bp out of 221 bp), it is likely that the organism isolated contains multiple copies of the 16S rDNA gene, a common trait in bacteria (17). The sulfate-reducing culture did not dechlorinate 2,3,4,5-CB when it was incubated in E-Cl medium containing 10 mM formate or 10 mM lactate (82 days of incubation with 350 μM PCB). These data further confirmed that OTUs 2 and 3 do not represent the organism that dechlorinates 2,3,4,5-CB in the DF culture.
FIG. 2.
DGGE resolution of PCR-amplified 16S rDNA fragments from an isolated sulfate-reducing (SR) vibrio and the DF culture incubated with 350 μM 2,3,4,5-CB in E-Cl medium. The duplicate lanes contained individual samples from duplicate cultures.
The DF culture has remained nonmethanogenic for more than 2 years and 10 transfers. To confirm that archaea were not present in the culture, genomic DNA was extracted by bead beating the culture and was then subjected to PCR amplification with primers specific for archaea. No PCR product was detected. These results confirm that methanogens and archaea play no role in the dechlorination of doubly flanked chlorines of PCBs by the DF culture.
Specificity of PCB dechlorination and effect of other PCB congeners on the microbial community profile of the DF culture.
The DF culture was incubated with different PCB congeners individually in order to further assess the dechlorinating specificity of the culture and to determine the effect that these congeners had on the microbial community. The congener 2,3,4-CB was meta dechlorinated to 2,4-CB (62 mol% in 84 days), and the congener 3,4,5-CB was para dechlorinated to 3,5-CB (87 mol% in 84 days). No dechlorination of 2,3,5-CB, 2,4,6-CB, or 2,3,5,6-CB was observed. These data are consistent with previous results (27) and confirm that the DF culture attacks only PCBs with doubly flanked chlorines.
The following two sets of DF cultures were examined further: (i) cultures that actively dechlorinated 2,3,4-CB and 3,4,5-CB and (ii) cultures that did not dechlorinate 2,3,5,6-CB. Cultures incubated with 2,3,4-CB were transferred into E-Cl medium with 2,3,4-CB or 2,3,4,5-CB to see if the ability to dechlorinate 2,3,4,5-CB is sustained after sequential transfers with 2,3,4-CB. After 56 days, the subcultures containing 2,3,4-CB had dechlorinated 62 mol% of this congener to 2,4-CB, and the subcultures containing 2,3,4,5-CB had dechlorinated 84 mol% of the tetrachlorobiphenyl to 2,3,5-CB. Similarly, the cultures sequentially transferred with 3,4,5-CB were transferred into E-Cl medium with 3,4,5-CB or 2,3,4,5-CB. Subcultures containing 3,4,5-CB dechlorinated 87 mol% of the trichlorobiphenyl to 3,5-CB, and subcultures containing 2,3,4,5-CB dechlorinated 82 mol% of the tetrachlorobiphenyl to 2,3,5-CB after 56 days. In contrast, dechlorination of 2,3,4,5-CB was not detected after 56 days in DF cultures that were first incubated with 2,3,5,6-CB, a congener without doubly flanked chlorines. These data indicate that the ability to dechlorinate 2,3,4,5-CB is sustained by cells grown on an alternative PCB congener with doubly flanked chlorines. The results also indicate that a culture loses the ability to dechlorinate congeners with doubly flanked chlorines when it is grown with a PCB substrate that cannot be dechlorinated (i.e., a congener without doubly flanked chlorines).
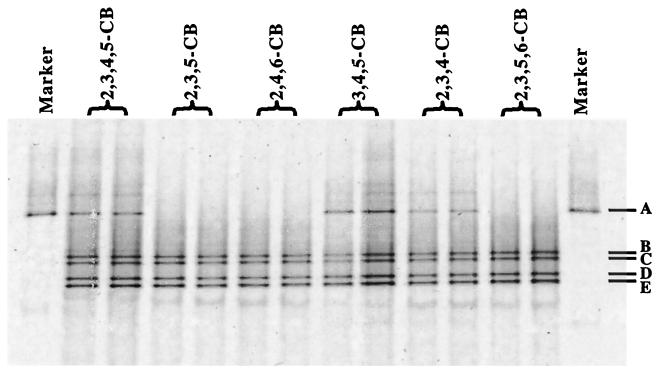
In order to determine whether substrate specificity and loss of dechlorination occur at the community level (i.e., by loss of species) or at the cellular level (e.g., by gene induction or repression), the microbial communities in DF cultures were examined by DGGE. Cultures incubated with 2,3,4-CB or 3,4,5-CB (Fig. 3) produced the same band pattern as cultures grown with 2,3,4,5-CB. When cultures incubated with 2,3,4-CB or 3,4,5-CB were transferred to E-Cl medium with 2,3,4,5-CB, DGGE analysis revealed the same operational taxonomic units in these subcultures as in the subcultures maintained with 2,3,4,5-CB (data not shown). DGGE analysis of the DF culture incubated with the congeners 2,3,5-CB, 2,4,6-CB, and 2,3,5,6-CB without doubly flanked chlorines revealed only OTUs 2 and 3 (Fig. 3). When cultures grown with 2,3,5,6-CB were transferred back to E-Cl medium with 2,3,4,5-CB, only bands B through E were detected (data not shown). These data further confirm that the microorganism represented by OTU 1 is responsible for the dechlorination of 2,3,4-CB, 3,4,5-CB, and 2,3,4,5-CB in the DF culture and that dechlorination and growth of OTU 1 are sustainable only with congeners with doubly flanked chlorines.
FIG. 3.
DGGE resolution of PCR-amplified 16S rDNA fragments from the DF culture incubated with 2,3,4-CB, 2,3,5-CB, 2,4,6-CB, 3,4,5-CB, 2,3,4,5-CB, or 2,3,5,6-CB (all congeners at 100 μM) for 84 days. Marker 1, excised and purified 16S rDNA from band A of DGGE gel. The duplicate lanes contained individual samples from duplicate cultures.
Phylogeny of OTU 1.
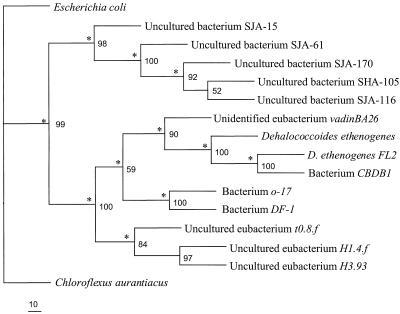
By using primers that flanked the DGGE primers, 1,214 bp of 16S rDNA sequence was obtained for OTU 1 and then compared with 16S rDNA sequences in the GenBank database. This longer sequence for OTU 1 exhibited high levels of similarity with 16S rDNA sequences of organisms affiliated with the green nonsulfur bacteria, including species of the genus Dehalococcoides (Fig. 4). The highest level of similarity (89%) was with bacterium o-17, which was found in a 2,3,5,6-CB-dechlorinating enrichment culture (9). The PCB-dechlorinating species represented by OTU 1 in the DF culture has been designated bacterium DF-1.
FIG. 4.
Neighbor-joining phylogenetic tree including bacterium DF-1 generated from analysis of 1,100 bp of the16S rDNA sequence. The asterisks indicate branches that were also found when the Fitch-Margoliash and maximum-parsimony methods were used. The numbers at the nodes are percentages that indicate the levels of bootstrap support, based on a neighbor-joining analysis of 1,000 resampled data sets. Scale bar = 10 substitutions per 100 nucleotide positions. Escherichia coli was used as an outgroup.
DISCUSSION
Bacterium DF-1 is the first microorganism identified that is capable of PCB dechlorination restricted to doubly flanked chlorines on the biphenyl. Based on the PCR-DGGE analysis of community 16S rDNA, it is clear that bacterium DF-1 grows in response to dechlorination of specific PCBs. Reductive dehalogenation of chlorinated organic compounds can be coupled to microbial growth through dehalorespiration (2; reviewed in references 14 and 21). There is evidence suggesting that PCB-dechlorinating microorganisms, which use PCBs as electron acceptors, may also derive energy from reductive PCB dechlorination. Kim and Rhee (16) observed that the number of PCB-dechlorinating microorganisms increased 188-fold in sediment microcosms amended with 300 ppm of Aroclor 1248. Conversely, the number decreased by 93% in samples without Aroclor 1248. These authors concluded that the growth of PCB dechlorinators requires the presence of PCBs. Recently, Wu et al. (26) reported that the number of microorganisms in Woods Pond sediment capable of dehalogenating 2,6-bromobiphenyl and PCBs increased nearly 1,000-fold after priming with 1,050 μM 2,6-bromobiphenyl plus 10 mM malate. These results demonstrate that halogenated biphenyls prime PCB dechlorination by stimulating the growth of PCB-dechlorinating microorganisms. However, these studies were conducted under undefined conditions (sediment), and the dechlorinating bacteria were not identified.
Here we established that the DF enrichment culture is a coculture of a sulfate-reducing vibrio and bacterium DF-1. The DGGE results and the inability of the vibrio to dechlorinate PCBs indicate that bacterium DF-1, not the vibrio, is the dechlorinator. However, it remains to be determined whether PCB-dechlorinating bacterium DF-1 relies on the vibrio for some unknown factor or function. The morphology of the other bacterium observed in the culture, a very small irregular coccus, is similar to the morphology of D. ethenogenes (20). The 16S rDNA sequence of bacterium DF-1 is most similar to the 16S rDNA sequences of a number of cultured and uncultured organisms that are associated with dechlorination, including Dehalococcoides spp. Bacterium DF-1 exhibits the highest level of similarity to bacterium o-17 (89%), an uncultured microorganism that ortho dechlorinates 2,3,5,6-CB and 2,3,5-CB (9). However, bacterium DF-1 does not dechlorinate 2,3,5,6-CB or 2,3,5-CB. In addition to the differences in dechlorinating specificity, these organisms can be distinguished by DGGE analysis (Fig. 1) and by the carbon and energy sources that support dechlorination by the different dechlorinating bacteria. Formate and H2-CO2 (80:20) support the growth and dechlorinating activity of bacterium DF-1, but acetate alone does not. In contrast, bacterium o-17 requires acetate for growth and for ortho dechlorination of 2,3,5,6-CB, but this bacterium cannot use formate or H2-CO2 for these activities (9). Indeed, addition of hydrogen (H2-CO2, 80:20) to the medium inhibits PCB dechlorination by bacterium o-17 even when acetate is added to the medium.
D. ethenogenes, an organism that uses tetrachloroethene as a terminal electron acceptor and dechlorinates this compound to ethene (19, 20), is in the same deeply branching phylogenetic cluster as bacterium DF-1. Bacterium CBDB1 (2), a chlorobenzene-dechlorinating microorganism, is also associated with this cluster of microorganisms. D. ethenogenes and bacterium CBDB1 have very similar 16S rDNA sequences (98% similarity). The 16S rDNA sequence of PCB-dechlorinating bacterium DF-1 is not as similar to the sequences of these other organisms (87% similarity with D. ethenogenes and 88% similarity with bacterium CBDB1). Other environmental sites and enrichment cultures are being investigated to determine the distribution and diversity of the microorganisms related to bacterium DF-1. In preliminary studies the dechlorinating activity expressed by bacterium DF-1 has been found at several sites (unpublished results), which suggests that bacterium DF-1 or very similar microorganisms may be widespread. Development of innovative approaches to isolate this species is ongoing, which should enable further research on the role of this unique group of bacteria in the global cycling of organic chlorine.
Acknowledgments
We thank Lisa A. May for technical assistance with the DGGE analysis.
This work was supported by the Office of Naval Research, U.S. Department of Defense (grants N00014-99-1-0978 and N00014-99-1-0575 to H.D.M. and grant N00014-99-1-0101 to K.R.S.).
REFERENCES
- 1.Abramowicz, D. A. 1990. Aerobic and anaerobic biodegradation of PCBs: a review. Bio/Technology 10:241-251. [Google Scholar]
- 2.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedard, D. L., and J. F. Quensen III. 1995. Microbial reductive dechlorination of polychlorinated biphenyls, p. 127-216. In L. Y. Young and C. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. Wiley-Liss Division, John Wiley & Sons, Inc., New York, N.Y.
- 6.Berkaw, M., K. R. Sowers, and H. D. May. 1996. Anaerobic ortho dechlorination of polychlorinated biphenyls by estuarine sediments from Baltimore Harbor. Appl. Environ. Microbiol. 62:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breznak, J. A., and R. N. Costilow. 1994. Physiochemical factors in growth, p. 137-154. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 8.Cutter, L. A., K. R. Sowers, and H. D. May. 1998. Microbial dechlorination of 2,3,5,6-tetrachlorobiphenyl under anaerobic conditions in the absence of soil or sediment. Appl. Environ. Microbiol. 64:2966-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutter, L. A., J. E. M. Watts, K. R. Sowers, and H. D. May. 2001. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,5,6-chlorobiphenyl. Environ. Microbiol. 3:699-709. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. E., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 11.de Souza, M. P., A. Amini, M. A. Dojka, I. J. Pickering, S. C. Dawson, N. R. Pace, and N. Terry. 2001. Identification and characterization of bacteria in a selenium-contaminated hypersaline evaporation pond. Appl. Environ. Microbiol. 67:3785-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, D. N., and N. R. Pace. 2001. Molecular-phylogenetic analyses of human gastrointestinal microbiota. Curr. Opin. Gastroenterol. 17:52-57. [DOI] [PubMed] [Google Scholar]
- 14.Holliger, C., G. Wohlfarth, and G. Diekert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 15.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, J., and G.-Y. Rhee. 1997. Population dynamics of polychlorinated biphenyl-dechlorinating microorganisms in contaminated sediments. Appl. Environ. Microbiol. 63:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maymo-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maymo-Gatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 21.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dehalogenation. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pulliam Holoman, T. R., M. A. Elberson, L. A. Cutter, H. D. May, and K. R. Sowers. 1998. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl. Environ. Microbiol. 64:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainey, F. A., M. Dorsch, H. W. Morgan, and E. Stackebrandt. 1992. 16S rDNA analysis of Spirochaeta thermophila: its phylogenetic position and implications for the systematics of the order Spirochaetales. Syst. Appl. Microbiol. 15:197-202. [Google Scholar]
- 24.Unterman, R. 1996. A history of PCB biodegradation, p. 209-253. In R. L. Crawford and D. L. Crawford (ed.), Bioremediation: principles and applications. University Press, Cambridge, Great Britain.
- 25.Wiegel, J., and Q. Wu. 2000. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol. 32:1-15. [DOI] [PubMed] [Google Scholar]
- 26.Wu, Q., D. L. Bedard, and J. Wiegel. 1999. 2,6-Dibromobiphenyl primes extensive dechlorination of Aroclor 1260 in contaminated sediment at 8-30°C by stimulating growth of PCB-dehalogenating microorganisms. Environ. Sci. Technol. 33:595-602. [Google Scholar]
- 27.Wu, Q., K. R. Sowers, and H. D. May. 2000. Establishment of a polychlorinated biphenyl-dechlorinating microbial consortium, specific for doubly flanked chlorines, in defined, sediment-free medium. Appl. Environ. Microbiol. 66:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]