Abstract
Sparse information is available on the virulence factors of Aeromonas strains isolated from diseased fish, from the environment, and from humans. In the present study, 52 Aeromonas isolates obtained from epizootic ulcerative syndrome (EUS) lesions in fish, from the aquatic environment, and from children with diarrhea in Bangladesh were identified by biochemical phenotyping (i.e., PhenePlate [PhP] typing) and DNA fingerprinting and then characterized with respect to certain putative virulence factors. The isolates from the fish exhibiting EUS symptoms were identified to be Aeromonas veronii biovar sobria by fatty acid methyl ester analysis and amplified fragment length polymorphism fingerprinting. Biochemical phenotyping revealed that all EUS-associated isolates belonged to a unique phenotype which was not identified among more than 1,600 environmental and diarrheal isolates in a previously collected database of PhP types of Bangladeshi Aeromonas isolates. The 52 Aeromonas isolates were investigated for the production of hemolysin and cytotoxin; for hemagglutination with erythrocytes from fish, human, and rabbit sources; for the presence of a cytolytic enterotoxin gene; and for adhesion to and invasion into fish cell lines. All of the EUS isolates produced all of the virulence factors investigated, as did also some of the environmental isolates, but the isolates from EUS were unique in their ability to agglutinate fish erythrocytes. Our results suggest that a clonal group of A. veronii biovar sobria is associated with, and may be a causative agent of, EUS in fish in Bangladesh.
Aeromonas spp. are ubiquitous inhabitants of aquatic ecosystems such as freshwater, coastal water, and sewage (36). They are increasingly being reported as important pathogens for humans and for lower vertebrates, including amphibians, reptiles, and fish (19). These bacteria have a broad host range, and have often been isolated from humans with diarrhea (17), as well as from fish with hemorrhagic septicemia (39). The pathogenesis, pathogenic mechanism, and virulence factors responsible for selected Aeromonas infection in different species are not well understood. Strains isolated from the environment do not seem to differ from strains isolated from cases of infection with respect to the prevalence of virulence factors (29). However, it has been shown that certain species are more frequently isolated from patients with diarrhea as well as from diseased fish than from the environment (25).
Epizootic ulcerative syndrome (EUS) is a fish disease characterized by the presence of severe, open dermal ulcers on the head, on the middle of the body, and on the dorsal regions of the fish (33). EUS has been characterized as an epizootic disease of freshwater fish in the Indo-Pacific region since 1980 (8) and was first reported in Bangladesh in 1988 (2). This disease is now frequently occurring in many fish farms in Bangladesh (6); the disease generally develops with ulcers that develop on the fish bodies, and the fish may die within a week of being infected. The disease has caused substantial economic loss to fish farmers and the fisheries sector. The etiological agent(s) of EUS in Bangladesh is still unknown; however, organisms belonging to the potentially fish-pathogenic genera Aeromonas, Vibrio, Plesiomonas, and Pseudomonas were often isolated from the lesions and blood samples of infected fish. Representatives of Aeromonas hydrophila and Aeromonas sobria were recovered most frequently, followed by Vibrio and Plesiomonas spp. (33).
In light of the increased incidence of EUS and the economic importance of these epizootic diseases and because of possible public health effects, it is of great importance to further study and characterize the etiologic agents of EUS. In the current study, we sought to identify bacteria recovered from EUS infections in Bangladesh and to investigate the virulence factors of the isolates associated with EUS. In addition, we also compared the virulence properties of the isolates from diseased fish to those of human diarrheal and environmental isolates from Bangladesh.
MATERIALS AND METHODS
Isolation and biochemical identification of bacterial isolates.
The fish isolates were isolated from different diseased fish (African catfish [Clarias gariepinus], rajputi [Puntius gonionotus], rui [Labeo rohita], catla [Catla catla], and shole [Channa striatus]) from different fish farms (the Bangladesh Agricultural University [BAU] fish farm; Jhalak Hatchery and Fish Farm; and Dhaka Fisheries, Ltd., Bangladesh, Bangladesh) by the fish disease laboratory at BAU in 1997 and 1998. The disease symptoms were deep hemorrhagic ulcers in the midbody and tail regions. Aeromonas selective agar, i.e., Aeromonas Medium Base (Oxoid, Ltd.), supplemented with ampicillin SR136 (5 μg ml−1), was used to obtain Aeromonas isolates from fish ulcer scraps. Fourteen randomly selected Aeromonas isolates (BDF1 to BDF14) were obtained from the BAU fish disease laboratory collection. For comparison, a number of water samples were collected from the same ponds as the fish within a 6-month time period. These environmental samples were processed by using the same selective medium, except that no ampicillin was added, due to the low-level ampicillin resistance among environmental isolates. Among the environmental isolates, 26 presumed Aeromonas isolates (BDE15 to BDE40) were randomly selected. Furthermore, 12 randomly selected human diarrheal isolates (BDD41 to BDD52) of Aeromonas spp. obtained from a previous study (30) on Bangladeshi children with diarrhea were included in the present study.
The presumed Aeromonas isolates were confirmed by oxidase and catalase test and by determining the sensitivity to the vibriostatic reagent 0/129 (150 μg ml−1; Sigma, St. Louis, Mo.). The isolates were identified to the species level by traditional biochemical methods (3), including tests for esculin hydrolysis, lysine decarboxylase, arginine dihydrolase, and ornithine decarboxylase; tests for acid production from arabinose, glucose, sucrose, and mannitol; and tests for susceptibility to ampicillin and cephalothine (30 μg ml−1) (4, 20, 21), and, when necessary, by using API 20NE and API 20E biochemical identification strips (bioMérieux, Marcy l'Etoile, France). All isolates were stored in 30% glycerol in brain heart infusion (BHI) broth at −70°C until further use, subsequently recultured on BHI agar plates (Becton Dickinson Microbiology Systems), and then incubated overnight at 37°C.
Biochemical phenotyping with the PhP system.
Bacterial isolates were typed through a biochemical phenotyping method, PhenePlate (PhP) system (PhPlate Microplate Techniques AB, Stockholm, Sweden), according to the manufacturer's instructions. The PhP typing was performed in previously prepared microplates and is based on the kinetics of fermentation of 48 dehydrated reagents especially selected to discriminate between individual bacterial strains (32, 35). The biochemical reactions of the isolates were compared pairwise, and a similarity matrix consisting of the correlation coefficient between all possible pairs was constructed. The similarity matrix was clustered according to unweighted pair-group method by using average linkages (UPGMA) (38). Isolates with a level of similarity of >0.97 were assigned to the same PhP phenotypes. All data processing, including optical reading and calculation of the correlation coefficient, as well as the clustering and printing of dendrograms, was performed with PhP software (PhPlate Microplate Techniques AB).
Identification of Aeromonas spp. by gas-liquid chromatographic analysis of FAME profile and AFLP fingerprinting.
Presumptive Aeromonas isolates were further identified to the genomic species level by using gas-liquid chromatographic analysis of cellular fatty acid methyl esters (FAMEs) as described previously (14). Unknown FAME profiles were compared to the laboratory-based identification library AER48C (13). Isolates that remained unidentified or yielded unreliable FAME identifications were further subjected to whole-genome fingerprinting by using amplified fragment length polymorphism (AFLP) analysis according to the method of Huys and Swings (15). The AFLP profiles of unknown isolates were compared with the laboratory-based identification library AEROLIB comprising AFLP profiles generated from a collection of well-characterized type and reference strains encompassing all currently recognized Aeromonas taxa (12).
Cytotoxic and hemolytic activity.
The cytotoxic activity of the isolates was tested on the fish cell line EPC (epithelioma papulosum of carp [Cyprinus carpio]) as described previously (9). Briefly, confluent monolayers of the cell were grown in 24-well tissue culture plates (Costar, Corning, N.Y.) in minimal essential medium (MEM; SVA, Uppsala, Sweden) supplemented with 10% fetal bovine serum, 1% (wt/vol) glutamine, and 1% (wt/vol) penicillin-streptomycin. The cells were incubated for 6 h at 18°C with 100 μl of sterile culture supernatant (containing 150 μg of protein ml−1 as determined by using the Bio-Rad protein assay) serially twofold diluted in supplemented MEM. The cytotoxic activity was measured as rounding up, detachment, and loss of viability of the cells, as seen under a light microscope within 6 h. The titer was determined as the highest dilution of supernatant affecting at least 50% of the cells. Isolates showing a cytotoxic effect at a dilution of 1/8 (final concentration) or more were regarded as positive.
Hemolytic activity of the isolates was measured on 1% (vol/vol) human and rabbit erythrocytes as described earlier (22). Isolates were considered positive for hemolysin production when the culture supernatant at a final concentration of 1/8 lysed at least 50% of the erythrocytes as determined by visual examination. Known positive (BD2 to BD9) and negative (BD12) isolates from an earlier investigation were included as controls for both the cytotoxin and hemolysin assays.
Hemagglutination.
Hemagglutination tests were performed on glass slides by mixing a loopful of bacteria with a 3% (vol/vol) suspension of erythrocytes from rabbit, humans, or fish (Labeo rohita) in phosphate-buffered saline (PBS). Visible agglutination within 5 min was considered a positive result. The agglutination inhibition test was performed by using a dilution of 1% (wt/vol) d-mannose, d-galactose, and l-fucose sugar in PBS (10).
Adhesion and invasion.
Two freshly seeded fish cell lines, EPC and RTG (for rainbow trout gonad), were cultured to monolayers on 24-well tissue culture plates with coverslips in MEM at 18°C and used for both kinds of tests. Adhesion and invasion were expressed as the percentage ± the standard error of the mean (SEM) of bacteria recovered after a careful washing.
Bacterial strains were cultured in BHI broth overnight at 22 or 37°C, harvested by centrifugation (3,000 rpm, 30 min, 22°C), and washed in PBS, and the concentration was adjusted before the adhesion assay was performed. At 2 h before an adhesion assay was done as described elsewhere (28), the cell medium was replaced by fresh medium without antibiotic. The monolayer was then incubated with a bacterial suspension of 107 CFU ml−1 for 1 h at 18°C and washed three times with PBS in order to eliminate nonattached bacteria. The coverslip with the monolayer and attached bacteria was removed from the tissue culture plates, fixed in methanol, stained with Giemsa, and examined for evidence of bacterial adhesion by using an inverted microscope. For a quantitative assay, the monolayer and attached bacteria of some of the coverslips were then lysed in 0.1% Triton in sterile water for 10 min. The ability of the bacteria to adhere to the cell lines was determined by spreading the Triton-induced cell lysate on nutrient agar plates and counting the number of colonies after incubation overnight at 37°C.
The invasion capacity of the isolates was determined by the gentamicin protection assay as described elsewhere (34). Briefly, after a 1-h incubation of the confluent cell monolayer with a bacterial suspension, as described above, the monolayer was washed three times with PBS and incubated for another hour in MEM containing gentamicin (50 μg ml−1) in order to kill extracellular bacteria. The monolayer was again washed three times with PBS and lysed with 0.1% Triton to release intracellular bacteria, and a viable count was made as described above. All experiments were repeated five times, and the mean value was calculated. Escherichia coli HB101, an adhesion and invasion negative strain, was used as a negative control in all experiments.
An adhesion inhibition assay was performed by using trypsin or vigorous stirring as described by Kirov et al. (24). In brief, bacteria were incubated with trypsin at a final concentration of 1 mg ml−1 for 30 min at 37°C, and the residual trypsin was removed by washing the bacteria in PBS. Vigorous stirring was performed in an Omnimixer (Labora AB, Sollentuna, Sweden) at high speed (1,400 rpm) in order to detach the fimbriae from the surface; detached fimbriae were then removed by washing the cells in PBS, and the bacterial concentration was adjusted before the adhesion assay was performed.
PCR detection of the cytolytic enterotoxin gene and/or extracellular hemolysin gene.
A cytolytic enterotoxin gene (AHCYTOEN) in A. hydrophila has been reported as a multivirulence gene involved in lethality (in mice), hemolysis, cytotoxicity, and enterotoxicity (5), i.e., activities that are established virulence factors of Aeromonas spp. The various Aeromonas isolates were therefore investigated by using the PCR primer combination strategy of Kingombe et al. (23) with the primers AHCF1 (5"-GAG AAG GTG ACC ACC AAG AAC A-3") and AHCR1 (5"-AAC TGA CAT CGG CCT TGA ACT C-3"). These primers would detect the presence of the cytolytic enterotoxin gene and/or the extracellular hemolysin gene in Aeromonas spp . The PCRs used here were slightly modified from those of Kingombe et al. (23) and were performed in a final volume of 50 μl containing 10 μl of DNA, 5 μl of a mixture containing 0.2 mM deoxyribonucleotide triphosphate, 2.5 μl of 50 mM MgCl2 solution, 5 μl of 10× PCR buffer, 2.5 μl of a 20 μM solution of each primer, 0.3 μl of Taq DNA polymerase (Life Technologies) at 5 U/μl, and 22.2 μl of double-distilled sterile water. We used 1 cycle of denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 66°C for 30 s, and extension at 72°C for 30 s, and a final extension round at 72°C for 7 min. The PCR amplicons were separated electrophoretically in a 1.5% agarose gel (Sigma type 1) and visualized after ethidium bromide staining. The specificity of the primer combination was corroborated with negative PCR results obtained by using E. coli reference strain DS17 (27).
RESULTS
Species identification and typing of isolates.
All 14 EUS-associated fish isolates were identified as A. veronii biovar sobria by traditional biochemical tests both in tubes and by using the API 20E and 20NE systems. Furthermore, in the PhP typing (based upon kinetic reading of 48 biochemical reagents) all of the fish isolates clustered with reference strains of A. veronii biovar sobria, the identity of which had been previously established by FAME analysis and AFLP fingerprinting (31, 32). Among the environmental isolates (n = 26), 42% (n = 11) clustered with the A. veronii biovar sobria, 19% (n = 5) clustered with the A. veronii biovar veronii, 31% (n = 8) clustered with A. trota, 4% (n = 1) clustered with the A. caviae complex, and one isolate did not cluster with any of the reference strains. These tentative identifications were fully confirmed by the traditional biochemical tests for Aeromonas spp.
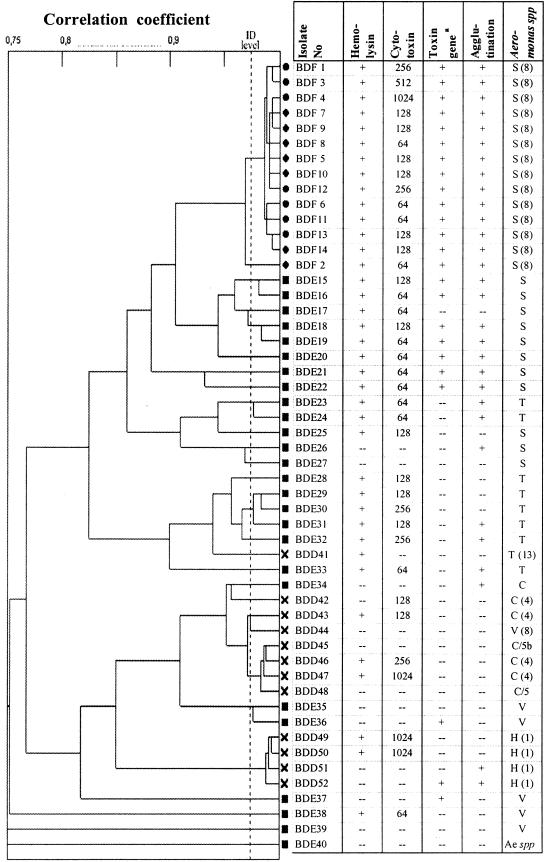
The PhP typing data of the 14 EUS-associated Aeromonas isolates were compared to and clustered with environmental (n = 26) and human diarrheal (n = 12) Aeromonas isolates (Fig. 1). It was found that the EUS-associated Aeromonas isolates showed a low level of diversity, and essentially all isolates were of the same PhP type. The environmental isolates, on the other hand, showed a higher diversity and belonged to a wide variety of PhP types. None of the environmental isolates or human diarrheal isolates was of an identical or even similar PhP type compared to the cluster containing the EUS-associated isolates. Furthermore, the PhP types representing the EUS-associated Aeromonas cluster were not found in a previously constructed laboratory-based PhP database comprising 1,200 environmental isolates and 400 diarrheal isolates from Bangladesh.
FIG. 1.
Dendrogram showing UPGMA clustering of PhP typing data obtained from 52 Aeromonas isolates. Circles indicate isolates from fish, squares indicate isolates from the environment, and crosses indicate isolates from humans with diarrhea. The results from assays of putative virulence properties are shown. The last column indicates the species codes. Some isolates were identified by FAME analysis, in which case the hybridization group (HG) is given, e.g., S(8) = A. veronii biovar sobria (hybridization group). Other isolates were identified by biochemical test: S, A. veronii biovar sobria; T, A. trota; C, A. caviae complex; H, A. hydrophila, V, A. veronii biovar veronii, etc. The column heading “toxin gene” refers to the cytolytic enterotoxin gene and/or extracellular hemolysin gene.
FAME and AFLP identification of fish Aeromonas isolates.
The FAME identification of the fourteen fish isolates was not conclusive but indicated that the isolates belonged to the species A. veronii. Upon AFLP analysis, on the other hand, the isolates were clearly determined to be A. veronii. According to the results of API analysis, all isolates lacked ornithine decarboxylase activity, indicating that these isolates belonged to A. veronii biovar sobria and not to A. veronii biovar veronii (1).
Production of cytotoxin and hemolysin.
A total of 73% (38 of 52) of the isolates showed cytotoxic activity to EPC cell lines and also produced hemolysin active on rabbit and human erythrocytes (Table 1). All A. veronii biovar sobria isolates from EUS (14 of 14) were positive for cytotoxin and hemolysin, whereas 69% (18 of 26) of the environmental isolates, including 9 of 11 A. veronii biovar sobria isolates, and 50% (6 of 12) of the human diarrheal isolates were also positive.
TABLE 1.
Frequency of putative virulence factors among isolates obtained from fish, from the environment, and from humans with diarrhea in Bangladesh
| Source of isolate | n | No. of isolates (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cytotoxin | Hemolysin | Toxin genea | Agglutination of erythrocytes from:
|
||||||
| Fish | Humans | Rabbits | |||||||
| Fish | 14 | 14 (100) | 14 (100) | 14 (100) | 14 (100) | 14 (100) | 14 (100) | ||
| Environment | 26 | 18 (69) | 18 (69) | 9 (35) | 0 | 14 (54) | 9 (35) | ||
| Human diarrhea | 12 | 6 (50) | 6 (50) | 1 (8) | 0 | 2 (17) | 2 (17) | ||
| Total | 52 | 38 (73) | 38 (73) | 24 (46) | 14 (27) | 30 (58) | 25 (48) | ||
Cytolytic enterotoxin gene and/or extracellular hemolysin gene.
PCR for cytolytic enterotoxin gene and/or extracellular hemolysin gene.
Among the isolates, 46% (24 of 52) isolates were positive in the PCR test for cytolytic enterotoxin gene and/or the extracellular hemolysin gene (Table 1). However, all of the 14 A. veronii biovar sobria isolates from fish were positive both for production of cytotoxin and in the PCR assay, and 7 of 11 environmental A. veronii biovar sobria isolates were also positive (Fig. 1).
Hemagglutination.
Aeromonas isolates from fish, the environment, and humans with diarrhea were tested for the ability to agglutinate fish, human, and rabbit erythrocytes. In all, 25 isolates (48%) from various sources showed agglutination with human and rabbit erythrocytes (Table 1). However, fish erythrocytes were only agglutinated by the EUS-associated fish isolates and not by any of the human or environmental isolates. All of the erythrocyte agglutinations were inhibited by d-mannose but not by d-galactose. The reaction of four of the environmental isolates was inhibited by l-fucose, indicating different mechanisms for the agglutination reactions among the isolates.
Adhesion and invasion.
One representative of the 14 EUS-associated isolates (isolate BDF4) that showed high cytotoxic activity against EPC cells was also tested for adhesion and invasion to fish cell lines. The values for adhesion and invasion experiments were averages from at least five independent experiments. A dose-dependent adhesion was observed, a poor adherence (0.8% ± 0.08%) was observed at 105 CFU ml−1, but increasing concentrations significantly increased the degree of adherence to a maximum of 5.3% ± 0.21% recovered at 108 CFU ml−1. However, at >108 CFU ml−1 the high concentration of bacteria caused a detachment of the cells from the chamber bottom, resulting in a lowered degree of adherence.
The adhesion of the BDF4 isolate was found to be similar in both cell lines, although a better adhesion was observed when bacteria were grown at 22°C compared to bacteria grown at 37°C (Table 2). Isolate BDF4 could also invade both fish cell lines (Table 2). The adhesion and invasion displayed by the strain were 140- and 85-fold greater, respectively, than that shown by the nonadherent E. coli HB101 strain used as control. Thus, the data indicate that the level of adhesion and invasion seemed to depend on the bacterial growth temperature, the concentration of the bacteria, and the cell line employed. Bacterial adhesion was inhibited by 20% ± 1.6% by vigorous stirring and inhibited by 24% ± 1.0% by trypsin treatment, irrespective of the cell line used (data not shown).
TABLE 2.
Adhesion and invasion of Aeromonas sp. strain BDF4 to different cell lines at different temperatures
| Adhesion or adherence | Fish cell line | Temp (°C) | Mean % bacteria ± SEM
|
|
|---|---|---|---|---|
| Aeromonas sp. strain BDF4 | E. coli HB101 (control) | |||
| Adherence | EPC | 22 | 3.1 ± 0.19a | 0.022 ± 0.004 |
| 37 | 2.8 ± 0.21 | NT | ||
| RTG | 22 | 3.0 ± 0.18 | 0.03 ± 0.003 | |
| 37 | 2.8 ± 0.25 | NT | ||
| Invasion | EPC | 22 | 0.27 ± 0.030b | 0.003 ± 0.0004 |
| 37 | 0.24 ± 0.027 | NT | ||
| RTG | 22 | 0.23 ± 0.031 | 0.004 ± 0.0004 | |
| 37 | 0.29 ± 0.030 | NT | ||
Mean percent bacteria (±SEM) recovered after washing. NT, not tested.
Mean percent bacteria recovered after gentamicin treatment.
DISCUSSION
In the present study, several of the Aeromonas isolates examined from various sources (i.e., from EUS in fish, from the environment, and from humans with diarrhea) were able to produce putative virulence factors, as reported earlier (18, 29, 30). It is thus possible that the environment contains reservoirs of Aeromonas strains that are capable of causing human and animal infections. So far, a major problem has been the lack of knowledge concerning the primary virulence factors causing EUS and the identification of the bacterial groups that cause the disease. It is not known, for example, whether these virulence factors are produced by all members of a given taxon or only by one or more groups of pathogenic clones within a given taxon. One major achievement would be the development of an inexpensive but accurate identification and typing methodology to allow the detection of pathogenic Aeromonas clones.
Phenotyping with the PhP system allowed us to detect that the Aeromonas isolates associated with EUS formed a separate and very homogeneous phenotypic cluster. The PhP type of the EUS-associated isolates was not identical to any of the environmental and human diarrheal Aeromonas isolates included in our previous and ongoing studies. We have previously reported that Aeromonas isolates from children with diarrhea, compared to isolates from other sources, showed a low level of diversity, indicating that these diarrheal isolates most probably represent a limited number of clonal groups. Similar to the previous study, wherein a specific PhP type was strongly associated with diarrhea (30), we conclude here that the EUS-associated isolates belong to a clonal group that is possibly pathogenic to fish. We assume that this clonal group carries specific, as-yet-uncharacterized properties that makes it a good colonizer of fish.
Aeromonas spp. have previously been isolated from EUS-diseased fish in the Indo-Pakistan region by Iqbal et al. (16). They found that 27% (12 of 44) Aeromonas isolates from fish with EUS in Malaysia, Thailand, and Bangladesh belonged to A. veronii biovar sobria and that 6 of the 11 isolates from Bangladeshi fish belonged to this species. In agreement with these findings, our EUS-associated fish isolates were also genotypically identified as A. veronii biovar sobria, indicating that this Aeromonas species may constitute an important causative agent of EUS in this geographic area.
We found that the production of cytotoxin and hemolysin was prevalent in more than 50% of the Aeromonas isolates of various species and various origins, as reported by several other investigators (29). However, the EUS-associated isolates were all hemolysin and cytotoxin positive. Furthermore, most of the environmental A. veronii biovar sobria isolates were also positive, which is in agreement with our previous report on environmental isolates in Bangladesh (30).
Recent studies have shown that the virulence genes of Aeromonas spp., e.g., the genes for cytolytic enterotoxin, aerolysin, and hemolysin, have 70 to 99% homology. By using primers from conserved region, it was possible to study the epidemiology of these genes (23). By a similar strategy, we found that all our isolates from EUS were positive for cytolytic enterotoxin and/or extracellular hemolysin genes. As expected, the production of cytotoxin and that of hemolysin were highly correlated, but the PCR assay did not detect all hemolysin- and cytotoxin-positive isolates (Table 1). Thus, of 38 Aeromonas isolates that were positive for hemolysin and cytotoxin production, only 24 possessed the target gene sequence. The prevalence of toxin gene-positive isolates (24 of 52 [46%]) is in agreement with recently published data of Kingombe et al. (23), who reported that of 61 Aeromonas reference isolates tested, only 30 (49%) were positive for the same primer combination (AHCF1-AHCR1). Using a different set of primers, Wang et al. (40) studied 41 isolates exhibiting hemolytic activity, and only 6 of these isolates possessed the target cytotoxin gene.
A majority of the Aeromonas isolates in the present study agglutinated human and rabbit erythrocytes. This finding is in agreement with previous reports by other investigators (17, 37). Interestingly, only the EUS-associated fish isolates could agglutinate fish erythrocytes. It is plausible that a specific ligand-receptor interaction is important for colonization in fish. The agglutination was inhibited by d-mannose, indicating that glycoproteins, such as bacterial cell lectins, may be involved in the adhesion mechanism, a phenomenon that has been described previously (7). The agglutination of erythrocytes by four of the environmental isolates was also inhibited by l-fucose, which indicates that a different adhesive component might be present in these isolates. Since many different agglutination patterns of Aeromonas spp. have been reported (11), these bacteria might have various colonization factors that are antigenically diverse and play important roles in evading the host immune system (26).
In the present study we also demonstrated the presence of adhesion and invasion ability in the EUS-associated isolates that may play a role in the pathogenesis at an early stage in the disease process. The adhesion and invasion appeared to be more pronounced when bacteria were grown at 22°C than when grown at 37°C, a finding that agrees with the observation that adhesive fimbriae are better expressed at 22 than at 37°C. Adhesion was inhibited by trypsin or vigorous stirring, which is in agreement with the observations of Kirov et al. (24) that fimbriae and other filamentous structures might play important roles in colonization.
Since the varieties of fish considered in the present study live in a vast body of water and represent hosts for various opportunistic bacteria and other microorganisms, it may be very difficult to define a single causative agent of EUS. However, our results suggest that the Aeromonas bacteria isolated from EUS in Bangladeshi fish likely belong to a certain clonal group of A. veronii biovar sobria and that this clonal group is not common in other sources, including humans with diarrhea or the environment. Isolates belonging to this clonal group displayed several putative virulence factors, including the unique property of being able to agglutinate fish erythrocytes, and so this clonal group can be considered a possible causative agent of EUS in fish in Bangladesh.
Acknowledgments
This work was supported by SIDA/SAREC grant SWE-1998-353 for research fellowships and the Karolinska Institutet. G.H. is a postdoctoral fellow of the Fund for Scientific Research (F.W.O.-Vlaanderen; Flanders, Belgium).
We thank M. B. R. Chowdhury and H. Kumar Pal at Bangladesh Agriculture University for providing the fish isolates; the staff at International Center for Diarrhea Diseases Research in Bangladesh for technical support; the National Veterinary Institute, Uppsala, Sweden, for providing the fish cell lines; and Lena Guldevall at Karolinska Institutet for laboratory support.
REFERENCES
- 1.Abbott, S., W. K. Cheung, S. K. Bystrom, T. Malekzadeh, and J. M. Janda. 1992. Identification of Aeromonas strains to the genospecies level in the clinical laboratory. J. Clin. Microbiol. 30:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barua, G., A. N. H. Banu, and M. H. Khan. 1991. An investigation into the prevalence of fish disease in Bangladesh during 1988-1989. Bangladesh J. Aquacult. 11-13:27-29. [Google Scholar]
- 3.Blazevic, D. J., and G. M. Ederer. 1975. Principles of biochemical tests in diagnostic microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Carnahan, A. M., S. Behram, and S. W. Joseph. 1991. Aerokey II: a flexible key for identifying clinical Aeromonas species. J. Clin. Microbiol. 29:2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra, A. K., C. W. Houston, J. W. Peterson, and G. F. Jin. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513-523. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhury, M. B. R. 1997. Bacteria involved in fish disease in Bangladesh. International Symposium on Diseases in Marine Aquaculture. Society of Fish Pathology, Hiroshima, Japan.
- 7.Fernandez, A. I. G., M. J. Perez, L. A. Rodriguez, and T. P. Nieto. 1995. Surface phenotypic characteristics and virulence of Spanish isolates of Aeromonas salmonicida after passage through fish. Appl. Environ. Microbiol. 61:2010-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Agriculture Organisation. 1986. Report of the expert consultation on fish diseases in the Asia-Pacific region. Food and Agriculture Organisation, Bangkok, Thailand.
- 9.Gentry, M. K., and J. M. Dalrymple. 1980. Quantative microtiter cytotoxicity assay for shigella toxin. J. Clin. Microbiol. 12:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunilla, K., and R. Möllby. 1979. Adhesion of Escherichia coli to human periurethral cells correlated to mannose-resistant agglutination of human erythrocytes. FEMS Microbiol. Lett. 5:295-299. [Google Scholar]
- 11.Hokama, K., and M. Iwanaga. 1991. Purification and characterization of Aeromonas sobria pili, a possible colonization factor. Infect. Immun. 59:3478-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huys, G., R. Coopman, P. Janssen, and K. Kersters. 1996. High resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int. J. Syst. Bacteriol. 46:572-580. [DOI] [PubMed] [Google Scholar]
- 13.Huys, G., I. Kersters, M. Vancanneyt, R. Coopman, P. Janssen, and K. Kersters. 1995. Diversity of Aeromonas spp. in Flemish drinking water production plants as determined by gas-liquid chromatographic analysis of cellular fatty acid methyl esters (FAMEs). J. Appl. Bacteriol. 78:445-455. [DOI] [PubMed] [Google Scholar]
- 14.Huys, G., M. Vancanneyt, R. Coopman, P. Jansson, E. Falsen, M. Altwegg, and K. Kersters. 1994. Cellular fatty acid composition as a chemotaxonomic marker for the identification of phenospecies and the hybridization groups in the genus Aeromonas. Int. J. Syst. Bacteriol. 44:651-658. [Google Scholar]
- 15.Huys, G., and J. Swings. 1999. Evaluation of a fluorescent amplified fragment length polymorphism (FAFLP) methodology for the genotypic discrimination of Aeromonas taxa. FEMS Microbiol. Lett. 177:83-92. [Google Scholar]
- 16.Iqbal, M. M., K. Tajima, and Y. Ezura. 1998. Phenotypic identification of motile Aeromonas isolated from fishes with epizootic ulcerative syndrome in Southeast Asian countries. Bull. Fac. Fish. Hokkaido Univ. 49:131-141. [Google Scholar]
- 17.Ishiguro, E. E., and T. J. Trust. 1981. Differentiating characteristics of virulent and attenuated strains of Aeromonas salmonicida. Dev. Biol. Stand. 49:163-168. [Google Scholar]
- 18.Janda, J. M. 1991. Recent advances in the study of the taxonomy, pathogenicity, and infectious syndromes associated with the genus Aeromonas. Clin. Microbiol. Rev. 4:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 20.Janda, J. M., and P. S. Duffey. 1988. Mesophilic aeromonads in human disease: current taxonomy, laboratory identification, and infectious disease spectrum. Rev. Infect. Dis. 10:980-997. [DOI] [PubMed] [Google Scholar]
- 21.Janda, J. M., and R. P. Kokka. 1991. The pathogenicity of Aeromonas strains relative to genospecies and phenospecies identification. FEMS Microbiol. Lett. 15:29-33. [DOI] [PubMed] [Google Scholar]
- 22.Kanclerski, K., and R. Mollby. 1987. A simple and exact two-point interpolation method for determination of haemolytic activity in microtiter plates. Acta Pathol. Microbiol. Immunol. Scand. Sect. B 95:175-179. [DOI] [PubMed] [Google Scholar]
- 23.Kingombe, C. I. B., G. Huys, M. Tonolla, M. J. Albert, J. Swings, R. Peduzzi, and T. Jemmi. 1999. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirov, S. M., L. J. Hayward, and M. A. Nerrie. 1995. Adhesion of Aeromonas sp. to cell lines used as models for intestinal adhesion. Epidemiol. Infect. 115:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirov, S. M., J. A. Hudson, L. J. Hayward, and S. J. Mott. 1994. Distribution of Aeromonas hydrophila hybridization groups and the virulence properties in Australasian clinical and environmental strains. Lett. Appl. Microbiol. 18:71-73. [DOI] [PubMed] [Google Scholar]
- 26.Kirov, S. M., and K. Sanderson. 1996. Characterization of a type IV bundle-forming pilus (SFP) from a gastroenteritis-associated strain of Aeromonas veronii biovar sobria. Microb. Pathog. 21:23-34. [DOI] [PubMed] [Google Scholar]
- 27.Kjell, T., K. Horlin, and S. B. Svenson. 1984. Epidemic outbreaks of acute pyelonephritis caused by nosocomial spread of P fimbriated Escherichia coli in children. J. Infect. Dis. 150:728-736. [DOI] [PubMed] [Google Scholar]
- 28.Krovacek, K., F. Ahmed, W. Ahne, and I. Mansson. 1987. Adhesion of Aeromonas hydrophila and Vibrio anguillarum to fish cells and to mucus-coated glass slides. FEMS Microbiol. Lett. 42:85-89. [Google Scholar]
- 29.Krovacek, K., V. Pasquale, S. B. Baloda, V. Soprano, M. Conte, and S. Dumontet. 1994. Comparison of putative virulence factors in Aeromonas hydrophila strains isolated from the marine environment and human diarrheal cases in southern Italy. Appl. Environ. Microbiol. 60:1379-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kühn, I., M. J. Albert, M. Ansaruzzaman, N. A. Bhuiyan, S. A. Alabi, M. S. Islam, P. K. Neogi, G. Huys, P. Janssen, K. Kersters, and R. Mollby. 1997. Characterization of Aeromonas spp. isolated from humans with diarrhea, from healthy controls, and from surface water in Bangladesh. J. Clin. Microbiol. 35:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kühn, I., G. Allestam, G. Huys, P. Janssen, K. Kersters, K. Krovacek, and T. X. Stenstrom. 1997. Diversity, persistence and virulence of Aeromonas strains isolated from drinking water distribution systems in Sweden. Appl. Environ. Microbiol. 63:2708-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kühn, I., T. Lindberg, K. Olsson, and T. A. Stenström. 1992. Biochemical fingerprinting for typing of Aeromonas strains from food and water. Lett. Appl. Bacteriol. 15:261-265. [Google Scholar]
- 33.McGarey, D. J., L. Milanesi, D. P. Foley, B. J. Reyes, L. C. Frye, and D. V. Lim. 1991. The role of motile aeromonads in the fish disease, ulcerative disease syndrome (UDS). Experientia Rev. 47:441-444. [PubMed] [Google Scholar]
- 34.Merino, S., X. Rubires, A. Aguilar, and J. M. Tomas. 1997. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol. Lett. 151:213-217. [DOI] [PubMed] [Google Scholar]
- 35.Möllby, R., I. Kühn, and M. Katouli. 1993. Computerized biochemical fingerprinting: a new tool for typing of bacteria. Rev. Med. Microbiol. 4:231-241. [Google Scholar]
- 36.Monfort, P., and B. Baleux. 1990. Dynamics of Aeromonas hydrophila, Aeromonas sobria, and Aeromonas caviae in a sewage treatment pond. Appl. Environ. Microbiol. 56:1999-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos, Y., I. Bandin, T. P. Nieto, J. L. Barja, and A. E. Toranzo. 1991. Cell-surface-associated properties of fish pathogenic bacteria. J. Aquat. Ann. Health 3:297-301. [Google Scholar]
- 38.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman and Co., New York, N.Y.
- 39.Thune, R. L., L. A. Stanley, and R. K. Cooper. 1993. Pathogenesis of gram-negative bacteria infections in warm-water fish. Fish. Annu. Rev. Fish Dis. 3:37-68. [Google Scholar]
- 40.Wang, G., K. D. Tyler, C. K. Munro, and W. M. Johnson. 1996. Characterization of cytotoxic, hemolytic Aeromonas caviae clinical isolates and their identification by determining the presence of a unique hemolysin gene. J. Clin. Microbiol. 34:3203-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]