Abstract
This paper presents the first report of bacteriophage isolated from commercial vegetable fermentations. Nine phages were isolated from two 90-ton commercial sauerkraut fermentations. These phages were active against fermentation isolates and selected Leuconostoc mesenteroides and Lactobacillus plantarum strains, including a starter culture. Phages were characterized as members of the Siphoviridae and Myoviridae families. All Leuconostoc phages reported previously, primarily of dairy origin, belonged to the Siphoviridae family.
Commercial cabbage fermentations in the United States typically are carried out by epiphytic lactic acid bacteria (LAB), without the benefit of an added starter culture (8). Four species of LAB have been identified as the primary species involved in sauerkraut fermentations: Leuconostoc mesenteroides, Lactobacillus brevis, Pediococcus pentosaceus, and Lactobacillus plantarum (15). Pederson and Albury (12, 13) determined that if the conditions of salt and temperature (2% NaCl and 18°C) are correct, a succession of these species will result in the consistent production of high-quality sauerkraut. For this reason, researchers have concluded that starter cultures are not needed for sauerkraut fermentations (14, 16).
Our interest in studying the microbial ecology of sauerkraut fermentations has developed from the current need of the vegetable fermentation industry to reduce waste chloride production. Low-salt fermentation procedures are currently being developed as a means to reduce the chloride waste for sauerkraut fermentations (Fleming et al., unpublished). These procedures may require the use of starter cultures because lower salt concentrations may not result in the production of high-quality sauerkraut (12).
Bacteriophages active against all major LAB genera have been isolated and characterized (1), including phages of dairy origin active against Lactobacillus, Lactococcus, and Streptococcus species. In contrast, phages infecting other species of lactic acid bacteria have received relatively little attention. To our knowledge, the phages active against Leuconostoc have been characterized as belonging to the Siphoviridae family (1). Phages active against Oenococcus oeni (formerly classified as Leuconostoc oenos) isolated from wine fermentations have also been classified as Siphoviridae (14).
The presence and ecology of bacteriophages in vegetable fermentations remain unexplored. Our objective was to identify bacteriophages in commercial cabbage fermentations, including one inoculated with a Leuconostoc mesenteroides starter culture.
Sauerkraut fermentations were carried out at a commercial processing plant and consisted of one inoculated and one uninoculated tank, both of 90-ton capacity. L. mesenteroides LA10 was used as a starter culture; cells were grown overnight at 30°C in 8 liters of MRS broth containing 1% NaCl. The culture (109 CFU/ml) was then diluted to 76 liters with tap water and sprayed (20 to 40 lb/in2) onto the sliced, salted cabbage as it was being conveyed on a belt to the fermentation tank. This resulted in ≈105 CFU/g of the culture being uniformly distributed in the fermentation tank. This fermentation was carried out with a final (equilibrated) NaCl concentration of 1.0 to 1.2%. The starter culture was used in an attempt to produce a normal fermentation under low-salt conditions.
An uninoculated tank, salted according to the normal industry procedure (equilibrated at 2.25% NaCl), was also sampled for phages. The fermentation temperature was not controlled, but the temperature in these commercial fermentations typically averages 18°C. Brine samples for the isolation of LAB and bacteriophage were obtained with a 1-cm-diameter, perforated stainless steel tube from a depth of ≈60 cm from the tops of the fermentation tanks, about 60 cm from the sides, at the time intervals indicated below. Samples were transported on wet ice by overnight mail in 15-ml plastic screwcap tubes (430791; Corning Inc., Corning, N.Y.) from the commercial facility to our laboratory. Brine samples were refrigerated and processed within 24 h of arrival.
Selected LAB cultures were used as potential phage hosts: Pediococcus acidilactici LA74 (ATCC 33314), Lactobacillus confusus LA277 (NRRL B-1064), L. mesenteroides LA10 (C33), Leuconostoc sp. strain BI116 (4G12, unidentified species), L. mesenteroides LA112 (ATCC 10880), L. plantarum LA280 (KCCM 11322), and L. plantarum LA89. These cultures were obtained from the U.S. Department of Agriculture Agricultural Research Service (USDA-ARS) Food Science Research Unit culture collection (our lab, Raleigh, N.C.). In addition, LAB colonies were isolated directly from brine samples using spread plates with 0.1 ml of brine on MRS agar (Difco Laboratories, Detroit, Mich.) supplemented 0.02% sodium azide (MMRS, to prevent the growth of yeasts [9]). All cultures were incubated at 30°C during growth in the laboratory.
Identification of LAB isolates was carried out by a PCR method (6) based on the length of the 16S-23S rDNA intergenic transcribed spacer (ITS-PCR). Primers G1-16S, GAAGTCGTAACAAGG, which hybridizes to the 3" end of the 16S rRNA gene, and L2-23S, GGGTTTCCCCATTCGGA (Genosys Biotechnologies Inc., The Woodlands, Tex.), complementary to the 5" end of the 23S rRNA gene, were used to amplify the spacer region. PCR products were separated on a 1.5-mm-thick 5% polyacrylamide (Sigma Chemical Co., St. Louis, Mo.) gel using a vertical gel electrophoresis box (BRL model 16; Life Technologies, Inc., Gaithersburg, Md.) and compared to an existing database of banding patterns (6; F. Breidt, unpublished).
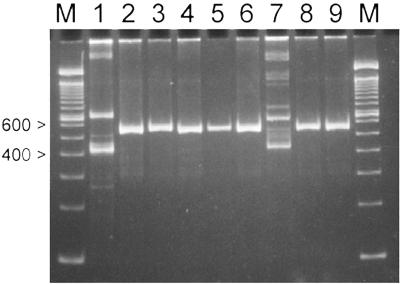
Five strains, designated LA291 through LA295, were isolated from the inoculated fermentation, and one strain, LA296, was isolated from the uninoculated fermentation. The ITS-PCR banding patterns shown in Fig. 1 indicated that the five LAB (LA291 to LA295) were Leuconostoc species, and one isolate from the control fermentation (LA296) was a Lactobacillus. All LAB isolates exhibited gas formation via heterofermentative metabolism during growth for 48 h at 30°C in 5 ml of MRS broth containing a Durham tube (Labsource, Chicago, Ill.).
FIG. 1.
Strain identification by ITS-PCR products of LAB isolates from sauerkraut fermentation. Lane 1, L. plantarum LA280; lane 2, L. mesenteroides LA10; lane 3, Leuconostoc sp. strain BI116; lane 4, Leuconostoc sp. strain LA291; lane 5, Leuconostoc sp. strain LA292; lane 6, Leuconostoc sp. strain LA294; lane 7, Lactobacillus sp. strain LA296; lane 8, Leuconostoc sp. strain LA295; lane 9, Leuconostoc sp. strain LA293; M, molecular weight markers (100-bp DNA ladder).
Bacteriophages were isolated from the fermentation brine by centrifugation in 15-ml plastic screwcap tubes (Corning) at 3,000 × g for 10 min (Sorvall model RC5-B, with a SS-34 rotor; Sorvall Products, Newtown, Conn.) to remove suspended matter. Supernatant (0.1 ml) was added to 10 ml of MRS broth along with 0.1 ml of 1 M CaCl2 (Sigma) and 0.5 ml of an early-log-phase culture of the host LAB, with an optical density of 0.3 to 0.4 (corresponding to approximately 107 CFU/ml). After overnight incubation at 30°C, the samples were centrifuged (as above) and the supernatant (phage lysate) was filtered using 0.45-μm filters (Pall Corp., Ann Arbor, Mich.). The phage-containing filtrate (approximately 9 ml) was treated with 0.1 ml of chloroform (Sigma) and stored at 4°C. The phage lysate samples (0.1 ml) were mixed with 3 ml of soft agar (warmed to 50°C), 0.1 ml of 100 mM CaCl2, and 0.1 ml of a mid-log culture of the cells and poured onto MRS agar plates. Plates were incubated overnight at 30°C for plaque assays (2).
Host strains for the initial phage isolation were identified by using the brine supernatant (described above) in a plaque assay with cycloheximide (50 μg/ml; Sigma) added to the soft agar in order to suppress yeast growth. All bacteriophage isolates were repurified using a single plaque from a plaque assay of the filtered lysate prior to further characterization.
Bacteriophages from the inoculated and uninoculated tanks are shown in Table 1. Phages active against the LA10 starter culture were present at greater than 103 PFU/ml in the initial enrichment broths during the first 3 days after the start of the fermentation in the inoculated tank. Interestingly, phages active against the starter culture, L. mesenteroides LA10, were also isolated from the day 2 and 3 samples of the uninoculated fermentation, though at lower numbers after enrichment. Viable phages were isolated from the fermentation tanks over a range of pH values (from 5.6 to 3.5) for up to 21 days after the start of the fermentation, although most phages were recovered in the first 3 days, including the selected isolates Y4, Y5, Y10, R1, R2, and R3. Other phages selected for further characterization, phages Y11, Y12, and Y20, were isolated from day 7.
TABLE 1.
Bacteriophage assay of brine samples from fermenting sauerkraut
| Timea (days) | Tank pH
|
Activity against selected hostc
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control tank
|
Inoculated tank
|
|||||||||||||
| Control | Inoculatedb | LA74 | LA277 | LA280 | LA10 | BI116 | LA74 | LA277 | LA280 | LA10 | BI116 | |||
| 1 | 5.6 | 5.4 | − | − | − | − | − | − | − | − | +++ | − | ||
| 2 | 5 | 4.3 | − | − | − | + | − | − | − | − | +++ | − | ||
| 3 | 4.3 | 4.2 | − | − | + | + | − | − | − | ++ | +++ | − | ||
| 7 | 3.8 | 3.8 | − | − | + | − | +++ | − | − | ++ | + | ++ | ||
| 14 | 3.5 | 3.7 | − | − | + | − | ++ | − | − | +++ | − | +++ | ||
| 21 | 3.5 | 3.7 | − | − | + | − | + | − | − | +++ | − | − | ||
| 28 | 3.4 | 3.6 | − | − | − | − | − | − | − | − | − | − | ||
Elapsed time from the start of the fermentation.
Inoculated with LA10.
+++, above 300 PFU/plate; ++, 100 to 200 PFU/plate; +, under 100 PFU/plate; −, no plaques. LA74, Pediococcus acidilactici; LA277, Lactobacillus confusus; LA280, Lactobacillus plantarum; LA10, Leuconostoc mesenterodes; BI116, Leuconostoc species (unidentified).
To generate high-titer phage lysates for electron microscopy, 1 ml of the phage lysate was transferred aseptically to 10 ml of the mid-log host cell culture (OD600 = 0.3) in MRS broth containing 10 mM CaCl2. The broth culture was incubated at 30°C until clearing occurred (typically 3 to 4 h). The phage supernatant was collected by centrifugation at 3,000 × g for 10 min in an SS-34 rotor (Sorvall). The fresh lysate (10 ml) was added to 100 ml of a mid-log culture containing 10 mM CaCl2 and incubated at 30°C until clearing occurred. The phage supernatant was again collected by centrifugation (Sorvall GSA rotor, 20 min, 8,000 rpm) and then filter-sterilized using a 0.45-μm bottle filter (Corning). NaCl and polyethylene glycol were added to the high-titer phage lysate at 2.9% and 10%, respectively, and incubated for 15 h at 4°C.
Phages were concentrated by centrifugation (8,000, 20 min, GSA rotor; Sorvall), and the phage pellet was resuspended in 2 ml of TE buffer (100 mM Tris [pH 7.6], 50 mM EDTA; Sigma). The 2-ml concentrated phage suspension was overlaid onto a three-step CsCl gradient containing 1 ml of 1.7-g/ml CsCl (molecular biology gradep; Sigma), 1 ml of 1.50-g/ml CsCl, and 1 ml of 1.40-g/ml CsCl in a 5-ml ultracentrifuge tube (Sorvall). Phage were centrifuged for 7 h at 600,000 × g in a mini-ultracentrifuge (RC-M150 GX; Sorvall). Phage-containing bands (blue-gray) were extracted through the wall of the centrifuge, and the CsCl was removed by dialysis (6,000- to 8,000-Da membrane; Baxter Diagnostics Inc., McGaw Park, Ill.) for 15 h with three changes of deionized water. The concentrated phages were negatively stained with 2% uranyl acetate (pH 4.0).
Electron microscopy was carried out using a Jeol model 100S electron microscope (Jeol USA, Peabody, Mass.), at 80 kV. Electron micrographs were taken at a magnification of 50,000× and printed at 80,000× (Valerie Knowlton, Center for Electron Microscopy, NC State University, Raleigh, N.C.). Bacteriophages were identified using the morphological criteria outlined by the International Committee of Taxonomy of Viruses (4, 12) and Ackermann (1).
The host ranges for these phages are shown in Table 2. Only phages R1 and R2 showed identical host ranges and could not be distinguished. These studies were carried out by spotting 5 μl of serial dilutions of the phage lysates on a lawn of the indicated host strain until individual plaques were visible. To ensure that false-positive results were not recorded, positive results were subsequently confirmed by plaque assay. Bacterial isolates LA291 to LA295 all showed unique patterns of infection by the bacteriophages, indicating that the isolates represented different bacterial strains. These isolates appear to be the same genus, however, because ITS-PCR banding patterns were identical. Further characterization of these bacterial isolates will be the subject of future work.
TABLE 2.
Host range of bacteriophages isolated from fermenting sauerkraut
| Host strain | Plaques formeda with bacteriophage:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Y1b | Y4 | Y5 | Y10 | Y11 | Y12 | Y20 | R1 | R2 | R3 | |
| Leuconostoc mesenteroides LA10c | − | − | − | − | − | − | − | − | − | − |
| Leuconostoc mesenteroides LA112 | − | +d | − | − | − | − | − | − | − | − |
| Lactobacillus plantarum LA280 | +d | − | − | − | − | − | − | − | − | − |
| Lactobacillus plantarum LA89 | + | − | − | − | − | − | − | − | − | − |
| Lactobacillus sp. LA296e | − | − | − | − | − | − | +d | − | − | − |
| Leuconostoc sp. strains | ||||||||||
| BI116 | − | − | +d | + | − | + | − | + | + | + |
| LA291f | − | − | + | +d | − | + | − | + | + | − |
| LA292 | − | − | ± | − | +d | + | − | − | − | − |
| LA293 | − | − | − | − | − | − | − | − | − | +d |
| LA294 | − | − | + | + | + | +d | − | + | + | − |
| LA295 | − | − | − | − | − | − | − | +d | +d | + |
+, clear plaques; ±, turbid plaques; −, no lysis.
Phage isolated from kimchi fermentation.
LA10 was the starter culture for the inoculated tank.
Initial host for the phage isolate.
LA296 was isolated from the control fermentation tank.
LA291 to LA295 were isolated from the inoculated tank.
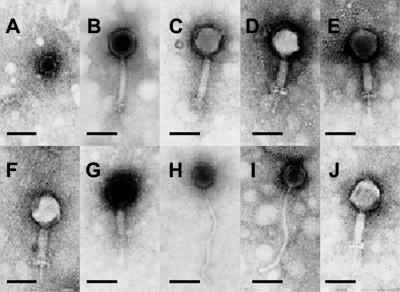
All phages isolated in this study could be assigned to the order Caudovirales and three virus families, Myoviridae, Siphoviridae, and Podoviridae, based on electron micrograph data (Fig. 2). All phages exhibited icosahedral head structures. Phages belonging to the Myoviridae family were further divided into subcategories, morphotype 1 and morphotype 2, on the basis of tail length, base plates, and terminal spikes. It is possible that the two Myoviridae morphotypes represent contracted or noncontracted tail structures for these phages. The reported sizes represent the means from the measure of nine more phages (Table 3).
FIG. 2.
Electron micrographs of bacteriophage particles photographed at a magnification of 80,000×. A, Y1; B, Y4; C, Y5; D, Y10; E, Y11; F, Y12; G, Y20; H, R1; I, R2; J, R3. Bars, 100 nm.
TABLE 3.
Morphological characteristics of bacteriophage isolatesa
| Family | Phages | Head diam (nm) | Tail
|
|
|---|---|---|---|---|
| Length (nm) | Width (nm) | |||
| Myoviridae 1 | Y4, Y5 | 89 (5) | 166 (25) | 20 (5) |
| Myoviridae 2 | Y10, Y11, Y12, R3, Y20 | 94 (6) | 118 (13) | 28 (4) |
| Siphoviridae | R1, R2 | 73 (4) | 348 (20) | 13 (2) |
The reported measurements represent the mean values determined for each group, followed by the standard deviation in parentheses. Myoviridae morphotypes are described in the text.
Within the morphotype 2 group, digestion of phage DNA with EcoRI and KpnI gave distinct banding patterns on agarose gels for these five phages (data not shown). Prior to this work, all reported Leuconostoc phages have been identified as Siphoviridae, and phages R1 and R2 were characterized as Siphoviridae, group B of Bradley's classification (3). Phage Y1 was characterized as Podoviridae, with a short or absent tail structure. This phage was isolated from kimchi, a Korean fermented cabbage product, in a previous study (16).
The burst size and latent period were determined for selected phages on the host used for primary isolation by a modification of the method of Ellis and Delbruck (7). Bacteriophages and the indicated mid-log host culture, at a multiplicity of infection between 0.05 and 0.1, were incubated for 10 min at 30°C. The phage-host suspension was diluted with 10 volumes of MRS, and cells were harvested by centrifugation and resuspended in 10 ml of fresh MRS for continued incubation. The titer was periodically determined by plaque assay. All assays were carried out with two or more replications. Burst size was calculated as the ratio of the final number of phage divided by the initial number of infected cells (equivalent to the initial phage titer). The latent period was determined as the time between challenge with phage and the initial rise in phage titer in the MRS broth.
The phage isolates exhibited a range of burst sizes from 11 ± 0 PFU/cell for phage Y5 to 74 ±10 PFU/cell for phage Y20. The latent periods for these phages were 46 ± 1 min and 19 ± 2 min, respectively, for Y5 and Y20. The other bacteriophage isolates were within these ranges for burst size and latent period with the exception of phage Y4, active against the L. mesenteroides starter culture, which had a burst size of greater than 200 PFU/cell, and phage Y11, which had an eclipse phase of 1.5 h (data not shown).
Previous research with starter cultures for sauerkraut from our laboratory (5, 10) has shown that LAB starter cultures may significantly influence the microbial ecology of small-scale laboratory fermentations. In the commercial-scale fermentations, we found that bacteriophages active against the starter culture, L. mesenteroides LA10, were present in brine samples from the first day after inoculation. In this study, it was not possible to follow the growth or population changes of the unmarked starter culture in the inoculated commercial fermentation due to the naturally present Leuconostoc strains. Further research to determine the effects of bacteriophages on starter cultures in low-salt fermentations is suggested by this study.
To our knowledge, these experiments represent the first isolation and characterization of phages from a commercial vegetable fermentation. The diversity of phage species found in the two commercial sauerkraut fermentations indicates that phages may play a significant role in the ecology of this natural fermentation. Additional studies are forthcoming to fully characterize these phage species and the impact of these phages on the LAB present in vegetable fermentations.
Acknowledgments
This investigation was supported in part by a research grant from Pickle Packers International, Inc., St. Charles, Ill.; NRICGP grant no. 97-35503-4368; and NCAES project no. NC06369.
We thank Evelyn Durmaz for technical assistance with phage purification and propagation procedures, Valerie Knowlton of the NC State University Electron Microscopy Center, and Dean Toler for excellent secretarial assistance. We also thank Bush Brothers & Company (Shiocton, Wis.) for cooperation on this project and especially King Pharr, Ron Lempke, Dorothy Curtiss, and other personnel of the company who assisted in our efforts.
Footnotes
Paper no. FSR0038 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh, NC 27695-7624.
REFERENCES
- 1.Ackermann, H.-W. 1996. Frequency of morphological phage descriptions in 1995. Arch. Virol. 141:209-218. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. H. 1959. Bacteriophages. Interscience Publishers, Inc., New York, N.Y.
- 3.Bradley, D. E. 1967. Ultrastructure of bacteriophages and bacteriocins. Bacteriol. Rev. 31:230-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., Jr., S. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomic differentations of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 5.Breidt, F., Jr., K. A. Crowley, and H. P. Fleming. 1995. Controlling cabbage fermentations with nisin and nisin-resistant Leuconostoc mesenteroides. Food Microbiol. 12:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breidt, F., Jr., and H. P. Fleming. 1996. Identification of lactic acid bacteria by ribotyping. J. Rapid Methods Autom. Microbiol. 4:219-233. [Google Scholar]
- 7.Ellis, E. L., and M. Delbruck. 1939. The growth of bacteriophage. J. Gen. Physiol. 22:365-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming, H. P., K. H. Kyung, and F. Breidt. 1995. Vegetable fermentations, p. 629-661. In H.-J. Rehm and G. Reed (ed.), Biotechnology, 2nd ed. VCH Publishers, Inc., New York, N.Y.
- 9.Fleming, H. P., R. F. McFeeters, and M. A. Daeschel. 1992. Fermented and acidified vegetables, p. 929-935. In C. Vanderzant and D. F. Splittstoesser (ed.), Compendium of methods for the microbiological examination of foods, 3rd ed. American Public Health Association, Washington, D.C.
- 10.Harris, L. J., H. P. Fleming, and T. R. Klaenhammer. 1992. Novel paired starter culture system for sauerkraut, consisting of a nisin-resistant Leuconostoc mesenteroides strain and a nisin-producing Lactococcus lactis strain. Appl. Environ. Microbiol. 58:1484-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. M. Martelli, and M. A. Mayo. 1995. Virus taxonomy: classification and nomenclature of viruses. Springer Verlag, Vienna, Austria.
- 12.Pederson, C. S., and M. N. Albury. 1954. The influence of salt and temperature on the microflora of sauerkraut fermentation. Food Technol. 8:1-5. [Google Scholar]
- 13.Pederson, C. S., and M. N. Albury. 1969. The sauerkraut fermentation. New York State Agric. Exp. Stn. Tech. Bull. 824. New York State Agricultural Experiment Station, Geneva, N.Y.
- 14.Poblet-Icart, M., A. Boudons, and A. Lonvaud-Funel. 1998. Lysogeny of Oenococcus oeni (syn. Leuconostoc oenos) and study of their induced bacteriophages. Curr. Microbiol. 36:365-369. [DOI] [PubMed] [Google Scholar]
- 15.Stamer, J. R. 1983. Lactic acid fermentation of cabbage and cucumbers, p. 365-378. In H. J. Rehm and G. Reed (ed.), Biotechnology. Verlag Chemie, Weinheim, Germany.
- 16.Yoon, S.-S., J.-W. Kim, F. Breidt, Jr., and H. P. Fleming. 2001. Characterization of a lytic Lactobacillus plantarum bacteriophage and molecular cloning of a lysin gene in Escherichia coli. Int. J. Food Microbiol. 65:63-74. [DOI] [PubMed] [Google Scholar]