Abstract
Antisense RNA complementary to a putative helicase gene (hel3.1) of a cos-type Streptococcus thermophilus bacteriophage was used to impede the proliferation of a number of cos-type S. thermophilus bacteriophages and one pac-type bacteriophage. The putative helicase gene is a component of the Sfi21-type DNA replication module, which is found in a majority of the S. thermophilus bacteriophages of industrial importance. All bacteriophages that strongly hybridized a 689-bp internal hel3.1 probe were sensitive to the expression of antisense hel3.1 RNA. A 40 to 70% reduction in efficiency of plaquing (EOP) was consistently observed, with a concomitant decrease in plaque size relative to that of the S. thermophilus parental strain. When progeny were released, the burst size was reduced. Growth curves of S. thermophilus NCK1125, in the presence of variable levels of bacteriophage κ3, showed that antisense hel3.1 conferred protection, even at a multiplicity of infection of approximately 1.0. When the hel3.1 antisense RNA cassette was expressed in cis from the κ3-derived phage-encoded resistance (PER) plasmid pTRK690::ori3.1, the EOP for bacteriophages sensitive to PER and antisense targeting was reduced to between 10−7 and 10−8, beyond the resistance conferred by the PER element alone (less than 10−6). These results illustrate the first successful applications of antisense RNA and explosive delivery of antisense RNA to inhibit the proliferation of S. thermophilus bacteriophages.
Lactic acid bacteria represent a heterogeneous family of nondifferentiating, gram-positive eubacteria that derive metabolic energy from the fermentation of carbohydrates to lactate. The dairy industry has utilized extensively species from the genera Lactococcus, Lactobacillus, and Streptococcus, as starter cultures or culture adjuncts for use in the manufacture of a variety of fermented dairy products. Bacteriophages specific for dairy starter cultures, notably lactococci and, recently Streptococcus thermophilus, have long been recognized as a significant problem for the dairy industry. The problem became more severe as cheese plants increased in size and product throughput became more mechanized (4). Pasteurized milk and lysogenic starter cultures serve as continuous reservoirs for virulent bacteriophages capable of disrupting product manufacture (6, 33). Loss of fermentative capacity associated with starter culture lysis can significantly retard or halt batch fermentations, thereby causing significant losses of time and production capital to the dairy industry each year.
In recent years, an increased incidence of bacteriophage-related problems have been observed for S. thermophilus strains, which are an essential component of starter systems for yogurt and Italian cheese varieties. Our understanding of S. thermophilus bacteriophages has progressed rapidly since the release of six bacteriophage whole-genome sequences: those for DT1 (42), φO1205 (40), Sfi11 (24), Sfi19 (25), Sfi21 (25), and φ7201 (41). In contrast to bacteriophages that infect Lactococcus species, S. thermophilus bacteriophages are closely related, at both the genetic and morphological levels, making differentiation difficult. Electron microscopy studies revealed that S. thermophilus bacteriophages, both temperate and lytic, are nearly identical and that all belong to the Siphoviridae family (morphotype B1), having small isometric heads, long noncontractile tails, and genomes comprised of double-stranded DNA (31). It has recently been shown that S. thermophilus strains are attacked by two groups of highly related bacteriophages that differ in their mechanism of genome encapsidation: cos type and pac type (23). These bacteriophages can be identified and differentiated by examination of their distinct capsid protein profiles or through the detection of the genes that encode those structural proteins (23).
Studies on native bacteriophage defense systems remain limited. S. thermophilus strains have been found to posses both chromosomal and plasmid-borne restriction and modification (R-M) systems (2, 15, 37, 38, 39). At this juncture, however, these native R-M systems have not yet been exploited as means of augmenting the intrinsic level of resistance of industrial starter strains. The plasmid-borne expression of LlaDCHI (formerly LlaII), a heterologous R-M system derived from Lactococcus lactis, does confer broad-range bacteriophage resistance in various strains of S. thermophilus (32). In addition, efforts to construct strains of S. thermophilus with passive resistance properties are also under way. Lucchini et al. (27) recently described the use of pG+host9::ISS1-mediated insertional mutagenesis to identify four distinct host-encoded loci involved in bacteriophage sensitivity. A putative transmembrane protein (Orf394) was discovered and proposed to be functionally analogous to the lactococcal Pip protein, which is essential for infection of L. lactis by c2-type bacteriophages (13).
The application of molecular biology and modern functional genomics is accelerating the development of novel bacteriophage resistance mechanisms through genetic engineering. Sequence analysis can be used to interpret the encoded genetic information, determine genetic features that are common to a variety of bacteriophages, and target potentially sensitive events in bacteriophage development. Recently, comparative genomic analyses have revealed that the genomes of S. thermophilus bacteriophages are molecular mosaics assembled upon a relatively simple scaffold consisting of four independently evolving segments (5, 26). The bacteriophage genome replication functions are clustered on the same genomic segment. To date, two distinct DNA replication modules have been identified among the various S. thermophilus bacteriophages: Sfi21 type and φ7201 type. Hybridization studies have demonstrated that the Sfi21-type module, which is present in five of the six completely sequenced bacteriophages (i.e., DT1, φO1205, Sfi11, Sfi19, and Sfi21), is also present in a majority of industrial bacteriophage isolates (3, 8). Computational analysis of the Sfi21-type replication module predicts a single origin of DNA replication (ori) and several open reading frames that encode a putative helicase, a putative primase, and a number of other proteins of undetermined function. This module is highly conserved at the nucleotide level, suggesting a recent acquisition via horizontal gene transfer followed by rapid dissemination (5). The replication module from bacteriophage φ7201, on the other hand, includes two distinct oris and encodes a probable single-stranded DNA binding protein, a putative replication protein, a putative DnaC homologue, and a number of other proteins of undetermined function (41).
Origin-conferred phage-encoded resistance (PER) was first reported to be effective in L. lactis (16, 28), and has since been demonstrated to be effective in S. thermophilus (12, 41). When a bacteriophage ori is provided in trans on a recombinant plasmid, the origin acts as a molecular decoy that competes for and titrates away bacteriophage-specific replication factors. The result is a reduction in the number of bacteriophage genomes replicated and a dramatic increase in plasmid copy number. The efficacy of PER is bacteriophage specific, and the conferred level of resistance correlates with the copy number of the false origin presented in trans (29, 34). Further, when an antisense RNA expression cassette was linked, in cis, to a PER vector, the result was the expression of an explosive dose of antisense RNA that effectively inhibited a lactococcal bacteriophage (43).
Genome replication functions in S. thermophilus bacteriophages are perhaps the most obvious targets for gene silencing by antisense RNA, since they are highly conserved among industrial bacteriophages and expressed early and transiently during the lytic life cycle. Recently, antisense RNAs, which were designed to target genes involved in DNA replication, have been found to be extremely effective at inhibiting a number of related lactococcal bacteriophages (29). In this study, the expression of a putative helicase, which is a component of the highly conserved Sfi21-type DNA replication module, was targeted for disruption by antisense RNA.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. All bacterial stocks were maintained at −80°C in fresh culture medium supplemented with 10% glycerol. All bacteriological media and components were purchased from Difco Laboratories (Detroit, Mich.). Escherichia coli strains were grown at 37°C with constant aeration in Luria-Bertani broth. Unless otherwise indicated, S. thermophilus derivatives were propagated aerobically at 42°C in Elliker broth supplemented with 1% (wt/vol) beef extract (Elliker-B). Chloramphenicol was added at 2.5 μg/ml for both E. coli and S. thermophilus when appropriate. For solid media, Bacto Agar was added at final concentrations of 1.5% (wt/vol) for base agar and 0.75% (wt/vol) for soft agar.
TABLE 1.
Bacterial strains, bacteriophages, and plasmids
| Bacterial strain, bacteriophage, or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| S. thermophilus strains | ||
| NCK1434 | Industrial isolate; sensitive to κ1; Cm2 | This study |
| NCK1125 | Industrial isolate; sensitive to κ3, κ4, κ5, κ6, κ9, κ10, and κ12; Cms | This study |
| NCK1435 | Industrial isolate; sensitive to κ2; Cms | This study |
| NCK1124 | Industrial isolate; sensitive to κ13; Cms | This study |
| SMQ495 | Industrial isolate; sensitive to DT1; Cms | 42 |
| E. coli MC1061 | Transformation host | 18 |
| Bacteriophages | ||
| κ1 | pac-type bacteriophage | This study |
| κ2 | No information available | This study |
| κ3 | cos-type bacteriophage | This study |
| κ4 | cos-type bacteriophage | This study |
| κ5 | cos-type bacteriophage | This study |
| κ6 | pac-type bacteriophage | This study |
| κ9 | cos-type bacteriophage | This study |
| κ10 | cos-type bacteriophage | This study |
| κ12 | pac-type bacteriophage | This study |
| κ13 | No information available | This study |
| DT1 | cos-type bacteriophage | 42 |
| Plasmids | ||
| pNZ123 | 2.8 kb; high-copy-number shuttle vector; Cmr | 9 |
| pTRK686 | 2.4 kb; deletion derivative of pNZ123; Cmr | This study |
| pTRK687 | 3.0 kb; pTRK686 containing the high-expression P6 promoter | This study |
| pTRK688::hel3.1-S | 4.5 kb; pTRK687 containing 1.4-kb sense hel3.1 cassette | This study |
| pTRK689::hel3.1-AS | 4.5 kb; pTRK687 containing 1.4-kb antisense hel3.1 cassette | This study |
| pTRK690::ori3.1 | 3.7 kb; pTRK687 containing 0.7-kb ori3.1 cassette; Per+ | This study |
| pTRK691::ori3.1::hel3.1-AS | 5.1 kb; pTRK690 containing 1.4-kb antisense hel3.1 cassette | This study |
Abbreviations: Cmr, resistant to chloramphenicol; Cms, sensitive to chloramphenicol; Per+, origin-conferred phage-encoded resistance.
Plasmids, bacteriophages, and propagation assays.
The plasmids and bacteriophages used in this study are listed in Table 1. Bacteriophages described in this study were isolated from mozzarella whey. Bacteriophages were propagated in Elliker-B broth supplemented with 10 mM CaCl2 (Elliker-BC) at 42°C and diluted in 0.1× Elliker-B broth supplemented with 10 mM CaCl2. For plaque assays, 20 ± 1 ml of M17-G base agar supplemented with 10 mM CaCl2 was dispensed using a Bellco Biotechnology (Vineland, N.J.) automatic medium dispenser to limit volume-dependant variations in plaque size and incubated aerobically at 37°C for 18 h prior to analysis. The efficiency of plaquing (EOP) was calculated by dividing the bacteriophage titer, in PFU per milliliter, of the test strain by the bacteriophage titer of the parental strain. Bacteriophages were characterized as cos- or pac-type bacteriophages as described by Le Marrec et al. (23). Lysis-in-broth assays were performed in Elliker-BC medium as described previously, except that samples were taken every 15 min for a period of 4 h (41).
Enzymes and reagents.
Restriction enzymes, Taq DNA polymerase, and deoxynucleoside triphosphates were obtained from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). T4 DNA ligase, the SuperScript reverse transcription kit, and DNA molecular weight markers were obtained from Gibco-BRL Life Technologies, Inc. (Gaithersburg, Md.). Pwo DNA polymerase was obtained from Roche (Indianapolis, Ind.), and all enzymes were used according to the manufacturer's specifications. All other chemicals were of analytical grade and obtained from Sigma Chemical Company (St. Louis, Mo.).
Bacterial transformation.
All electroporations were performed using a Gene Pulser (Bio-Rad Laboratories, Hercules, Calif.) apparatus configured to 25 μF, 2.5 kV, and 200 Ω. Preparation of electrocompetent E. coli strain MC1061 (18) was conducted as described by Sambrook et al. (36), and electrocompetent S. thermophilus strains were prepared by a method modified from that of Holo and Nes (17). A stationary-phase culture of S. thermophilus was diluted 100-fold into 42°C Elliker-B broth, incubated at 42°C, and allowed to reach an optical density at 600 nm of 0.2 prior to the addition of 1/10 volume of 42°C 15% (wt/vol) glycine and 1/10 volume of 42°C 2× Elliker-B broth. Cells were harvested by centrifugation at an optical density at 600 nm of between 0.6 and 0.8, washed three times with 2 volumes of sterile deionized water, washed once with 2 volumes of SG buffer (0.5 M sucrose and 10% glycerol), resuspended in 0.003 volume of SG buffer, and incubated on ice prior to use. Plasmid DNA (1 μg) was mixed with 40 μl of cells in a chilled Gene Pulser 0.2-cm cuvette. Following electroporation, cells were immediately resuspended in 960 μl of recovery medium (Elliker-B broth supplemented with 20 mM MgCl2 and 2 mM CaCl2) and incubated for 2 h at 42°C, before being spread onto Elliker-B base agar supplemented with chloramphenicol (5.0 μg/ml).
Preparation of plasmid and genomic DNAs.
Small-scale preparations of plasmid DNA were isolated from E. coli (36) and S. thermophilus (35) as described previously. Large-scale preparations of plasmid DNA were isolated using a Midi Kit (Qiagen, Chatsworth, Calif.) according to the manufacturer's instructions. PCR products were purified by using the Qiagen PCR Purification Kit prior to further manipulation. When required, DNA was extracted from agarose gels by using the QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer's instructions.
Southern hybridizations.
Bacteriophage genomic DNA was isolated using the Qiagen Lambda Kit according to the manufacturer's instructions. Alkaline transfer of HindIII-digested genomic DNA fragments from electrophoresed 0.8% agarose gels to 0.45-μm-pore-size Magnacharge nylon membranes (Micron Separations, Inc., Westborough, Mass.) was performed as described by Sambrook et al. (36). A 689-bp internal hel3.1 fragment was amplified by PCR in the presence of digoxigenin (DIG)-11-UTP (Roche Molecular Biochemicals) using primers JMSp4 and JMSp5 (Table 2) and was used as a hybridization probe. Southern hybridizations (with 30% formamide and at 42°C) were performed using the Roche Molecular Biochemicals DIG-based nonradioactive nucleic acid labeling and detection system according to the manufacturer's instructions.
TABLE 2.
Primers used in this study
| Primer name | Nucleic acid sequence (5" nucleotide position)a |
|---|---|
| JMSp1 | 5"-AAACTGCAGGCTTGCAAGATTGAAGACC-3" (25911) |
| JMSp2 | 5"-AAACTGCAGCCGTCTTTGATAGATCCG-3" (27341) |
| JMSp3 | 5"-GGAGCGTGATTTTTATGG-3" |
| JMSp4 | 5"-GTTAAAGCTAAGACCTACC-3" (26275) |
| JMSp5 | 5"-CCCTTTAGTGACCATTCACGG-3" (26963) |
| JMSp6 | 5"-GGAATTCCAGTTAGGTTCTTGTGG-3" (29810) |
| JMSp7 | 5"-GGAATTCCCCATAATCTTCGTCGGTCC-3" (30486) |
PstI (5"-CTGCAG-3") and EcoRI (5"-GAATTC-3") restriction sites are underlined. The position of the 5" nucleotide (in boldface) relative to the DT1 genomic sequence is indicated when appropriate.
PCR and DNA sequencing.
PCR was performed in a Hybaid Ltd. (Middlesex, United Kingdom) PCR Express thermal cycler with either Taq or Pwo DNA polymerase. DNA primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). When appropriate, restriction endonuclease recognition sites were incorporated into the 5" ends of DNA primers to facilitate the cloning of PCR products. Primers utilized in this study are listed in Table 2. Cycle sequencing reactions and DNA sequence determination were performed by the University of California-Davis automated DNA sequencing facility, using an ABI Prism 377 DNA sequencer with a 96-lane upgrade (Applied Biosystems, Foster City, Calif.). DNA sequences were analyzed using the Genetics Computer Group, Inc. (Madison, Wis.) sequence analysis package version 10.0 and Clone Manager version 6.0 (Scientific and Educational Software, Durham, N.C.). Searches for nucleic acid (BLASTN) and protein (BLASTX) homology were performed with the basic local alignment search tool (1), using the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/).
RNA isolation and strand-specific RT-PCR.
RNA was isolated from S. thermophilus at various times during the bacteriophage infection cycle by using the TRIzol reagent (Gibco-BRL) according to the procedure described by Dinsmore and Klaenhammer (10). Strand-specific reverse transcription-PCR (RT-PCR) was performed using the SuperScript reverse transcriptase kit (Gibco-BRL).
Nucleotide sequence accession numbers.
The DNA sequences for the κ3-derived putative helicase (hel3.1), κ3-derived origin of replication (ori3.1) region, and bacteriophage DT1 are available through GenBank under nucleotide sequence accession numbers AF442520, AF442521, and AF085222, respectively.
RESULTS
Amplification of hel-containing fragments from S. thermophilus bacteriophages.
The nucleic acid sequences for the DNA replication modules of bacteriophages DT1 (GenBank accession number NC_002072), φO1205 (U88974), Sfi11 (NC_002214), Sfi18 (AF158601), Sfi19 (NC_000871), and Sfi21 (NC_000872) were aligned, and a consensus sequence was generated (data not shown). PstI-tagged primers JMSp1 and JMSp2, designed upstream and downstream of the consensus putative helicase gene, respectively, were used to amplify the putative helicase genes (hel) from the S. thermophilus bacteriophages listed in Table 1, using DT1 genomic DNA as a positive control. The expected 1.4-kb fragment, which contained the upstream putative ribosome binding site (RBS), was successfully amplified from DT1, κ1, κ3, κ4, κ5, κ9, and κ10 but failed to be amplified from κ2, κ6, κ12, and κ13. These results indicated that the local syntony and nucleotide sequence proximal to the nested primer sites are highly conserved among the cos- and pac-type bacteriophages that encode the Sfi21-type DNA replication module.
Sequence and phylogenetic analyses of the κ3-derived hel3.1 cassette.
The 1,431-bp PCR fragment amplified from bacteriophage κ3 was sequenced and revealed a single open reading frame of 1,331 bp (GenBank accession number AF442520). This open reading frame, designated hel3.1, begins with a 5"-ATG-3" translation initiation codon, ends with a 5"-TAA-3" stop codon, and is preceded by a putative RBS (5"-AAATTTGGTGA-3"). The putative Hel3.1 protein is 443 amino acids long and has a predicted molecular mass of 50.5 kDa. Conserved structural motifs that are characteristic of ATP-dependent helicases were found in the deduced primary sequence, including the nucleoside triphosphate binding (SPPRSGKT), nucleoside triphosphate hydrolysis (DEAH), and variant zinc finger (CDECYATFWSAERICPLC) motifs (14).
The coding region of hel3.1 was compared to the putative helicase genes from S. thermophilus bacteriophages DT1, φO1205, Sfi11, Sfi18, Sfi19, and Sfi21. BLASTN sequence analysis revealed the coding region of the bacteriophage κ3 putative helicase gene to have 99% sequence similarity to that of bacteriophage DT1 and 90% similarity to those of bacteriophages Sfi11, Sfi18, Sfi19, Sfi21, and φO1205. Therefore, hel3.1 was more closely related to the helicase gene from bacteriophage DT1 than to the helicase alleles from the other bacteriophages. Over the entire length of the proteins, BLASTX analysis indicated the following amino acid similarities: DT1, 99%; φO1205, 98%; Sfi11, 97%; Sfi18, 97%; Sfi19, 97%; and Sfi21, 97%.
Southern hybridization.
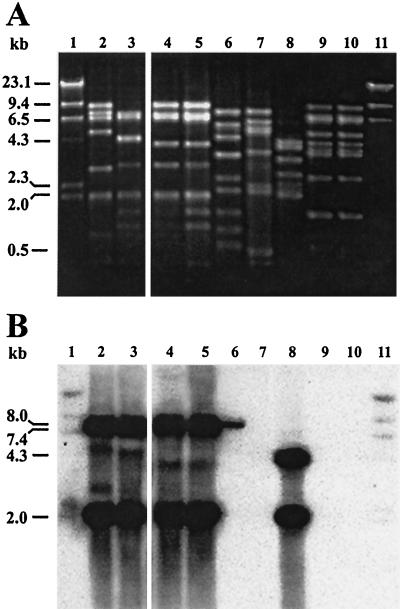
A 689-bp internal hel3.1 fragment generated by PCR using primers JMSp4 and JMSp5 was used as a probe during Southern hybridization experiments to confirm the conservation of the helicase gene among the bacteriophages listed in Table 1. Genomic DNAs were isolated from bacteriophages DT1, κ1, κ2, κ3, κ5, κ6, κ9, κ10, κ12, and κ13; digested with HindIII; and probed under low-stringency conditions (Fig. 1). The 5" ends of primers JMSp4 and JMSp5 were designed 346 bp upstream and 337 bp downstream, respectively, of the HindIII site located near the midpoint of the hel3.1 gene. Two strong bands of hel3.1 hybridization (7.4 and 2.0 kb) were present in the cos-type bacteriophages DT1, κ3, κ5 (data not shown), κ9, and κ10. The pac-type bacteriophage κ1 also showed two strong hybridization bands (4.3 and 2.0 kb). Three bacteriophages (i.e., the pac-type bacteriophages κ6 and κ12 and the uncharacterized bacteriophage κ2) failed to hybridize the internal hel3.1 probe. These results indicated that the putative helicase gene was present in both cos- and pac-type S. thermophilus bacteriophages but is not universally conserved. The 2.0-kb hel3.1-hybridizing fragment present in bacteriophages DT1, κ1, κ3, κ5 (data not shown), κ9, and κ10 is a component of the Sfi21-type DNA replication module that is highly conserved among the S. thermophilus bacteriophages (5, 8). In contrast, bacteriophage κ13 (Fig. 1, lanes 6) showed a single, weaker band of homology (8.0 kb) and loss of an internal HindIII site. This suggested that the κ13-derived fragment was divergent from the κ3-derived hel3.1 allele. These results indicated that a majority of the bacteriophages listed in Table 1 possessed the Sfi21-type DNA replication module.
FIG. 1.
(A) Restriction profiles of HindIII-restricted bacteriophage genomic DNAs. (B) Southern hybridization using an internal hel3.1 probe. Lanes 1 and 11, DIG-labeled λ DNA molecular size marker; lanes 2, DT1; lanes 3, κ3; lanes 4, κ9; lanes 5, κ10; lanes 6, κ13; lanes 7, κ2; lanes 8, κ1; lanes 9, κ6; lanes 10, κ12.
Construction of a basal antisense RNA expression vector (pTRK687).
Of the vector systems tested in our laboratory to date, only those that replicate via a rolling-circle mechanism are transformable into strains of S. thermophilus (unpublished observations). As a result, antisense RNA expression systems were assembled on a stable derivative of the high-copy-number plasmid pNZ123 (9). This vector, which was recovered from E. coli as a deletion derivative and designated pTRK686, was completely sequenced (2,410 bp). This plasmid transforms S. thermophilus NCK1124, which is plasmidless, and NCK1125, which contains two native plasmids, at frequencies of between 104 and 105 transformants per μg of supercoiled plasmid DNA. The 0.6-kb BglII expression cassette from pTRK593 (43), containing the Lactobacillus acidophilus ATCC 4356 P6 promoter (11) and a mini-multiple cloning site with a downstream coliphage T7 transcriptional terminator, was cloned into the Sau3AI site of pTRK686. The resulting 3,018-bp plasmid, designated pTRK687, was used as a basal RNA expression vector.
Construction of a DNA helicase-based antisense RNA expression system.
The 1.4-kb hel3.1-containing fragment amplified from cos-type bacteriophage κ3 was digested with PstI and cloned, in either orientation (i.e., sense or antisense) relative to the P6 promoter, into the PstI site of pTRK687. The resulting sense (hel3.1-S) and antisense (hel3.1-AS) constructs, designated pTRK688::hel3.1-S and pTRK689::hel3.1-AS, respectively (Fig. 2), were electroporated into S. thermophilus NCK1124, NCK1125, and NCK1434 to determine their impact on bacteriophage infection during standard plaque assays. The plasmid pTRK688::hel3.1-S was included in this study as a sense RNA control to exclude the possibility that any observed drop in EOP or plaque size might be attributed to the increased metabolic burden associated with RNA expression from these high-copy-number expression vectors.
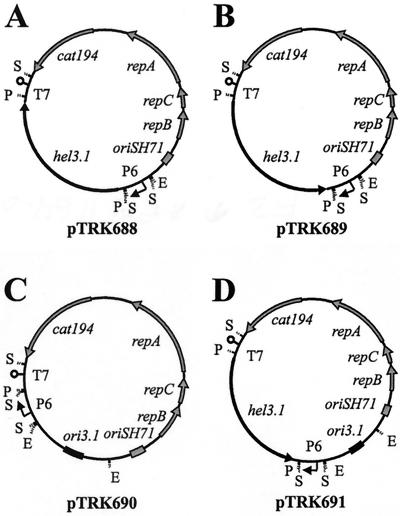
FIG. 2.
(A) Sense RNA control plasmid pTRK688::hel3.1-S; (B) antisense RNA plasmid pTRK689::hel3.1-AS; (C) PER plasmid pTRK690::ori3.1; (D) explosively replicated antisense RNA expression plasmid pTRK691::ori3.1::hel3.1-AS. Abbreviations: T7, coliphage T7 transcription terminator; P6, L. acidophilus P6 promoter; repBCA, genes encoding plasmid replication factors; cat194, chloramphenicol resistance gene; oriSH71, plasmid origin of DNA replication; ori3.1, bacteriophage κ3-derived origin of DNA replication; hel3.1, bacteriophage κ3-derived putative helicase. Restriction endonuclease recognition sites: E, EcoRI; S, Sau3AI; P, PstI.
Effect of antisense hel3.1 expression on EOP and plaque size.
S. thermophilus NCK1125, NCK1434, and their derivatives harboring both sense and antisense constructs were challenged with cos- or pac-type bacteriophages during standard plaque assays (Table 3). Antisense hel3.1 expression consistently caused a 40 to 70% reduction in EOP with a concomitant decrease in plaque size (relative to the parent strains) when challenged with bacteriophages that harbored strong bands of hel3.1 hybridization (i.e., bacteriophages κ1, κ3, κ4, κ5 [data not shown], κ9, and κ10). Bacteriophages picked from plaques formed on NCK1125(pTRK689::hel3.1-AS) and NCK1434(pTRK689::hel3.1-AS) were seemingly unchanged and remained equally sensitive upon reinfection of antisense hel3.1-expressing hosts.
TABLE 3.
Effects of antisense hel3.1 RNA and ori-conferred PER on various S. thermophilus bacteriophages
| Bacteriophage isolate | Strain (construct) | EOP (mean ± SD) | ΔPSa | Bacteriophage isolate | Strain (construct) | EOP (mean ± SD) | ΔPSF |
|---|---|---|---|---|---|---|---|
| κ1 | NCK1434 | 1.0 | — | κ9 | NCK1125 | 1.0 | — |
| NCK1434(pTRK687) | 0.8 ± 0.2 | SE | NCK1125(pTRK687) | 0.9 ± 0.1 | SE | ||
| NCK1434(hel3.1-S) | 0.8 ± 0.2 | SE | NCK1125(hel3.1-S) | 1.0 ± 0.1 | SE | ||
| NCK1434(hel3.1-AS) | 0.3 ± 0.2 | PP | NCK1125(hel3.1-AS) | 0.5 ± 0.2 | PP | ||
| NCK1434(ori3.1) | NTb | NT | NCK1125(ori3.1) | <10−6 | IP | ||
| NCK1434(ori3.1::hel3.1-AS) | NT | NT | NCK1125(ori3.1::hel3.1-AS) | <10−8 | IP | ||
| κ3 | NCK1125 | 1.0 | — | κ10 | NCK1125 | 1.0 | — |
| NCK1125(pTRK687) | 0.9 ± 0.2 | SE | NCK1125(pTRK687) | 0.9 ± 0.2 | SE | ||
| NCK1125(hel3.1-S) | 0.9 ± 0.2 | SE | NCK1125(hel3.1-S) | 1.1 ± 0.2 | SE | ||
| NCK1125(hel3.1-AS) | 0.5 ± 0.1 | PP | NCK1125(hel3.1-AS) | 0.4 ± 0.1 | PP | ||
| NCK1125(ori3.1) | <10−6 | IP | NCK1125(ori3.1) | <10−6 | IP | ||
| NCK1125(ori3.1::hel3.1-AS) | <10−8 | IP | NCK1125(ori3.1::hel3.1-AS) | <10−8 | IP | ||
| κ4 | NCK1125 | 1.0 | — | κ12 | NCK1125 | 1.0 | — |
| NCK1125(pTRK687) | 1.1 ± 0.1 | SE | NCK1125(pTRK687) | 0.8 ± 0.1 | SE | ||
| NCK1125(hel3.1-S) | 1.2 ± 0.1 | SE | NCK1125(hel3.1-S) | 0.8 ± 0.2 | SE | ||
| NCK1125(hel3.1-AS) | 0.6 ± 0.2 | PP | NCK1125(hel3.1-AS) | 0.9 ± 0.2 | SE | ||
| NCK1125(ori3.1) | <10−6 | IP | NCK1125(ori3.1) | 0.8 ± 0.2 | SE | ||
| NCK1125(ori3.1::hel3.1-AS) | <10−7 | IP | NCK1125(ori3.1::hel3.1-AS) | 0.8 ± 0.2 | SE | ||
| κ6 | NCK1125 | 1.0 | — | κ13 | NCK1124 | 1.0 | — |
| NCK1125(pTRK687) | 0.8 ± 0.1 | SE | NCK1124(pTRK687) | 0.8 ± 0.2 | SE | ||
| NCK1125(hel3.1-S) | 0.9 ± 0.1 | SE | NCK1124(hel3.1-S) | 0.8 ± 0.2 | SE | ||
| NCK1125(hel3.1-AS) | 0.8 ± 0.1 | SE | NCK1124(hel3.1-AS) | 0.9 ± 0.2 | SE | ||
| NCK1125(ori3.1) | 0.9 ± 0.2 | SE | NCK1124(ori3.1) | NT | SE | ||
| NCK1125(ori3.1::hel3.1-AS) | 1.0 ± 0.1 | SE | NCK1124(ori3.1::hel3.1-AS) | NT | SE |
ΔPS, change in plaque size. —, reference plaque size; SE, slightly enlarged; PP, pinpoint; IP, irregularly shaped pinpoint plaques.
NT, not tested.
The expression of antisense hel3.1 by NCK1125(pTRK689::hel3.1-AS) failed to affect bacteriophages that lacked fragments of hel3.1 hybridization, (i.e., the pac-type bacteriophages κ6 and κ12) (Table 3). When expressed from NCK1124(pTRK689::hel3.1-AS), antisense hel3.1 was also ineffective against bacteriophage κ13, which weakly hybridized the hel3.1-internal probe (Table 3).
When cos-type bacteriophages κ3, κ4, κ5 (data not shown), κ9, and κ10 were plaqued on vector control strains, including pTRK687 and pTRK688::hel3.1-S, they generally gave rise to slightly enlarged plaques and did not exhibit large reductions in EOP (Table 3). The plaque size of pac-type bacteriophages κ6 and κ12 was similarly affected by the presence of the control plasmids.
The bacteriophage sensitivity data correlated well with the data obtained from both the hel3.1-specific Southern hybridization and JMSp1- and JMSp2-derived PCR amplification experiments. Bacteriophages sensitive to the antisense technology hybridized the hel3.1-specific probe and gave rise to hel-containing amplicons during PCR. Conversely, bacteriophages that failed to generate hel-containing amplicons and either failed to hybridize or weakly hybridized the hel3.1-specific probe were insensitive to antisense hel3.1 RNA.
Lysis-in-broth assays.
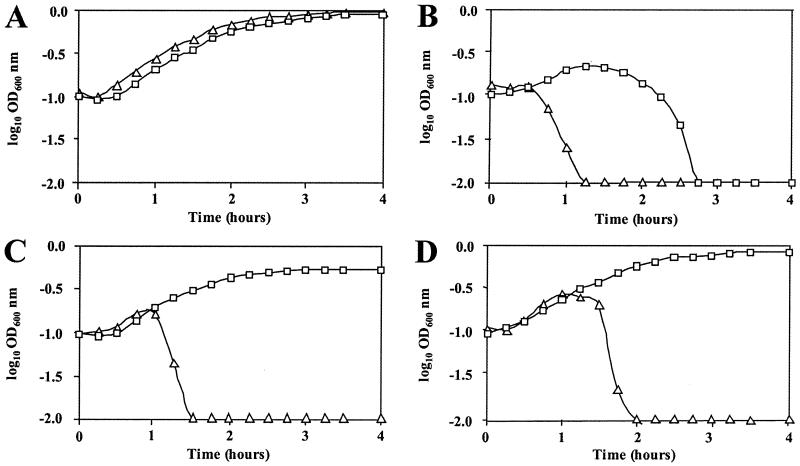
The growth curves of S. thermophilus NCK1125 and the antisense hel3.1 construct were evaluated in the presence and absence of bacteriophage κ3 at various multiplicities of infection (MOI) (i.e., 0 [negative control], ≈5, ≈1, and ≈0.1) (Fig. 3). The control strains, NCK1125, NCK1125(pTRK687) (data not shown), and NCK1125(pTRK688::hel3.1-S) (data not shown), were all lysed within 120 min at all experimental MOIs tested. Expression of antisense hel3.1 from the plasmid pTRK689::hel3.1-AS conferred significant protection from κ3-mediated lysis. Bacteriophage κ3 failed to lyse the antisense-expressing culture at initial MOIs of approximately 1 and 0.1, although the rate of growth and accumulated cell mass were slightly below those of the unchallenged parent strain at the higher MOI.
FIG. 3.
Effect of bacteriophage κ3 on the growth of NCK1125 (▵) and NCK1125(pTRK688::hel3.1-AS) (□) at various MOI. (A) Growth in the absence of bacteriophage κ3 (i.e., MOI = 0). (B to D) Growth in the presence of bacteriophage κ3 at MOIs of ≈5 (B), ≈1 (C), and ≈0.1 (D). OD600, optical density at 600 nm.
Strand-specific RT-PCR.
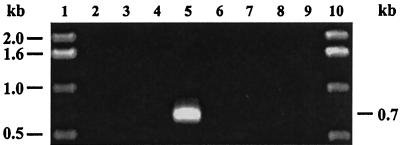
Nonquantitative, strand-specific RT-PCR was performed to confirm that hel3.1 antisense RNA was expressed in the appropriate strains. Primer JMSp4 was used during first-strand (RT) synthesis of cDNA, and primers JMSp4 and JMSp5 were used during the subsequent PCR amplification (Fig. 4). The expected 0.7-kb fragment was amplified only from total RNA isolated from NCK1125(pTRK689::hel3.1-AS), indicating that (i) antisense hel3.1 RNA was being expressed from pTRK689::hel3.1-AS and (ii) antisense hel3.1 RNA was not expressed in strains that did not harbor the plasmid.
FIG. 4.
DNase-treated total RNA preparations were subjected to RT-PCR (lanes 2 to 5) and control PCRs (without cDNA synthesis; lanes 6 to 9) in order to detect hel3.1 antisense RNA expression in NCK1125 (lanes 2 and 6), NCK1125(pTRK687) (lanes 3 and 7), NCK1125(pTRK688::hel3.1-S) (lanes 4 and 8), and NCK1125(pTRK689::hel3.1-AS) (lanes 5 and 9). Lanes 1 and 10, 1-kb ladder (Gibco-BRL Life Technologies).
A second experiment was carried out to ensure the absence of residual plasmid DNA during the confirmation of antisense expression. The primer JMSp1, which is located 346 bp 5" of JMSp4, was substituted for JMSp4 during PCR amplification (while still using JMSp4 for first-strand cDNA synthesis). In this case, a 1.0-kb fragment was obtained only when higher concentrations of RNA were used for cDNA synthesis (i.e., concentrations of more than 1 μg of total RNA per 20-μl reaction mixture). These results demonstrated that P6 promoter-driven transcription from pTRK689::hel3.1-AS yields a population of multimeric RNA transcripts. In all cases, the PCR control reactions performed on DNase-treated, non-reverse-transcribed samples failed to generate an amplification product, indicating the absence of detectable levels of plasmid DNA in the total RNA preparations.
Construction of PER and explosive antisense RNA vectors.
EcoRI-tagged primer JMSp6 was designed from the DT1-, φO1205-, Sfi11-, Sfi19-, and Sfi21-derived consensus region upstream of the putative origin of DNA replication (ori) consensus sequence, while EcoRI-tagged primer JMSp7 was designed exclusively from the DT1 genomic sequence. Regions downstream of the putative ori diverged among these bacteriophages, so preference was given to DT1 because it was derived from North American cheese fermentations. Primers JMSp6 and JMSp7 were used to amplify the putative ori from the S. thermophilus bacteriophages listed in Table 1, except that κ1 was not tested. The expected 0.7-kb amplicon was generated only from bacteriophages DT1, κ3, and κ5. The 677-bp κ3-derived fragment, designated ori3.1, was sequenced (GenBank accession number AF442521), digested with EcoRI, and cloned into the EcoRI site of pTRK687, which is located upstream of the P6 promoter. The resulting PER plasmid, designated pTRK690::ori3.1 (Fig. 2), served as a base vector for the construction of an explosive antisense RNA expression system. The hel3.1 fragment described above was subsequently cloned into the PstI site of pTRK690::ori3.1 to yield pTRK691::ori3.1::hel3.1-AS (Fig. 2). The plasmids pTRK690::ori3.1 and pTRK691::ori3.1::hel3.1-AS were then electroporated into S. thermophilus NCK1125 to determine their impact on the infection of bacteriophages κ3, κ4, κ6, κ9, κ10, and κ12 during standard plaque assays (Table 3). The presence of the κ3-derived origin alone on the PER plasmid pTRK690::ori3.1 had a significant impact on the proliferation of bacteriophages κ3, κ4, κ9, and κ10 but did not affect the replication of κ6 or κ12. The pTRK690::ori3.1 construct reduced the EOP of sensitive bacteriophages to less than 10−6 relative to that of the NCK1125 parental strain and gave rise to irregularly shaped pinpoint plaques. The addition of the antisense hel3.1 cassette to the PER plasmid further impeded bacteriophage κ3 replication significantly beyond the level with the PER parent plasmid alone (Table 3). NCK1125(pTRK691::ori3.1::hel3.1-AS) lowered the EOP of sensitive bacteriophages to less than 10−7 and 10−8 and gave rise to irregularly shaped pinpoint plaques, often with faint halos, that were difficult to enumerate. Bacteriophages picked from these plaques failed to propagate to detectable levels, even after multiple propagations on NCK1125. No antisense hel3.1 RNA-resistant bacteriophages have been isolated to date.
DISCUSSION
In this study, comparative computational analyses of the genomes of six S. thermophilus bacteriophages were used to choose genetic targets suitable for the construction of antisense RNA and explosive RNA expression strategies. When the bacteriophage κ3-derived putative helicase gene (hel3.1) was cloned in the antisense orientation behind the strong , L. acidophilus P6 promoter and expressed from a high-copy-number vector, hel3.1 antisense RNA consistently mediated a 50% reduction in EOP and reduction in plaque size against bacteriophage κ3. The proliferation of other hel-containing S. thermophilus bacteriophages, (i.e., κ1, κ4, κ5 [data not shown], κ9, and κ10) was similarly impeded by the expression of hel3.1 antisense RNA, causing a 40 to 70% reduction in EOP with a concomitant reduction in plaque size. Antisense hel3.1 failed to affect the proliferation of bacteriophages κ13 or κ6 and κ12, which either weakly hybridized or failed to hybridize a hel3.1-specific probe during Southern hybridization experiments, respectively.
The magnitude of bacteriophage inhibition via antisense RNA expression is similar to results reported previously for L. lactis. Kim et al. (21) found that antisense expression of two polycistronic open reading frames, designated gp18C and gp24C, inhibited the P335-type bacteriophage φ7-9, as measured by a 55% reduction in EOP. Interestingly, the reduction in EOP dropped to 30% if the RBS and coding region for the first 15 amino-terminal residues of Gp18C were omitted from the antisense construct. In both cases, the plaque size was also reduced by approximately 10-fold. Chung et al. (7) obtained variable reductions in EOP, which ranged between 0.5 and 0.8, as they expressed different lengths of the φF4-1 major coat protein (mcp) gene. Kim and Batt (20) found that antisense expression of the full-length, bacteriophage φ7-9-derived gp15C mediated a reduction in EOP of φ7-9 (and other gp15C-containing bacteriophages) to 10−2. More recently, McGrath et al. (29) targeted DNA replication functions and found a 50% to a 10−6-log-unit reduction in EOP, depending on the targeted gene and bacteriophage tested.
The results from these and other studies clearly indicate that certain genes are better targets than others for silencing by antisense RNA. Polzin et al. (K. M. Polzin, L. J. Collins, M. W. Lubbers, and A. W. Jarvis, Abstr. 5th Symp. Lactic Acid Bacteria, abstr. F2, 1996) found that the antisense expression of four early open reading frames (including e5, encoding a putative subunit of DNA polymerase; e12, encoding a putative transcription regulator; and e15, encoding a putative recombinase) and of four late open reading frames (including l7, encoding a major tail protein, and l12, encoding a putative terminase) was in all cases ineffective in inhibiting the replication of the L. lactis prolate-headed bacteriophage c2, regardless of the gene dosage tested. In addition, Walker and Klaenhammer (43) found that two middle-expressed open reading frames, including orf1 and orf2, and four late-expressed open reading frames, orf3 through orf6, were also ineffective at inhibiting the L. lactis P335-type bacteriophage φ31 when expressed from the L. acidophilus P6 promoter on the high-copy-number vector pTRKH2. In order to increase the ratio of antisense RNA to sense RNA, those authors cloned the aforementioned antisense expression cassettes into pTRK360, a low-copy-number vector containing the bacteriophage φ31 putative origin of DNA replication (ori31). Following bacteriophage φ31 invasion, the expression of bacteriophage-derived DNA replication factors triggered the explosive replication of pTRK360 from ori31 and produced inhibiting levels of antisense RNA during the later stages of the lytic cycle.
Mechanistically, antisense RNA hybridizes to the sense RNA strand and creates a translationally inactive double-stranded RNA (dsRNA) molecule (19). Formation of the dsRNA duplex molecule silences gene expression through the cooperative action of one or more intermolecular mechanisms. If the antisense RNA includes sequences complementary to the RBS, then the formation of dsRNA may mask the RBS, preventing efficient ribosome loading and reducing translation of the gene of interest. Formation of dsRNA downstream of the RBS may also interfere with translation by sterically impeding, to some degree, the procession of the mRNA through the ribosome. In addition, the formation of dsRNA may destabilize the sense mRNA by promoting the action of dsRNA-specific ribonucleases. Lastly, if the gene of interest is transcribed on a polycistronic mRNA, then antisense targeting may also negatively affect the expression of translationally coupled genes located downstream, causing pleiotropic effects that might further inhibit bacteriophage proliferation.
The variation in efficacy of antisense RNA-mediated gene silencing raises questions about the key characteristics of an ideal target gene or locus. Essentiality of the target RNA(s) for the replication, maturation, or release of progeny bacteriophages is perhaps the most obvious criterion. Unfortunately, essentiality must be derived empirically, cannot be garnered from the analysis of genomic data, and, often, should not be extrapolated from prior observations in heterologous systems. Analysis of a bacteriophage's transcriptome could provide additional insights into the choice of targets. In theory, optimal candidates will be genes that are transiently expressed, expressed at a very low level, and/or coded for by unstable, inefficiently translated mRNA species. In addition, the secondary structure(s) of potential mRNA species may also be examined and should be able to form structures that are conducive to recognition of the expressed antisense RNA molecule(s).
With regard to the practical efficacy of antisense cassettes in the dairy environment, identification of target genes that are effective against a variety of industrially relevant bacteriophages is of utmost importance. In this case, the use of nucleic acid hybridization in conjunction with the genomic analyses was employed to identify potential targets, although these processes cannot guarantee antisense RNA functionality. We found that the genes associated with the Sfi21-type DNA replication module are excellent candidates for targeting with antisense RNA. Five of the six model bacteriophages presently in the database (i.e., DT1, Sfi11, Sfi19, Sfi21, and φO1205) encode the 2.0-kb HindIII fragment. According to the consensus sequence of the DT1-, φO1205-, Sfi11-, Sfi19-, and Sfi21-derived replication modules, the conserved 2.2-kb HindIII hybridization signal (fragment B) of bacteriophage Sfi21, reported by Desiere et al. (8), actually corresponds to a conserved 2,027-bp HindIII restriction fragment. This 2.0-kb fragment is part of a larger, Sfi21-type DNA replication module that is highly conserved among S. thermophilus bacteriophages of industrial importance (8).
When using nucleotide sequence similarity as an indication of evolutionary descent, it was clear that the putative helicase genes of S. thermophilus bacteriophages can be divided into two groups based on either (i) the fermented product (i.e., yogurt versus cheese) or (ii) geographic location (i.e., North America versus Europe). Phylogenetically, one group contained bacteriophages isolated from North American cheese plants (DT1 and κ3), while the other included bacteriophages that were isolated from European yogurt plants (Sfi11, Sfi19, Sfi21, and φO1205).
Multiplex PCR strategies have been used to identify 936, c2, and P335 species of lactococcal bacteriophages, the three principal species encountered in worldwide dairy fermentations (22). While this technology has not yet been applied to S. thermophilus bacteriophages, composite oligonucleotide primers (Table 2) derived from a consensus DNA replication module were initially used in this study to amplify the putative helicase genes from a heterogeneous collection of bacteriophages. In other systems, molecular beacons have been used successfully in PCR to provide real-time, direct detection of pathogenic E. coli O157:H7 in food products (30). The combination of these two technologies would result in a real-time multiplex PCR strategy that could detect the presence or absence of antisense-targeted bacteriophage genes or loci, such as the putative helicase gene, among a heterogeneous bacteriophage population. If such a technology was applied to fermentation substrates prior to the addition of starter cultures, it would allow for the rapid design of culture rotation strategies that allow operators to choose which antisense RNA-expressing strains to deploy in order to maximize the integrity and quality of the fermentation.
In conclusion, we have exploited the conservation of the Sfi21-type DNA replication module and constructed effective antisense RNA expression strategies that are effective against S. thermophilus bacteriophages. Both cos- and pac-type S. thermophilus bacteriophages that attack two different strains of S. thermophilus are sensitive to these technologies. The combination of a bacteriophage origin of replication with a high-expression antisense RNA cassette provided significant protection from specific bacteriophages at levels higher than for each mechanism alone. Work is continuing to target other components of the putative DNA replication module.
Acknowledgments
This study was supported in part by the USDA-NRICGP under project number 97-35503-4468 and by Rhodia, Inc., Madison, Wis. Joseph Sturino was supported by a National Institutes of Health Biotechnology Training Program Fellowship.
We extend our gratitude to Sylvain Moineau for kindly providing bacteriophage DT1 and its propagating host S. thermophilus SMQ495.
Footnotes
Paper number FSR01-25 of the Department of Food Science, Southeast Dairy Foods Research Center, North Carolina State University, Raleigh, NC 27695-7624.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Benbadis, L., J. R. Garel, and D. L. Hartley. 1991. Purification, properties, and sequence specificity of SslI, a new type II restriction endonuclease from Streptococcus salivarius subsp. thermophilus. Appl. Environ. Microbiol. 57:3677-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brüssow, H., A. Probst, M. Fremont, and J. Sidoti. 1994. Distinct Streptococcus thermophilus bacteriophages share an extremely conserved DNA fragment. Virology 200:854-857. [DOI] [PubMed] [Google Scholar]
- 4.Brüssow, H., A. Bruttin, F. Desiere, S. Lucchini, and S. Foley. 1998. Molecular ecology and evolution of Streptococcus thermophilus bacteriophage—a review. Virus Genes 16:95-109. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 6.Bruttin, A., F. Desiere, N. d'Amico, J. P. Guerin, J. Sidoti, B. Huni, S. Lucchini, and H. Brüssow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, D. K., S. K. Chung, and C. A. Batt. 1992. Antisense RNA directed against the major capsid protein of Lactococcus lactis subsp. cremoris bacteriophage F4-1 confers partial resistance to the host. Appl. Microbiol. Biotechnol. 37:79-83. [DOI] [PubMed] [Google Scholar]
- 8.Desiere, F., S. Lucchini, A. Bruttin, M. C. Zwahlen, and H. Brüssow. 1997. A highly conserved DNA replication module from Streptococcus thermophilus phages is similar in sequence and topology to a module from Lactococcus lactis phages. Virology 234:372-382. [DOI] [PubMed] [Google Scholar]
- 9.De Vos, W. 1997. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46:281-295. [Google Scholar]
- 10.Dinsmore, P. K., and T. R. Klaenhammer. 1997. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense system AbiA. J. Bacteriol. 179:2949-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djordjevic, G., B. Bojovic, N. Miladinov, and L. Topisirovic. 1997. Cloning and molecular analysis of promoter-like sequences isolated from the chromosomal DNA of Lactobacillus acidophilus ATCC 4356. Can. J. Microbiol. 43:61-69. [DOI] [PubMed] [Google Scholar]
- 12.Foley, S., S. Lucchini, M. C. Zwahlen, and H. Brüssow. 1998. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology 250:377-387. [DOI] [PubMed] [Google Scholar]
- 13.Garbutt, K. C., J. Kraus, and B. L. Geller. 1997. Bacteriophage resistance in Lactococcus lactis engineered by replacement of a gene for a bacteriophage receptor. J. Dairy Sci. 80:1512-1519. [Google Scholar]
- 14.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 15.Guimont, C., P. Henry, and G. Linden. 1993. Restriction/modification in Streptococcus thermophilus: isolation and characterization of a type II restriction endonuclease Sth455I. Appl. Microbiol. Biotechnol. 39:216-220. [DOI] [PubMed] [Google Scholar]
- 16.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1990. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J. Bacteriol. 172:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh, T. V., R. A. Young, and R. W. Davis. 1985. Construction and screening cDNA libraries in λgt10 and λgt11, p. 49-78. In D. M. Glover (ed.), DNA cloning, vol. I. IRL Press Ltd., Oxford, United Kingdom. [Google Scholar]
- 19.Inouye, M. 1988. Antisense RNA: its functions and applications in gene regulation—a review. Gene 72:25-34. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. H., and C. A. Batt. 1991. Antisense RNA-mediated bacteriophage resistance in Lactococcus lactis. Appl. Environ. Microbiol. 57:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. G., Y. C. Bor, and C. A. Batt. 1992. Bacteriophage resistance in Lactococcus lactis ssp. lactis using antisense ribonucleic acid. J. Dairy Sci. 75:1761-1767. [DOI] [PubMed] [Google Scholar]
- 22.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Marrec, C., D. van Sinderen, L. Walsh, E. Stanley, E. Vlegels, S. Moineau, P. Heinze, G. Fitzgerald, and B. Fayard. 1997. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucchini, S., F. Desiere, and H. Brüssow. 1998. The structural gene module in Streptococcus thermophilus bacteriophage φSfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology 246:63-73. [DOI] [PubMed] [Google Scholar]
- 25.Lucchini, S., F. Desiere, and H. Brüssow. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232-243. [DOI] [PubMed] [Google Scholar]
- 26.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virology 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchini, S., J. Sidoti, and H. Brüssow. 2000. Broad-range bacteriophage resistance in Streptococcus thermophilus by insertional mutagenesis. Virology 275:267-277. [DOI] [PubMed] [Google Scholar]
- 28.McGrath, S., J. F. Seegers, G. F. Fitzgerald, and D. van Sinderen. 1999. Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl. Environ. Microbiol. 65:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKillip, J. L., and M. Drake. 2000. Molecular beacon polymerase chain reaction detection of Escherichia coli O157:H7 in milk. J. Food Prot. 63:855-859. [DOI] [PubMed] [Google Scholar]
- 31.Mercenier, A., P. H. Pouwels, and B. M. Chasy. 1994. Genetic engineering of lactobacilli, leuconostocs, and Streptococcus thermophilus, p. 253-293. In M. J. Gasson and W. M. DeVos (ed.), Genetics and biotechnology of lactic acid bacteria. Blackie Academic and Professional, Glasgow, United Kingdom.
- 32.Moineau, S., S. A. Walker, B. J. Holler, E. R. Vedamuthu, and P. A. Vandenbergh. 1995. Expression of a Lactococcus lactis phage resistance mechanism by Streptococcus thermophilus. Appl. Environ. Microbiol. 61:2461-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moineau, S., M. Borkaev, B. J. Holler, S. A. Walker, J. K. Kondo, E. R. Vedamuthu, and P. A. Vandenbergh. 1996. Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J. Dairy Sci. 79:2104-2111. [Google Scholar]
- 34.O'Sullivan, D. J., C. Hill, and T. R. Klaenhammer. 1993. Effect of increasing the copy number of bacteriophage origins of replication in trans on incoming-phage proliferation. Appl. Environ. Microbiol. 59:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Solaiman, D. K. Y., and G. A. Somkuti. 1990. Isolation and characterization of a type II restriction endonuclease from Streptococcus thermophilus. FEMS Microbiol. Lett. 67:261-266. [DOI] [PubMed] [Google Scholar]
- 38.Solaiman, D. K. Y., and G. A. Somkuti. 1992. A type II restriction endonuclease of Streptococcus thermophilus ST117. FEMS Microbiol. Lett. 80:75-80. [DOI] [PubMed] [Google Scholar]
- 39.Solow, B. T., and G. A. Somkuti. 2001. Molecular properties of Streptococcus thermophilus plasmid pER35 encoding a restriction modification system. Curr. Microbiol. 42:122-128. [DOI] [PubMed] [Google Scholar]
- 40.Stanley, E., G. F. Fitzgerald, M. C. Le Marrec, B. Fayard, and D. van Sinderen. 1997. Sequence analysis and characterization of φO1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology 143:3417-3429. [DOI] [PubMed] [Google Scholar]
- 41.Stanley, E., L. Walsh, A. van der Zwet, G. F. Fizgerald, and D. van Sinderen. 2000. Identification of four loci isolated from two Streptococcus thermophilus phage genomes responsible for mediating bacteriophage resistance. FEMS Microbiol. Lett. 182:271-277. [DOI] [PubMed] [Google Scholar]
- 42.Tremblay, D. M., and S. Moineau. 1999. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255:63-76. [DOI] [PubMed] [Google Scholar]
- 43.Walker, S. A., and T. R. Klaenhammer. 2000. An explosive antisense RNA strategy for inhibition of a lactococcal bacteriophage. Appl. Environ. Microbiol. 66:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]