Abstract
The genome sequence of the hyperthermophilic bacterium Thermotoga maritima encodes a number of glycosyl hydrolases. Many of these enzymes have been shown in vitro to degrade specific glycosides that presumably serve as carbon and energy sources for the organism. However, because of the broad substrate specificity of many glycosyl hydrolases, it is difficult to determine the physiological substrate preferences for specific enzymes from biochemical information. In this study, T. maritima was grown on a range of polysaccharides, including barley β-glucan, carboxymethyl cellulose, carob galactomannan, konjac glucomannan, and potato starch. In all cases, significant growth was observed, and cell densities reached 109 cells/ml. Northern blot analyses revealed different substrate-dependent expression patterns for genes encoding the various endo-acting β-glycosidases; these patterns ranged from strong expression to no expression under the conditions tested. For example, cel74 (TM0305), a gene encoding a putative β-specific endoglucananse, was strongly expressed on all substrates tested, including starch, while no evidence of expression was observed on any substrate for lam16 (TM0024), xyl10A (TM0061), xyl10B (TM0070), and cel12A (TM1524), which are genes that encode a laminarinase, two xylanases, and an endoglucanase, respectively. The cel12B (TM1525) gene, which encodes an endoglucanase, was expressed only on carboxymethyl cellulose. An extracellular mannanase encoded by man5 (TM1227) was expressed on carob galactomannan and konjac glucomannan and to a lesser extent on carboxymethyl cellulose. An unexpected result was the finding that the cel5A (TM1751) and cel5B (TM1752) genes, which encode putative intracellular, β-specific endoglucanases, were induced only when T. maritima was grown on konjac glucomannan. To investigate the biochemical basis of this finding, the recombinant forms of Man5 (Mr, 76,900) and Cel5A (Mr, 37,400) were expressed in Escherichia coli and characterized. Man5, a T. maritima extracellular enzyme, had a melting temperature of 99°C and an optimun temperature of 90°C, compared to 90 and 80°C, respectively, for the intracellular enzyme Cel5A. While Man5 hydrolyzed both galactomannan and glucomannan, no activity was detected on glucans or xylans. Cel5A, however, not only hydrolyzed barley β-glucan, carboxymethyl cellulose, xyloglucan, and lichenin but also had activity comparable to that of Man5 on galactomannan and higher activity than Man5 on glucomannan. The biochemical characteristics of Cel5A, the fact that Cel5A was induced only when T. maritima was grown on glucomannan, and the intracellular localization of Cel5A suggest that the physiological role of this enzyme includes hydrolysis of glucomannan oligosaccharides that are transported following initial hydrolysis by extracellular glycosidases, such as Man5.
Genome sequence information for hyperthermophiles has provided significant insights into the metabolic features of these microorganisms (40). With respect to the physiology of growth of these organisms on specific carbon and energy sources, nutritional patterns can be inferred from the presence or absence of genes encoding specific types of enzymes in the genome. For example, some hyperthermophiles, such as Archaeoglobus fulgidus (32), do not appear to have the enzymatic capability to use externally provided oligosaccharides for nutritional purposes (Table 1) and seem to rely instead on protein-based substrates for heterotrophic growth. This is entirely consistent with this organism's growth physiology (53). On the other hand, another hyperthermophilic heterotroph, Pyrococcus furiosus (22), has been shown to grow on a range of α- and β-linked polysaccharides, including starch, laminarin, and barley β-glucan (16). The capacity to utilize these substrates is reflected by the glycosyl hydrolase inventory inferred from the genome sequence of P. furiosus (16). Glycosyl hydrolases cleave the glycosidic bond between two or more saccharides or between a carbohydrate and a noncarbohydrate moiety (13). The synergistic activities of several of these enzymes may be needed to degrade polysaccharides in vitro. In combination, β-glucosidase (Cel1) (4, 30), laminarinase (Lam16) (24), and endoglucanase (Cel13) (2) from P. furiosus rapidly hydrolyze barley β-glucan to smaller oligosaccharides and ultimately glucose (15). Because two endo-acting glycosidases appear to be secreted by P. furiosus, as shown by the presence of putative signal peptides, and the β-glucosidase is cytoplasmic, an oligosaccharide transport system must also be involved in the utilization of barley β-glucan as a carbon and energy source.
TABLE 1.
Hyperthermophilic glycosyl hydrolases
| Type of glycosidase | Protein(s) in:
|
||||
|---|---|---|---|---|---|
| P. furiosus | P. horikoshii | T. maritima | Aquifex aeolicus | Methanococcus jannaschii | |
| Cellulase | Cel1, Cel12 | Cel1, Cel5 | Cel3, Cel5A, Cel5B, Cel12A, Cel12B, Cel74 | Cel8 | |
| Xylanase | Xyl3, Xyl10A, Xyl10B, Xyl31 | ||||
| Mannanase | Man1 | Man1, Man38 | Man2, Man5, Man38A, Man38B | ||
| Laminarinase | Lam16 | Lam16 | |||
| Chitinase | Chi18A, Chi18B | ||||
| Amylase | Amy13, Amy57A, Amy57B | Amy57 | Amy4A, Amy4B, Amy4C, Amy4D, Amy13A, Amy13B | Amy57 | Amy15, Amy57 |
| Pullulanase | Pul57 (Amy57B) | Pul13 | |||
| Galactosidase | GH35, Gal57 | Gal35, Gal57 | Gal2, Gal36, Gal42A, Gal42B, Gal53 | ||
| Others | GH65 | GH2, GH3, GH4, GH23A, GH23B, GH28, GH29, GH32, GH51, GH67, GH73 | GH23A, GH23B, GH77 | ||
The following proteins are putative proteins: GH35 in P. furiosus; Cel1, Cel5, Man1, Man38, Amy57, Gal35, and Gal57 in P. horikoshii; Cel5B, Xyl3, Man38A, Man38B, Lam16, Amy4B, Amy4C, Amy4D, Amy13B, Gal42B, Gal53, GH2, GH23A, GH23B, GH28, GH29, GH51, and GH73 in T. maritima; Cel8, Amy57, GH23A, GH23B, and GH77 in A. aeolicus; and Amy15 in M. jannaschii. No hyperthermophilic glycosyl hydrolases have been found in A. fulgidus.
Among the hyperthermophiles for which genome sequence data are available, Thermotoga maritima, a bacterium that grows optimally at 80°C (28), contains the largest number of identifiable glycosyl hydrolases (Table 1). T. maritima appears to possess the enzymatic capability to degrade a variety of α- and β-linked glucans, as well as several hemicelluloses, such as xylan, laminarin, and mannan (6, 9, 12, 17, 20, 23, 34-38, 44, 45, 49, 55). In some cases, expression of genes associated with individual glycosyl hydrolases has been examined (7, 41). However, how T. maritima orchestrates collective expression of functional subsets of the glycosyl hydrolase inventory in response to the presence of various saccharides in the environment is less clear. Furthermore, because of the broad substrate specificity of many glycosyl hydrolases, it is difficult to determine from genome sequence information which of these enzymes are needed for synergistically breaking down certain complex polysaccharides into their monosaccharide components for subsequent use in central metabolism. To study this, transcription of the genes encoding endo-acting glycosidases in T. maritima was examined by using various complex carbohydrates as primary carbon and energy sources. Among the findings of this study was the fact that while certain genes were expressed on all of the growth substrates examined, other genes were induced only in the presence of specific substrates. Furthermore, the physiological functions of glycosyl hydrolases were determined best by taking into account the results of sequence analyses and biochemical characteristics in conjunction with gene expression information.
MATERIALS AND METHODS
Growth of microorganisms.
T. maritima cells were grown anaerobically at 80°C on artificial seawater supplemented with 0.1% (wt/vol) yeast extract and 0.5% (wt/vol) tryptone (10). The medium contained (per liter) 15.0 g of NaCl, 2.0 g of Na2SO4, 2.0 g of MgCl2·6H2O, 0.50 g of CaCl2·2H2O, 0.25 g of NaHCO3, 0.10 g of K2HPO4, 50 mg of KBr, 20 mg of H3BO3, 20 mg of KI, 3 mg of Na2WO4·2H2O, and 2 mg of NiCl2·6H20. K2HPO4 was added after sterilization. The polysaccharides used as carbon sources (0.25%, wt/vol) included barley β-glucan, carboxymethyl cellulose, carob galactomannan, konjac glucomannan, and potato starch. Growth was monitored by determining the optical density at 600 nm, and final cell densities were determined by epifluorescent microscopy as described previously (12). Specific growth rates were determined from the slopes of semilog plots of exponential cell growth versus time.
Total RNA extraction.
Glassware was baked at 180°C for 16 h prior to use. Diethylpyrocarbonate (0.1%, wt/vol)-treated water was used in all procedures. All labware was treated with RNAseZap (Ambion, Austin, Tex.) prior to use. Total RNA was extracted from 350-ml cultures grown to the early to mid-exponential phase on the various growth substrates. Total RNA was extracted with an RNAqueous kit (Ambion). Cells were pelleted by centrifuging at 10,000 × g for 22 min and then frozen by using dry ice. Frozen cells were disrupted by using a mortar and pestle and 12 volumes of lysis/binding solution (Ambion). Total RNA was then extracted by using the manufacturer's instructions and the columns provided. RNA concentrations and integrity were determined by measuring the absorbance at 260 and 280 nm, as well as by 1% native agarose gel analysis.
Northern analyses.
Total RNA (15 μg) was separated on a 1.3% agarose-formaldehyde denaturing gel. RNA Millenium Markers (6 μg; Ambion) were run on the same gel. Following electrophoresis, the RNA was transferred to a BrightStar-Plus nylon membrane (Ambion) by passive capillary transfer (50). The transferred RNA was cross-linked to the nylon membrane by exposure to UV light at 260 nm. Blots were stained with methylene blue to ensure that the transfer was complete and that the amounts of RNA were equal in all lanes. Hybridization was carried out overnight in Ultrahyb (Ambion) at 42°C. Blots were developed with a Storage Phosphor Screen (Kodak, Rochester, N.Y.) and were scanned with a Molecular Imager FX (Bio-Rad, Hercules, Calif.). Probes were generated by PCR amplification of genomic DNA from T. maritima. The PCR products were purified by using Qiaquick purification columns (Qiagen, Valencia, Calif.) and were labeled with [α-32P]dATP by nick translation. The probes used are shown in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Locus | Glycosyl hydrolase gene | Primer sequences | Probe length (bp) |
|---|---|---|---|
| TM0024 | lam16 | 5"-AACGGCTGTCTTGTGATTGAGG-3", 5"-GAATGAGGAAGAACGGATGGTCG-3" | 493 |
| TM0061 | xyl10A | 5"-ACGGAGGTCAACAGAGAAGACG-3", 5"-TGCCTCAACTATGTATCCTCCTTCG-3" | 627 |
| TM0070 | xyl10B | 5"-GGTAGAGAATGGACAAAGGAAGAACTT-3", 5"-ATACTCCTCCGAACCACTGAGAG-3" | 465 |
| TM0305 | cel74 | 5"-TGGAGCAACTCATATGTGGAAATCGGTGG-3", 5"-GAAGGAGGAATGATTTCTATGAAACCCCGT-3" | 2,064 |
| TM1227 | man5 | 5"-TTACGTTCTCCATATGAGTGACGAGTTCGTG-3", 5"-AGTGAGAAGCTTGTCATATTCACATACCTCCTG-3" | 1,956 |
| TM1524 | cel12A | 5"-CGAACCGTCTCTGCCTTTGAAC-3", 5"-TTCAAACTTCCACCCGAACTGTG-3" | 427 |
| TM1525 | cel12B | 5"-AGGTGGTTCTCACGAGCGTTG-3", 5"-TTCGCCGCTGTTGTGTTCG-3" | 724 |
| TM1751 | cel5A | 5"-TGATCTTGATAATGCGAGGTATTCATATGGGTGTTGATCC-3", 5"-ACGCCATCTTGGATCCGTGTTATTCAATGCTATC-3" | 951 |
| TM1752 | cel5B | 5"-CTCAAGAAGCGTTCATTCATCACTGG-3", 5"-CCTTACTATCCACCCATTCCGCTT-3" | 324 |
Cloning and purification.
The cel5A and man5 genes were cloned in the Lambda ZAP II vector (Stratagene Cloning Systems, La Jolla, Calif.) as described elsewhere (12). The recombinant enzymes were purified by fast protein liquid chromatography (Pharmacia, Uppsala, Sweden). Heat-treated fractions were loaded on a DEAE-Sepharose column (Pharmacia), equilibrated in sodium phosphate buffer (pH 7), and eluted with a single-step NaCl salt gradient. Fractions containing β-mannanase and/or β-glucanase activity were pooled and stored at 4°C. Enzyme purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein concentrations were determined by a dye-binding method (8); bovine serum albumin was used as the standard.
Sequencing and analysis.
Amino acid sequences of Cel5A and Man5 were deduced from the genes encoding the enzymes. All sequencing reactions were performed with an Applied Biosystems dye primer or dye terminator cycle sequencing kit and a model 373A automated DNA sequencer (Perkin-Elmer Corp., Norwalk, Conn.). N-terminal amino acid sequences were determined by using the Edman degradation reaction at the Sequencing Facility at the University of Georgia (Athens, Ga.). The amino acid sequences of both proteins were compared with the amino acid sequences of other proteins in the GenBank database at http://www.ncbi.nlm.nih.gov/ by using the BLAST (1) program available at the same website. Multiple sequence alignments were carried out by using CLUSTALW (54) available at http://www2.ebi.ac.uk/clustalw/. The signal peptide sequences were identified by using the program SignalP available at http://www.cbs.dtu.dk/services/SignalP/ (42).
Assay of enzyme activity on glucan- and mannan-based polysaccharides.
The polysaccharide substrates barley β-glucan, carboxymethyl cellulose, konjac glucomannan, carob galactomannan, carob β-mannan, icelandic moss lichinen, and tamarind xyloglucan were obtained from Megazyme (Wicklow, Ireland). Potato starch, Laminaria digitata laminarin, and birch wood xylan were obtained from Sigma (St. Louis, Mo.). Guar (Uniguar 150) was obtained from Rhodia (Washington, Pa.). The ratio of d-mannose residues to d-galactose residues in carob galactomannan (3.5:1) was higher than the ratio in guar galactomannan (2:1). The chromogenic polysaccharides carob azo-galactomannan and azo-carboxymethyl cellulose were obtained from Megazyme. All polysaccharides except carob β-mannan were dissolved in water at a final concentration of 10 mg/ml (1%, wt/vol) as recommended by the manufacturer. β-Mannan was dissolved at a concentration of 10 mg/ml in 10% sodium hydroxide and neutralized with 50% acetic acid. The chromogenic substrates were prepared as described previously (12). Unless indicated otherwise, enzyme assays were done in triplicate at 80°C (Cel5A) or at 90°C (Man5) in 0.5-ml reaction mixtures containing 50 mM sodium phosphate buffer (pH 7.0) and 0.8% (wt/vol) solutions of soluble polysaccharide substrates. The standard deviations for triplicate assays were less than 10%. For chromogenic polysaccharides, enzymatic activity was measured by monitoring the release of soluble oligosaccharide fragments containing the dye Remazolbrilliant Blue R (12). One unit of activity was defined as the amount of enzyme that increased the absorbance by 1 U in 1 min (change in optical density at 590 nm per minute per milligram). For other polysaccharides, enzymatic activity was measured by monitoring the release of reducing sugars (11). One unit of enzyme activity was defined as the amount of enzyme required to release 1 μmol of glucose (or mannose) equivalent reducing groups per min. Nonenzymatic hydrolysis of the substrates at elevated temperatures was corrected with the appropriate blanks.
Estimation of temperature and pH optima.
The temperature dependence of Cel5A was determined by measuring the specific activity of the enzyme with a 0.8% (wt/vol) barley β-glucan solution in 100 mM sodium phosphate buffer (pH 6.0) at various temperatures. Similarly, the temperature dependence of Man5 was determined by measuring the specific activity of the enzyme with a 0.8% (wt/vol) carob galactomannan solution in 100 mM sodium phosphate buffer (pH 7.0) at various temperatures. The pH dependence of both enzymes was investigated by determining the specific activities of the enzymes with their substrates at pH values between 3.6 and 5.6 with 100 mM sodium acetate buffer, at pH values between 5.4 and 8.4 with 100 mM sodium phosphate buffer, and at pH values between 8.6 and 10.0 with 100 mM glycine-NaOH. Thermostability was determined by incubating the purified enzymes for various lengths of time at 80 or 90°C in 100 mM sodium phosphate buffer (pH 7.0) and determining the residual activity.
Determination of enzyme kinetics.
Kinetic parameters were determined under optimal conditions for both enzymes by using barley β-glucan as the substrate for Cel5A and carob galactomannan for Man5. Reaction rates were determined for substrate concentrations that ranged from approximately 0.4 to 6.0 times the Km. Km and Vmax values were determined from these rates by performing a nonlinear regression analysis with DataFit (Oakdale Engineering, Oakdale, Pa.).
Differential scanning microcalorimetry.
Melting temperatures for Man5 and Cel5A were determined with a NanoDifferential scanning calorimeter (Calorimetry Sciences, Salt Lake City, Utah). Both enzymes were dialyzed against 10 mM sodium phosphate buffer (pH 7.0). A sample added to a cell was maintained at a pressure of 3.0 atm to allow operation at temperatures greater than 100°C. The dialyzed enzymes were scanned at temperatures ranging from 25 to 125°C by using a scan rate of 1°C/min. Enzyme scans were corrected with a buffer-buffer baseline.
RESULTS
Analysis of the T. maritima genome with respect to glycosyl hydrolases.
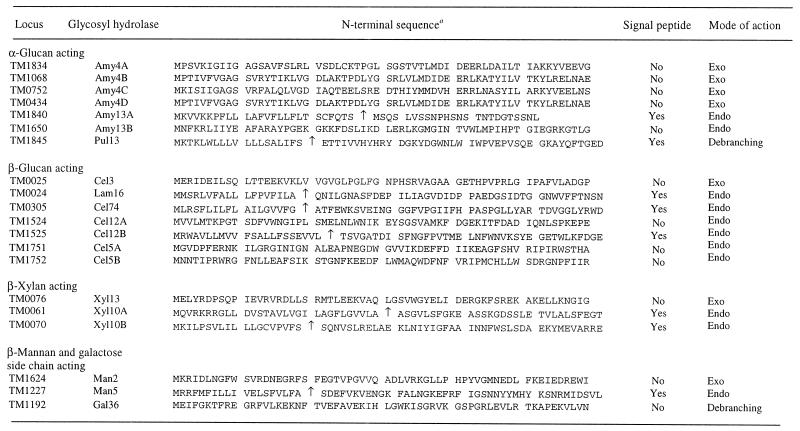
Examination of the genomic sequences of hyperthermophiles has shown that the glycosyl hydrolases (glycosidases) are widely distributed in these organisms. To date, hyperthermophilic glycosyl hydrolases have been found in at least 34 of the 77 known families (families 1 to 5, 8 to 13, 15, 16, 18, 26, 28, 29, 31, 32, 35, 36, 38, 39, 42 to 44, 48, 51, 53, 57, 65, 67, 74, and 77) (26). Table 1 shows the distribution of these enzymes in various hyperthermophilic microorganisms; glycosyl hydrolases have been classified according to the nomenclature scheme suggested by Henrissat et al. (27). Since many hyperthermophilic organisms utilize complex carbohydrates as carbon and energy sources, it is apparent that multienzyme systems are needed to hydrolyze polysaccharides that are too large to be transported across the cell membrane (3, 16). As a result, many endo-acting glycosidases are cell membrane associated or completely secreted (14). Oligosaccharides with various degrees of polymerization produced by endo-acting enzymes are transported into the cell for further processing (14). Once the oligosaccharides are inside the cell, other endo-acting and exo-acting glycosyl hydrolases within the cytoplasm play essential roles in assimilation and catabolism of these compounds to provide saccharides (e.g., glucose and galactose) to metabolic pathways (14, 31). From this perspective, putative polysaccharide-degrading enzyme systems can be identified from genome sequence data on the basis of the type of glycosidic linkages potentially cleaved by specific glycosyl hydrolases. For instance, in the case of T. maritima, glycosidases involved in the degradation of starch (an α-glucan) could potentially include two endo-acting enzymes, Amy13A (TM1840) and Amy13B (TM1650), along with exo-acting enzymes, such as Amy4A to Amy4D (TM1834, TM1068, TM0752, and TM0434). Table 3 shows putative polysaccharide-degrading enzyme systems in T. maritima associated with degradation of glycosides containing the following linkages: α-glucan, β-glucan, β-xylan, and β-mannan. Table 3 also shows sequences of identifiable N-terminal signal peptides that are associated with some of the endo-acting glycosidases in T. maritima. The data indicate that this organism is capable of exporting endo-acting glycosyl hydrolases for degradation of starch and pullulan (Amy13A, Pul13), β-glucan (Cel12B, Cel74), laminarin (Lam16), xylan (Xyl10A, Xyl10B), and mannan (Man5). It is interesting that none of the identifiable exo-acting glycosyl hydrolases have signal peptides, suggesting that oligosaccharides must be imported prior to processing to monosaccharides. Also, there are a number of intracellular endo-acting glycosyl hydrolases (Amy13B, Cel12A, Cel5A, and Cel5B) that could be involved in hydrolysis of transported oligosaccharides. However, it is not clear how T. maritima uses its glycosyl hydrolase inventory strategically to recruit carbohydrates as carbon and energy sources or the extent to which particular enzymes are utilized. In particular, the functions of Cel5A and Cel5B, which have no apparent extracellular counterparts, are difficult to assess based on genome sequence information.
TABLE 3.
Polysaccharide-degrading enzyme systems in T. maritima
An arrow indicates the predicted cleavage site of the signal peptide.
Growth of T. maritima on complex carbohydrates and induction of endo-acting glycosidase genes.
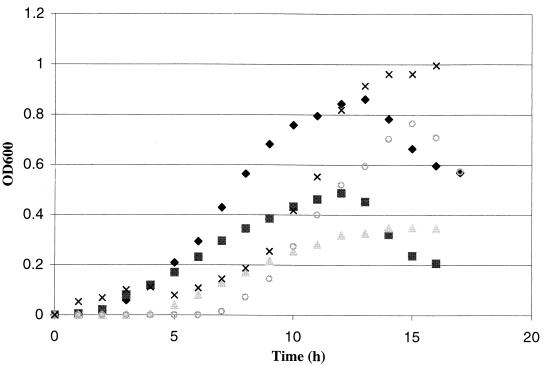
To investigate the potential roles of various endo-acting glycosyl hydrolases in the utilization of polysaccharides, T. maritima was grown on a range of substrates that should be suitable carbon and energy sources based on the information in Table 3. These substrates included konjac glucomannan, a linear polysaccharide containing β-1,4-linked d-mannopyranose and d-glucopyranose units as backbone residues; carob galactomannan, a heteropolysaccharide consisting of a β-1,4-linked d-mannopyranose backbone decorated with α-1,6-linked galactose residues; barley β-glucan, a linear polymer of d-glucopyranose residues linked by two types of linkages (β-1,4 and β-1,3); carboxymethyl cellulose, a soluble form of cellulose (made by chloroacetic acid treatment of cellulose) and a linear homopolysaccharide made up of β-1,4-linked d-glucopyranose residues; and starch, a mixture of the linear α-1,4-linked d-glucanpyranose homopolysaccharide amylose (15 to 25%) and branched amylopectin (75 to 85%) containing α-1,6-glycosidic linkages (at every 17 to 26 glucose residues) in addition to the α-1,4 bonds. The results of batch growth experiments at 80°C are shown in Fig. 1. As expected based on the information in Table 3, T. maritima exhibited significant growth on all of the polysaccharides tested, reaching peak cell densities near or greater than 109 cells/ml. The growth rates were highest on the linear polysaccharides barley β-glucan (72 min) and konjac glucomannan (74 min), followed by carboxymethyl cellulose (78 min), carob galactomannan (85 min), and potato starch (119 min).
FIG. 1.
Growth curves for T. maritima on various polysaccharide substrates added at initial concentrations of 0.25% (wt/vol). Symbols: ♦, carob galactomannan; ▪, konjac glucomannan; ▴, carboxymethyl cellulose; ○, barley β-glucan; ×, potato starch. OD600, optical density at 600 nm.
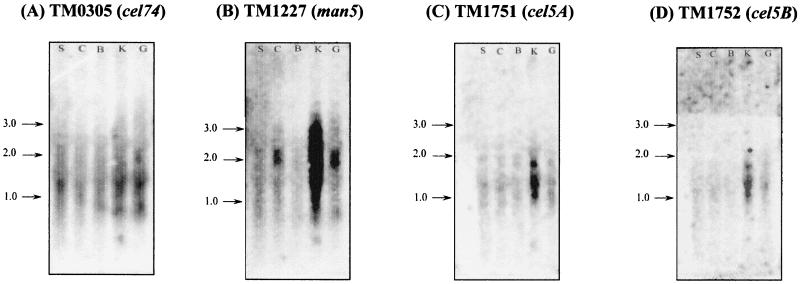
In an effort to elucidate the patterns of expression of the various glycosidases, total RNA was extracted from cells (early to mid-log phase) grown on all polysaccharide substrates, and Northern blot analysis was used to monitor the patterns of expression of several endo-β-glycosidase genes in T. maritima, including cel5A, cel5B, cel12A, cel12B, cel74, man5, lam16, xyl10A, and xyl10B. Northern blots showing the expression of these genes are shown in Fig. 2, and the results are summarized in Table 4. It was observed that a putative endoglucanase gene, cel74, was expressed on all substrates tested, including starch. On the other hand, lam16, xyl10A, xyl10B, and cel12A were not expressed on any of the substrates tested, and the other family 12 endoglucanase gene, cel12B, was expressed when carboxymethyl cellulose was the primary carbon source (data not shown). Expression of man5, which encodes an extracellular mannanase, was induced on carob galactomannan and konjac glucomannan and to some extent on carboxymethyl cellulose. An unexpected result was that the cel5A and cel5B genes encoding intracellular β-specific endoglucanases were expressed only when T. maritima was grown on konjac glucomannan. The observed transcript sizes for cel5A (1 kb), cel5B (1 kb), man5 (2 kb), cel12B (0.9 kb), and cel74 (2 kb) were 1.5, 1.5, 2, 0.9, and 1.5kb, respectively. Unlike the sizes of the man5 and cel12A transcripts, the observed sizes of the cel5A and cel5B transcripts were larger than the genes. In case of cel74, the observed transcript size was smaller than the gene. A discrepancy between the sizes of transcripts and the corresponding genes has also been observed for exoglycosidases from Thermotoga neapolitana, in which smaller-than-expected transcript sizes were attributed to selective degradation of unstable transcripts by RNases (41).
FIG. 2.
Northern blots showing expression of glycosidase genes under different growth conditions. Total RNA was extracted from T. maritima cells grown on the following polysaccharide substrates: potato starch (lanes S), carboxymethyl cellulose (lanes C), barley β-glucan (lanes B), konjac glucomannan (lanes K), and carob galactomannan (lanes G). Blots for the following genes are not shown since these genes were not expressed under the conditions tested: xyl10A (TM0061), xyl10B (TM0070), lam16 (TM0024), and cel12B (TM1524). Arrows indicate transcript sizes (in kilobases).
TABLE 4.
Summary of Northern analysis results
| Probe | Gene length (bp) | Activity | Growth substratesa
|
||||
|---|---|---|---|---|---|---|---|
| Carob galactomannan | Konjac glucomannan | Barley β-glucan | Carboxymethyl cellulose | Potato starch | |||
| TM0024 (lam16) | 1,926 | Laminarinase | − | − | − | − | − |
| TM0061 (xyl10A) | 3,177 | Endoxylanase | − | − | − | − | − |
| TM0070 (xyl10B) | 1,041 | Endoxylanase | − | − | − | − | − |
| TM0305 (cel74) | 2,121 | Endoglucanase | + | + | + | + | + |
| TM1227 (man5) | 2,007 | Mannanase | + | + | − | + | − |
| TM1524 (cel12A) | 774 | Endoglucanase | − | − | − | − | − |
| TM1525 (cel12B) | 822 | Endoglucanase | − | − | − | + | − |
| TM1751 (cel5A) | 951 | Endoglucanase/mannanase | − | + | − | − | − |
| TM1752 (cel5B) | 987 | Endoglucanase | − | + | − | − | − |
+, gene expression; −, no gene expression.
Sequence analysis and biochemical properties of recombinant Cel5A and Man5.
To further examine the potential biochemical basis for expression of cel5A and cel5B on konjac glucomannan, the recombinant form of Cel5A was produced in Escherichia coli. Since Man5 was also active on konjac glucomannan, it was used as a basis for comparison. The nucleotide sequence of the cel5A gene (TM1751) corresponds to a 951-bp open reading frame that encodes a 317-amino-acid protein with a predicted molecular mass of 37.3 kDa (39). The Cel5A sequence exhibited the highest level of homology (38% amino acid sequence identity) to the sequence of a family 5 endoglucanase (CelD) from Clostridium cellulolyticum, which has an optimum temperature of 50°C (52). An alignment of the Cel5A sequence with the sequences of other members of glycosyl hydrolase family 5 is shown in Fig. 3, and the data indicate that the following eight amino acid residues in Cel5A that are characteristic of family 5 are conserved: Arg-51, His-95, Asn-135, Glu-137 His-196, Tyr-198, Glu-253, and Trp-286 (positions 375, 420, 459, 460, 520, 522, 577, and 610, respectively). The catalytic residues Glu-137 and Glu-253 act as the proton donor and a nucleophile, respectively. Mutation of any one of these residues resulted in a complete loss of catalytic activity (Chhabra and Kelly, unpublished results). The nucleotide sequence of the man5 gene (TM1227) (39) corresponded to a 2,007-bp open reading frame that encodes a 669-amino-acid protein with a predicted molecular mass of 76.9 kDa (12). The Man5 sequence exhibited the highest level of similarity (46% amino acid sequence identity) to the sequence of a β-mannanase (ManF) from Bacillus stearothermophilus, a multidomain family 5 enzyme which has a molecular mass of 76 kDa (19). Conserved residues in Man5 that are characteristic of family 5 include Arg-83, His-164, Asn-209, Glu-211, His-290, Tyr-292, Glu-329, and Trp-362 (12). In Man5, mutation of Glu-329 (nucleophile) results in a complete loss of activity (Chhabra and Kelly, unpublished results).
FIG. 3.
Multiple sequence alignment for T. maritima Cel5A and other proteins. CelD___Cc, endoglucanase CelD from C. cellulolyticum; CelH___Ct, endoglucanase CelH from C. thermocellum; Cel5A___Tm, endoglucanase Cel5A from T. maritima. The highlighted regions are conserved regions in family 5 glycosidases as suggested by Ethier et al. (19). An asterisk indicates identical or conserved residues; a colon indicates conserved substitutions; a period indicates semiconserved substitutions.
The recombinant versions of Cel5A and Man5 expressed in E. coli were purified by heat treatment in addition to column chromatography. Recombinant Cel5A, purified twofold from the heat-treated E. coli crude extract by using ion-exchange chromatography, had a specific activity of 17 U/mg on azo-carboxymethyl cellulose. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the recombinant protein resulted in a band at 37 kDa. Purification of Man5 has been reported elsewhere (45). Cel5A has a temperature optimum of 80°C and a pH optimum of 6. Under these conditions, Cel5A had a half-life of 18 h and a melting temperature of 90°C. The enzyme followed Michaelis-Menten kinetics on barley β-glucan. The apparent kcat/Km on this substrate was estimated to be 1,112 ml s−1 mg−1. As reported previously, Man5 had an apparent kcat/Km of 150 ml s−1 mg−1 on carob galactomannan under optimal conditions (90°C and pH 7) (45). At 90°C, Man5 has a half-life of 3 h. Properties of both enzymes are summarized in Table 5.
TABLE 5.
Properties of recombinant glycosidases from T. maritima
| Enzyme | Locus | Glycosyl hydrolase family | Putative catalytic residues | Calculated molecular mass (Da) | Optimum temp (°C) | Optimum pH | Half-life (h) | Melting temp (°C) | Apparent kcat/Km (ml s−1 mg−1) | Substrate used |
|---|---|---|---|---|---|---|---|---|---|---|
| Cel5A | TM1751 | 5 | Glu-137, Glu-253 | 37,383 | 80 | 6 | 18 | 90 | 1,112 | β-Glucan |
| Man5 | TM1227 | 5 | Glu-211, Glu-329 | 76,913 | 90 | 7 | 3 | 99 | 150 | Galactomannan |
The specific activities of both enzymes were determined for a number of polysaccharide substrates (Table 6). These polysaccharides differed in their backbone compositions, types of glycosidic linkages, and side chain residues. Man5 exhibited activity on mannan-based substrates, including the linear polysaccharides β-mannan and konjac glucomannan and the branched galactomannans from carob and guar. However, no activity was detected on glucan-based substrates. The specific activities on all mannan-based substrates were the same order of magnitude, suggesting that the galactose side chain interaction was not important in galactomannan hydrolysis by this enzyme. Cel5A exhibited activity on both mannan-based and glucan-based polysaccharides. This enzyme exhibited approximately threefold-higher activity on guar and carob galactomannan than on linear β-mannan, suggesting that there was a positive interaction with galactose side chains. However, the degree of galactose substitution did not seem to affect the specific activities significantly. Among the glucan-based substrates tested, the highest activity was observed on barley β-glucan, but Cel5A did not have any detectable activity on L. digitata laminarin or potato starch. Interestingly, Cel5A had higher activity than Man5 on konjac glucomannan.
TABLE 6.
Substrate specificity data for Cel5A and Man5
| Polysaccharide (source) | Molecular mass (Da) | Backbone
|
Side chain
|
Sp acta
|
|||
|---|---|---|---|---|---|---|---|
| Residue(s) | Linkage(s) | Residue(s) | Linkage | Cel5A | Man5 | ||
| Mannan (carob) | 3,500 | Mannose | β-1,4 | None | 22 | 72 | |
| Galactomannan (carob) | 40,000 | Mannose | β-1,4 | Galactose | α-1,6 | 71 | 83 |
| Azo-galactomannan (carob) | NAb | Mannose | β-1,4 | Galactose | α-1,6 | 9 | 17 |
| Galactomannan (guar) | 2,000,000 | Mannose | β-1,4 | Galactose | α-1,6 | 69 | 90 |
| Glucomannan (konjac) | 100,000 | Mannose (60%), glucose (40%) | β-1,4 | None | 377 | 97 | |
| β-Glucan (barley) | 250,000 | Glucose | β-1,4; β-1,3 | None | 2,345 | NDc | |
| Carboxymethyl cellulose | 90,000 | Glucose | β-1,4 | None | 616 | ND | |
| Azo-carboxymethyl cellulose | NA | Glucose | β-1,4 | None | 17 | ND | |
| Xyloglucan (tamarind) | NA | Glucose | β-1,4 | Xylose, galactose | α-1,6 | 270 | ND |
| Lichenan (icelandic moss) | NA | Glucose | β-1,4; β-1,3 | None | 76 | ND | |
| Laminarin (L. digitata) | 5,000 | Glucose | β-1,3 | None | ND | ND | |
| Starch (potato) | NA | Glucose | α-1,6 | None | ND | ND | |
| Xylan (birch wood) | NA | Xylose | β-1,4 | None | ND | ND | |
For all polysaccharides except azo-galactomannan and azo-carboxymethyl cellulose specific activity is expressed in units (number of reducing sugar equivalents released per minute) per milligram. For azo-galactomannan and azo-carboxymethyl cellulose specific activity is expressed in absorbance units per minute.
NA, not available.
ND, activity not detected.
DISCUSSION
T. maritima utilizes many simple and complex carbohydrates, including glucose, sucrose, maltose, starch, galacto- and glucomannans, carboxymethyl cellulose and xylan, as growth substrates. The recently released genome sequence of T. maritima revealed the presence of a large number of glycosidases, and the percentage of predicted coding sequences involved in sugar metabolism in T. maritima is more than twice the percentages seen in the other eubacterial and archaeal species sequenced to date (39). Based on comparative genomics, it was suggested that there has been extensive lateral gene transfer between this bacterium and members of the archaeal domain, particularly Pyrococcus horikoshii (39, 56). In a recent study of transport systems encoded in the genomes of 18 prokaryotic organisms, it was found that T. maritima possesses a large number of transporters for sugars and for oligopeptides and comparatively few transporters for amino acids (46).
In order to understand the regulatory networks in organisms whose genome sequences are available, it is necessary to couple insights from bioinformatic approaches with the results of physiological and biochemical studies. For T. maritima, the propensity to utilize specific carbohydrates as growth substrates and not to utilize others can be understood from this perspective. T. maritima has been found to grow on a range of soluble complex polysaccharides. Although the T. maritima genome encodes a number of β-glucan-hydrolyzing enzymes, there is no indication that there is a complex cellulose degradation system, such as the cellulosome found in certain Clostridium species (21) and in the anaerobic rumen bacterium Ruminococcus albus (43). The cellulosome, which is typically composed of between 14 and 26 polypeptides that are presumably coexpressed, synergistically hydrolyzes crystalline cellulose, whereas its constituent polypeptides, alone or in mixtures, do not. Many of these polypeptides are catalytically active with soluble substrates and can be characterized as endoglucanases, xylanases, and cellodextrinases (5). The T. maritima intracellular endoglucanase, Cel5A, exhibits sequence homology to endoglucanases CelD and CelH from C. cellulolyticum (51) and Clostridium thermocellum (21), respectively, which are components of the cellulosome (5, 21). However, cellulases that can bind to and initiate hydrolysis of insoluble forms of cellulose are apparently not present in T. maritima. The process by which fermentative anaerobes such as C. thermocellum and C. cellulolyticum developed the capacity to utilize insoluble forms of cellulose is unclear.
The acquisition and processing of complex carbohydrates by T. maritima likely involve a range of glycosyl hydrolases and transporters, and the latter have not been identified and characterized yet. The results of the present study indicate that glucomannan induces the β-glucan endoglycosidase genes cel5A (TM1751) and cel5B (TM1752) and the putative β-specific endoglucanase gene cel74 (TM0305), as well as the β-mannan endoglycosidase gene man5 (TM1227), in T. maritima. Man5 and Cel74 are extracellular enzymes (Table 3) that could degrade glucomannan into smaller subunits, which are transported into the cell for further degradation by intracellular Cel5A and Cel5B. The endoglucanase genes cel12A (TM1524) and cel12B (TM1525) are not induced by glucomannan even though in vitro the glycosidases are active on β-glucans (34). Analysis of the genes in the vicinity of cel5A (TM1751), cel5B (TM1752), and cel74 (TM0305) indicates that multiple oligopeptide ABC transporters (TM1746 to TM1750 and TM0300 to TM0304) are present (39). In the case of man5 (TM1227), in addition to multiple oligopeptide ABC transporter subunits (TM1219 to TM1223) downstream of the gene, there are also a number of sugar ABC transporter subunits (TM1232 to TM1234) upstream of the gene (39). In a recent study of sugar transporters in Sulfolobus solfataricus, it was found that the maltose and cellobiose transporters exhibit significant sequence similarity to oligopeptide-dipeptide transporters (18). Oligopeptide transporters have also been found in the vicinity of glycosidase genes in P. horikoshii (29) and Thermoplasma acidophilum (48). This suggests that oligopeptide transporters in the vicinity of endoglycosidase genes in T. maritima are most likely involved in sugar transport.
Growth of T. maritima on the polysaccharides carob galactomannan and barley β-glucan resulted in expression of man5 and cel74, respectively. It remains to be seen whether the degradation products of these polysaccharides induce expression of the intracellular exoglycosidases man2 (TM1624) and gal36 (TM1192) for galactomannan oligosaccharide hydrolysis and cel3 (TM0025) for cellooligosaccharide hydrolysis. Recent studies performed with T. neapolitana cultures grown on galactomannan indicated that all three activities (β-mannanase [GenBank accession no. AY033477], β-mannosidase [GenBank accession no. AY033395], and α-galactosidase [GenBank accession no. AF011400]) were induced (45). The biochemical and physiological properties of Cel74 are not available yet. Preliminary studies of Cel74 have indicated that this enzyme exhibits activity on the polysaccharides barley β-glucan and carboxymethyl cellulose (Chhabra and Kelly, unpublished results), but expression of Cel74 in the presence of the polysaccharides examined here indicated that it may be active on a broad range of substrates.
The source of polysaccharides in the thermophilic environment of T. maritima has not been established yet. However, extracellular polysaccharides generated by hyperthermophiles (25, 33, 47) could be utilized as growth substrates. Transporters for the export of exopolysaccharides (the PST family of transporters) have been identified in a number of other hyperthermophiles (46). For instance, 9% of the total transporters in P. horikoshii are involved in macromolecular efflux (46). The inducers of polysaccharide synthesis and export in hyperthermophiles have not been examined yet, nor has the potential relationship of these processes to polysaccharide utilization been examined. To do this, a comprehensive study that involves the use of microarrays to examine regulation patterns involving polysaccharide degradation by glycosidases from T. maritima is under way.
Acknowledgments
This work was supported in part by grants from the National Science Foundation and the U.S. Department of Energy, Energy Biosciences Program.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, M. W., L. E. Driskill, W. Callen, M. A. Snead, E. J. Mathur, and R. M. Kelly. 1999. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes β-1,4 bonds in mixed-linkage (1 → 3),(1 → 4)-β-d-glucans and cellulose. J. Bacteriol. 181:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, M. W., L. E. Driskill, and R. M. Kelly. 1998. Glycosyl hydrolases from hyperthermophilic microorganisms. Curr. Opin. Biotechnol. 9:141-145. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, M. W., and R. M. Kelly. 1998. The family 1 β-glucosidases from Pyrococcus furiosus and Agrobacterium faecalis share a common catalytic mechanism. Biochemistry 37:17170-17178. [DOI] [PubMed] [Google Scholar]
- 5.Belaich, J. P., C. Tardif, A. Belaich, and C. Gaudin. 1997. The cellulolytic system of Clostridium cellulolyticum. J. Biotechnol. 57:3-14. [DOI] [PubMed] [Google Scholar]
- 6.Bibel, M., C. Brettl, U. Gosslar, G. Kriegshauser, and W. Liebl. 1998. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol. Lett. 158:9-15. [DOI] [PubMed] [Google Scholar]
- 7.Bok, J., D. A. Yernool, and D. E. Eveleigh. 1998. Purification, characterization and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neapolitana. Appl. Environ. Microbiol. 64:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Bronnenmeier, K., A. Kern, W. Liebl, and W. L. Staudenbauer. 1995. Purification of Thermotoga maritima enzymes for the degradation of cellulosic materials. Appl. Environ. Microbiol. 61:1399-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, S. H., C. Sjoholm, and R. M. Kelly. 1993. Purification and characterization of a highly thermostable glucose isomerase produced by the extremely thermophilic eubacterium Thermotoga maritima. Biotechnol. Bioeng. 41:878-886. [DOI] [PubMed] [Google Scholar]
- 11.Chaplin, M. 1994. Monosaccharides, p. 3-4. In M. F. Chaplin and J. F. Kennedy (ed.), Carbohydrate analysis: a practical approach. OIRL Press, Oxford, United Kingdom.
- 12.Chhabra, S., K. N. Parker, D. Lam, W. Callen, M. A. Snead, E. J. Mathur, J. M. Short, and R. M. Kelly. 2001. β-Mannanases from Thermotoga species. Methods Enzymol. 330:224-238. [DOI] [PubMed] [Google Scholar]
- 13.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 14.de Vos, W. M., S. W. M. Kengen, W. G. B. Voorhorst, and J. van der Oost. 1998. Sugar utilization and its control in hyperthermophiles. Extremophiles 2:201-205. [DOI] [PubMed] [Google Scholar]
- 15.Driskill, L. E., M. W. Bauer, and R. M. Kelly. 1999. Synergistic interactions among β-laminarinase, β-1,4-glucanase, and β-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus during hydrolysis of β-1,4-, β-1,3-, and mixed-linked polysaccharides. Biotechnol. Bioeng. 66:51-60. [PubMed] [Google Scholar]
- 16.Driskill, L. E., K. Kusy, M. W. Bauer, and R. M. Kelly. 1999. Relationship between glycosyl hydrolase inventory and growth physiology of the hyperthermophile Pyrococcus furiosus on carbohydrate-based media. Appl. Environ. Microbiol. 65:893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffaud, G. D., C. M. McCutchen, P. Leduc, K. N. Parker, and R. M. Kelly. 1997. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and α-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl. Environ. Microbiol. 63:169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elferink, M. G. L., S. V. Albers, W. N. Konings, and A. J. M. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494-1503. [DOI] [PubMed] [Google Scholar]
- 19.Ethier, N., G. Talbot, and J. Sygusch. 1998. Gene cloning, DNA sequencing, and expression of thermostable beta-mannanase from Bacillus stearothermophilus. Appl. Environ. Microbiol. 64:4428-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, B. R., A. K. Gilman, K. Cordray, and J. Woodward. 2000. Mechanism of substrate hydrolysis by a thermophilic endoglucanase from Thermotoga maritima. Biotechnol. Lett. 22:735-740. [Google Scholar]
- 21.Felix, C. R., and L. G. Ljungdahl. 1993. The Cellulosome--the exocellular organelle of Clostridium. Annu. Rev. Microbiol. 47:791-819. [DOI] [PubMed] [Google Scholar]
- 22.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 23.Gabelsberger, J., W. Liebl, and K. H. Schleifer. 1993. Cloning and characterization of β-galactoside and β-glucoside hydrolyzing enzymes of Thermotoga maritima. FEMS Microbiol. Lett. 109:131-137. [Google Scholar]
- 24.Gueguen, Y., W. G. B. Voorhorst, J. van der Oost, and W. M. de Vos. 1997. Molecular and biochemical characterization of an endo-β-1,3-glucanase of the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 272:31258-31264. [DOI] [PubMed] [Google Scholar]
- 25.Hartzell, P. L., J. Millstein, and C. LaPaglia. 1999. Biofilm formation in hyperthermophilic archaea. Methods Enzymol. 310:335-349. [DOI] [PubMed] [Google Scholar]
- 26.Henrissat, B., and P. M. Coutinho. 2001. Classification of glycoside hydrolases and glycosyltransferases from hyperthermophiles. Methods Enzymol. 330:183-201. [DOI] [PubMed] [Google Scholar]
- 27.Henrissat, B., T. T. Teeri, and R. A. J. Warren. 1998. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 425:352-354. [DOI] [PubMed] [Google Scholar]
- 28.Huber, R., T. A. Langworthy, H. Konig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 29.Kawarabayasi, H., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 30.Kengen, S. W. M., and A. J. M. Stams. 1994. An extremely thermostable β-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus--a comparison with other glycosidases. Biocatalysis 11:79-88. [Google Scholar]
- 31.Kengen, S. W. M., A. J. M. Stams, and W. M. de Vos. 1996. Sugar metabolism of hyperthermophiles. FEMS Microbiol. Rev. 18:119-137. [Google Scholar]
- 32.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. X. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. Dandrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 33.LaPaglia, C., and P. L. Hartzell. 1997. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 63:3158-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebl, W. 2001. Cellulolytic enzymes from Thermotoga species. Methods Enzymol. 330:290-300. [DOI] [PubMed] [Google Scholar]
- 35.Liebl, W., P. Ruile, K. Bronnenmeier, K. Riedel, F. Lottspeich, and I. Greif. 1996. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology 142:2533-2542. [DOI] [PubMed] [Google Scholar]
- 36.Liebl, W., I. Stemplinger, and P. Ruile. 1997. Properties and gene structure of the Thermotoga maritima α-amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. J. Bacteriol. 179:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCutchen, C. M., G. D. Duffaud, P. Leduc, A. R. H. Petersen, A. Tayal, S. A. Khan, and R. M. Kelly. 1996. Characterization of extremely thermostable enzymatic breakers (α-1,6-galactosidase and β-1,4-mannanase) from the hyperthermophilic bacterium Thermotoga neapolitana 5068 for hydrolysis of guar gum. Biotechnol. Bioeng. 52:332-339. [DOI] [PubMed] [Google Scholar]
- 38.Miller, E. S., K. N. Parker, W. Liebl, D. Lam, W. Callen, M. A. Snead, E. J. Mathur, J. M. Short, and R. M. Kelly. 2001. α-d-Galactosidases from Thermotoga species. Methods Enzymol. 330:246-260. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, L. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, K. E., I. T. Paulsen, J. F. Heidelberg, and C. M. Fraser. 2000. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 18:1049-1054. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen, T. N., K. M. Borges, A. H. Romano, and K. M. Noll. 2001. Differential gene expression in Thermotoga neapolitana in response to growth substrate. FEMS Microbiol. Lett. 195:79-83. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Ohara, H., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci. Biotechnol. Biochem. 64:254-260. [DOI] [PubMed] [Google Scholar]
- 44.Parker, K. N., S. Chhabra, D. Lam, M. A. Snead, E. J. Mathur, and R. M. Kelly. 2001. β-Mannosidase from Thermotoga species. Methods Enzymol. 330:238-246. [DOI] [PubMed] [Google Scholar]
- 45.Parker, K. N., S. R. Chhabra, D. Lam, W. Callen, G. D. Duffaud, M. A. Snead, J. M. Short, E. J. Mathur, and R. M. Kelly. 2001. Galactomannanases Man2 and Man5 from Thermotoga species: growth physiology on galactomannans, gene sequence analysis and biochemical properties of recombinant enzymes. Biotechnol. Bioeng. 75:322-333. [DOI] [PubMed]
- 46.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 47.Rinker, K. D., and R. M. Kelly. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng. 69:537-547. [DOI] [PubMed] [Google Scholar]
- 48.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretle, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 49.Ruile, P., C. Winterhalter, and W. Liebl. 1997. Isolation and analysis of a gene encoding α-glucuronidase, an enzyme with a novel primary structure involved in the breakdown of xylan. Mol. Microbiol. 23:267-279. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Shima, S., Y. Igarashi, and T. Kodama. 1991. Nucleotide sequence analysis of the endoglucanase encoding gene, celCCD, of Clostridium cellulolyticum. Gene 104:33-38. [DOI] [PubMed] [Google Scholar]
- 52.Shima, S., Y. Igarashi, and T. Kodama. 1993. Purification and properties of 2 truncated endoglucanases produced in Escherichia coli harboring Clostridium cellulolyticum endoglucanase gene celCCD. Appl. Microbiol. Biotechnol. 38:750-754. [DOI] [PubMed] [Google Scholar]
- 53.Stetter, K. O. 1988. Archaeoglobus fulgidus, gen. nov., sp. nov., a new taxon of extremely thermophilic archaebacteria. Syst. Appl. Microbiol. 10:172-173. [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wassenberg, D., H. Schurig, W. Liebl, and R. Jaenicke. 1997. Xylanase XynA from the hyperthermophilic bacterium Thermotoga maritima: structure and stability of the recombinant enzyme and its isolated cellulose-binding domain. Protein Sci. 6:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worning, P., L. J. Jensen, K. E. Nelson, S. Brunak, and D. W. Ussery. 2000. Structural analysis of DNA sequence: evidence for lateral gene transfer in Thermotoga maritima. Nucleic Acids Res. 28:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]