Abstract
The sensitivity of 12 Frankia strains to heavy metals was determined by a growth inhibition assay. In general, all of the strains were sensitive to low concentrations (<0.5 mM) of Ag1+, AsO21−, Cd2+, SbO21−, and Ni2+, but most of the strains were less sensitive to Pb2+ (6 to 8 mM), CrO42− (1.0 to 1.75 mM), AsO43− (>50 mM), and SeO22− (1.5 to 3.5 mM). While most strains were sensitive to 0.1 mM Cu2+, four strains were resistant to elevated levels of Cu2+ (2 to 5 mM and concentrations as high as 20 mM). The mechanism of SeO22− resistance seems to involve reduction of the selenite oxyanion to insoluble elemental selenium, whereas Pb2+ resistance and Cu2+ resistance may involve sequestration or binding mechanisms. Indications of the resistance mechanisms for the other heavy metals were not as clear.
Frankia, a member of the order Actinomycetales, forms a symbiotic nitrogen-fixing association with a variety of woody dicotyledonous plants (for reviews see references 3 and 28). The members of this bacterial genus are known to be associated with over 200 species of plants representing eight plant families. These bacteria fix N2 from the atmosphere and produce a significant amount of the fixed nitrogen on the planet. Actinorhizal plants are ecologically important as pioneer community plants and have economic value in land reclamation, reforestation, and soil stabilization.
The lack of a well-established genetic system is a major obstacle in the elucidation of the mechanism of actinorhizal nitrogen fixation and plant-microbe interactions (for reviews see references 22, 23, and 25). There is a paucity of genetic markers for Frankia. Some of the most useful genetic markers include resistance to antibiotics, resistance to antimetabolites, and resistance to heavy metals. These directly selectable traits provide a mechanism for positive selection in genetic studies and are also useful in the development of cloning vectors. For example, metal resistance has been useful in the development of cloning vectors for Rhodococcus, another member of the Actinomycetales (8). Previously, we developed a growth inhibition assay that was used to screen several Frankia strains for resistance to antibiotics (27). Although several important antibiotic resistance markers were described in that study, we were interested in identifying other selectable genetic markers. Since actinorhizal plants have been used for land reclamation in strip-mined areas, we reasoned that it is possible that the bacteria are resistant to heavy metals. The purpose of this study was to extend the use of our growth inhibition assay to identify new selectable markers, resistance to heavy metals.
Frankia strains ACN1AG (14), CcI3 (29), Cc1.17 (18), CN3 (20), CpI1 succinate variant (CpI1-S) (5, 26), CpI1 propionate variant (CpI1-P) (5, 26), DC12 (1), EI5c (17), EAN1pec (16), EuI1c (2), EUN1f (14), and QA3 (12) were grown and maintained in basal growth medium with NH4Cl as the nitrogen source, as described previously (26, 27). For the heavy metal sensitivity assays, the basal growth medium contained 20 mM glucose and 20 mM succinate as the carbon and energy sources (glucose-succinate medium). For strains EUN1f, Cc1.17, CpI1-P, and QA3, the growth medium contained 5 mM propionate instead of glucose and succinate. For comparative purposes, Escherichia coli W3110, Bacillus subtilis 168, Micromonospora echinospora ATCC 15836, and Streptomyces viridochromogenes NRRL B-1511 were also used in this study.
The sensitivities of Frankia strains to heavy metals were determined by a growth inhibition plate assay that was developed initially to test for antibiotic sensitivity (27). For Cu2+, AsO21−, AsO43−, and Pb2+, the standard growth medium was replaced with a low-phosphate growth medium that contained 1 mM K2HPO4. Metal ions, including AgNO3, Na2HAsO4, NaAsO2, CdCl2, CoCl2, K2CrO4, CuCl2, NiCl2, Pb(NO3)2, K(SbO)C4H4O6, and Na2SeO2, were added to the growth media at concentrations of 0.005, 0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 1.0, 2.0, 4.0, 5.0, 6.0, 8.0, 10.0, and 20.0 mM; AsO43− was also tested at concentrations of 25 and 50 mM. For these experiments, 14- to 21-day-old cultures were used as the inocula, and the hyphae were fragmented by using a tissue homogenizer. Each inoculum (0.5 ml) was added to a 10-ml sterile Falcon tube, and 3.0 ml of 0.8% Bacto Agar (Difco) at 45°C was also added to the tube. The contents of the tube were mixed by agitation with a vortex mixer and were poured onto the surface of a plate. Most plates were incubated at 30°C; the exceptions were the plates that contained strains DC12 and EAN1pec, which were incubated at 25°C, and the plates that contained strain Cc1.17, which were incubated at 28°C.
Growth was scored after 2 weeks and was monitored for an additional 6 weeks. When this assay was used, growth could be easily scored visually. Growth was scored relative to the growth of the control by using a scale ranging from + to ++++. With some Frankia strains, growth on the 100 control plates resulted in a thick bacterial lawn. After dilution and plating, the total number of hyphal fragments was also estimated. An average of about 107 fragments was plated for each assay. Plates that had the same amount of growth as the undiluted control were scored as ++++, while plates that showed a 2-log decrease were scored as +++. Thus, the +++, ++, and + growth scores were equivalent to the growth on the 10−2, 10−4, and 10−6 control plates, respectively. A negative result indicated that there was no growth. For E. coli and B. subtilis, cells were incubated at 37°C and growth was scored on days 1 and 2. For S. viridochromogenes and M. echinospora, cells were incubated at 28°C and growth was scored on days 7 and 14.
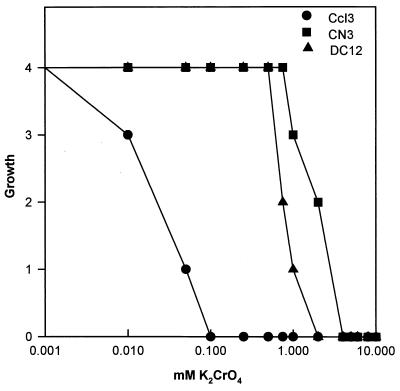
Growth was quantified relative to the growth of a control containing no metals as described above. To evaluate the levels of resistance, the following two parameters were used: MIC and maximum tolerable concentration (MTC). The MTC is the highest concentration of metal which does not affect the viable count and is used with selective media designed to promote the growth of a resistant bacterium by providing efficient counterselection for undesired bacteria (19). The values were determined by inspection after the relative growth was plotted as a function of the log of the metal concentration. The MIC was determined by determining the intersection of the survival curve with the horizontal axis (Fig. 1). In each experiment each metal was tested in duplicate, and each metal was tested in three to seven replicate experiments. In all cases, similar results were obtained in the experiments performed.
FIG. 1.
Frankia growth as a function of heavy metal (chromate) concentration. Growth was scored on a scale from 0 to ++++ as described in the text.
Heavy metal sensitivity.
Strains that showed resistance to metals as determined by the plate assay (Tables 1 and 2) were also resistant to the same metals in broth cultures (data not shown). All of the Frankia strains were sensitive to Ag1+, Ni2+, Cd2+, SbO21−, and AsO31−. Compared to the control bacteria, a few Frankia strains had high MICs (≥0.4 mM) for Co2+ and were considered cobalt resistant. Most Frankia strains were resistant to AsO43−, Pb2+, SeO22−, and CrO42−. Several of these metal resistance properties and the effect of Cu2+ are described below.
TABLE 1.
MICs of 10 heavy metals for 12 Frankia strains and other organisms
| Strain or organism | MIC (mM) ofa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AgNO3 | Na2HAsO4 | NaAsO2 | CdCl2 | CoCl2 | K2CrO4 | NiCl2 | Pb(NO3)2 | K(SbO)C4H4O6 | Na2SeO2 | |
| ACN1AG | <0.05 | >50 | 0.5 | 0.25 | 0.5 | 1.0 | 0.3 | 8.0 | 0.75 | 0.5 |
| Cc1.17 | <0.05 | >50 | 0.2 | 0.1 | 1.0 | 1.0 | 0.5 | 6.0 | 0.25 | 2.0 |
| CcI3 | <0.05 | 50 | 0.1 | 0.3 | 0.3 | 0.1 | 0.3 | 6.0 | 0.25 | 0.3 |
| CN3 | <0.05 | 40 | 0.1 | 0.4 | 0.4 | 1.75 | 0.3 | 8.0 | 0.3 | 3.5 |
| CpI1-P | <0.05 | 50 | 0.25 | 0.25 | 0.5 | 1.75 | 0.3 | 7.0 | 0.25 | 0.5 |
| CpI1-S | <0.05 | 30 | 0.1 | 0.25 | 0.3 | 1.5 | 0.3 | 5.0 | 0.5 | <0.5 |
| DC12 | <0.05 | 50 | 0.1 | 0.4 | 0.5 | 1.5 | 0.3 | 6.0 | 0.25 | 1.5 |
| EAN1pec | <0.05 | 30 | 0.1 | 0.4 | 0.3 | 1.0 | 0.2 | 8.0 | 0.25 | 0.3 |
| EI5c | <0.05 | 25 | 0.1 | <0.1 | 0.3 | 1.0 | 0.3 | 5.0 | 0.25 | 2.5 |
| EuI1c | <0.05 | 5 | 0.05 | 0.25 | 0.3 | 1.0 | 0.2 | 8.0 | 0.5 | 2.0 |
| EUN1f | <0.05 | >50 | 0.1 | 0.3 | 0.3 | 1.0 | 0.3 | 6.0 | 0.25 | 2.0 |
| QA3 | <0.05 | >50 | 0.5 | <0.1 | 0.1 | 0.5 | 0.3 | 8.0 | 0.25 | 0.3 |
| B. subtilis | <0.05 | 30 | 3.0 | <0.1 | 0.2 | 0.3 | 0.2 | 3.0 | 1.0 | 1.0 |
| E. coli | <0.05 | 10 | 3.0 | 0.3 | 0.2 | <0.05 | 0.2 | 2.0 | 2.0 | 0.5 |
| M. echinospora | <0.05 | 10 | 0.5 | 0.1 | 0.2 | 1.0 | 0.2 | 3.0 | 0.25 | 1.5 |
| S. viridochromogenes | <0.05 | 5 | 0.3 | 0.1 | 0.2 | 1.5 | 0.3 | 3.0 | 0.25 | 0.5 |
MICs were determined as described in the text.
TABLE 2.
MTCs of nine heavy metals for 12 Frankia strains and other organisms
| Strain or organism | MTC (mM) ofa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Na2HAsO4 | NaAsO2 | CdCl2 | CoCl2 | K2CrO4 | NiCl2 | Pb(NO3)2 | K(SbO)C4H4O6 | Na2SeO2 | |
| ACN1AG | 0.5 | 0.1 | 0.1 | 0.1 | 0.5 | 0.1 | 6.0 | 0.1 | 0.1 |
| Cc1.17 | 25 | 0.05 | 0.05 | 0.1 | 0.5 | 0.1 | 4.0 | 0.1 | 0.1 |
| CcI3 | 10 | <0.01 | 0.1 | 0.1 | <0.01 | 0.1 | 4.0 | 0.1 | <0.1 |
| CN3 | 10 | 0.01 | 0.1 | 0.1 | 1.0 | 0.1 | 4.0 | 0.1 | 0.5 |
| CpI1-P | 10 | 0.05 | <0.1 | 0.1 | 1.0 | 0.1 | 3.0 | 0.1 | 0.1 |
| CpI1-S | 1.0 | 0.01 | <0.1 | 0.1 | 0.5 | 0.1 | 2.0 | 0.1 | 0.1 |
| DC12 | 10.0 | <0.01 | <0.01 | 0.1 | 0.5 | 0.1 | 4.0 | <0.1 | 0.5 |
| EAN1pec | 10.0 | 0.05 | 0.05 | 0.1 | 0.5 | 0.1 | 4.0 | 0.1 | 0.1 |
| EI5c | 5.0 | 0.05 | <0.1 | 0.1 | 0.1 | 0.1 | 3.0 | <0.1 | 0.5 |
| EuI1c | <1.0 | <0.01 | 0.1 | <0.1 | 0.05 | 0.1 | 5.0 | 0.1 | 0.5 |
| EUN1f | 10.0 | 0.05 | <0.1 | 0.1 | 0.1 | 0.1 | 4.0 | 0.1 | 0.1 |
| QA3 | 1.0 | 0.1 | <0.05 | <0.05 | 0.25 | <0.1 | 6.0 | <0.01 | 0.1 |
| B. subtilis | 25.0 | >1.0 | <0.1 | 0.1 | 0.05 | 0.1 | 2.0 | 0.5 | 0.5 |
| E. coli | 5.0 | >1.0 | <0.1 | 0.1 | <0.05 | 0.1 | 1.0 | 1.5 | 0.1 |
| M. echinospora | 5.0 | 0.1 | <0.1 | <0.1 | 0.5 | <0.1 | 1.0 | 0.1 | 1.0 |
| S. viridochromogenes | <1.0 | 0.1 | <0.1 | <0.1 | 0.5 | 0.1 | 1.0 | 0.1 | 0.1 |
MTCs were determined as described in the text.
CrO42− resistance.
Most of the isolates were resistant to elevated levels of CrO42− (Table 1). The MICs for two isolates, CcI3 and EAN1pec, were <0.5 mM, and these isolates were considered chromate sensitive. For the chromate-resistant isolates the CrO42− MICs ranged from 1.0 to 1.75 mM. The control bacteria E. coli and B. subtilis were chromate sensitive, while the actinomycetes M. echinospora and S. viridochromogenes were chromate resistant. When observed by phase-contrast microscopy, these bacteria exhibited no obvious morphological or structural changes (data not shown). As a mechanism of resistance to CrO42− some organisms reduce the highly soluble oxyanion Cr(VI) to the less toxic cationic form Cr(III), which readily precipitates (6, 19, 24). No precipitate was observed with resistant strains that were grown in medium containing chromate. The amount of Cr(VI) in the spent growth medium was determined by measuring the absorbance at 380 nm. Cr(VI) absorbs light at 380 nm, while Cr(III) does not. The measurements showed that there was no change in the amount of Cr(VI) during growth. These results suggest that CrO42− resistance does not occur via a chromate reduction mechanism.
SeO22− resistance.
One-half of the Frankia isolates tested were resistant to elevated levels of SeO22− (Table 2). Isolates for which this metalloid had an MIC of <0.5 mM were considered selenite sensitive, while the SeO22− MICs for selenite-resistant isolates ranged from 1.5 to 3.5 mM. The control bacteria E. coli and S. viridochromogenes were selenite sensitive, while B. subtilis and M. echinospora were selenite resistant. Frankia isolates that were resistant to elevated levels of selenite formed red colonies on the surface of the growth medium. The red form is elemental selenium, an insoluble, generally less toxic state of the element. In liquid cultures, a red precipitate was found to be associated with cells (data not shown). Frankia strains resistant to SeO22− formed globular structures associated with their hyphae. These results suggest that the selenium oxyanion is reduced to Se0, which remains associated with the cells. Selenite resistance is probably due to reduction of colorless soluble SeO22− to an insoluble red form of Se0, which is much less toxic than selenite. This is a common mechanism of selenite resistance. In B. subtilis, an inducible detoxification system reduces selenite to elemental selenium (9). Rhodobacter sphaeroides and Rhodospirillum rubrum exhibit intracellular oxyanion reduction. R. sphaeroides deposits the metal in the cytoplasmic membrane (21), while R. rubrum expels the elemental selenium across the cytoplasmic membrane and the cell wall (13). From our results, it is not clear whether selenite reduction occurs intracellularly, extracellularly, or at the Frankia membrane.
Cu2+ resistance.
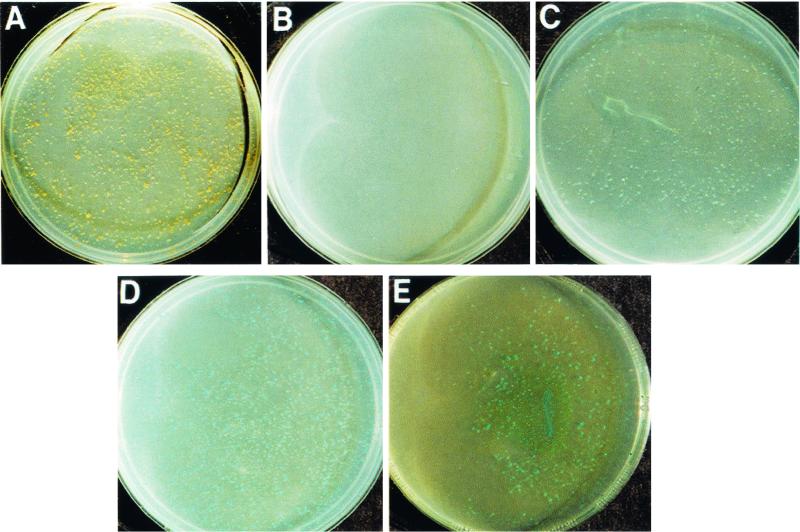
An unusual but consistent pattern of resistance was observed with Cu2+ (Fig 2). With strains DC12, EuI1c, and CN3, growth occurred in the presence of elevated levels of Cu2+ (2 to 5 mM and even up to 20 mM with strain CN3), but no growth was observed in the presence of moderate levels of Cu2+ (0.5 to 1.0 mM for EuI1c and CN3; 1.0 mM for DC12). None of the other Frankia strains tested grew in the presence of >0.1 mM CuCl2 (data not shown). For B. subtilis, S. viridochromogenes, and M. echinospora the MIC of Cu2+ was 0.2 mM, while for E. coli the MIC was 0.3 mM. Similar patterns of sensitivity and resistance were observed with CuSO4 (data not shown). Blue colonies formed on the surface of medium containing Cu2+ (Fig. 2). In some cases, the growth medium surrounding the colonies appeared to be lighter blue or white. This observation suggests that colonies bound or absorbed the Cu2+ in the medium, possibly by producing a diffusible binding compound. Several bacteria have this type of Cu2+ resistance mechanism, and the genes encoding resistance are plasmid borne or located on the chromosome (4, 7, 10, 24). In broth medium, the Cu2+ was also found to be associated with Frankia cells. As determined by phase-contrast microscopy, Cu2+-resistant Frankia strains formed unusual globular structures that were associated with their hyphae (data not shown). All of the Cu2+-resistant strains either formed ineffective nodules (EuI1c) or were members of the atypical Frankia group (DC12 and CN3) containing organisms which are unable to reinfect host plants.
FIG. 2.
Frankia strain CN3 colonies grown on succinate-glucose-NH4Cl low-phosphate medium containing CuCl2. Plates containing CuCl2 at the following concentrations were incubated for 35 days at 30°C: no CuCl2 (control) (A), 0.5 mM CuCl2 (B), 2.0 mM CuCl2 (C), 5.0 mM CuCl2 (D), and 10.0 mM CuCl2 (E).
Pb2+ resistance.
Several Frankia isolates were resistant to elevated levels of Pb(NO3)2, and the range of MICs for these strains was modest (Table 1). Compared to the four other bacteria tested, Frankia strains were more resistant to Pb2+. The MIC of Pb(NO3)2 for the most sensitive Frankia isolates was 5 mM, while for the most resistant isolates 8 mM Pb(NO3)2 was required to inhibit growth. Most of the Frankia lead-resistant strains had MTC values of ≥4 mM, while two strains (ACN1AG and QA3) had a Pb2+ MTC of 6 mM. In broth media, Pb(NO3)2 forms a cloudy solution. Growth of the resistant isolates cleared the media. When Pb2+ was present in the growth medium, resistant Frankia isolates produced an amorphous matrix that was associated with the hyphae.
Our results suggest that Pb2+ resistance and Cu2+ resistance may result from a sequestration or binding mechanism. Cu2+ and Pb2+ appear to bind to material on the cell surface. Polysaccharides or proteins are potential binding sites. Pb2+-resistant strains of Staphylococcus aureus and Citrobacter freundii accumulate lead as Pb2+ phosphate (15, 16). Cells of Streptomyces sp. also accumulate Pb2+ (11). Cu2+ is accumulated in the periplasm of Pseudomonas by periplasmic CopA and CopB proteins, which also causes the bacterial colonies to turn blue (7, 24). Our results also suggest that Cu2+ is bound to the cells. Cu1+ rapidly oxidizes to Cu2+ under aerobic conditions. With some systems, Cu1+ resistance is linked with Ag1+ resistance (7, 24), but Frankia isolates were very sensitive to Ag1+.
Most of the Frankia strains were resistant to elevated levels of several heavy metals. In genetic studies, these heavy metal resistance traits should be extremely valuable as positive selection markers. Frankia strains grow slowly and require long incubation times. Compared to antibiotics, heavy metals are more stable and do not degrade over long incubation times. The high MTCs of Pb2+, CrO42−, and SeO22− (Table 2) suggest that these metals could be used as selective agents for some Frankia strains. We are currently testing the use of heavy metal resistance as a counterselection marker in conjugation experiments.
Acknowledgments
This investigation was supported in part by Hatch grant 377, by a grant from The University of New Hampshire Vice President for Research Discretionary Funds, by a faculty summer fellowship from the Graduate School of The University of New Hampshire, and by the College of Life Science and Agriculture, The University of New Hampshire-Durham. A summer undergraduate research fellowship from the University of New Hampshire-Durham supported M.S.C.
We thank Robert Mooney for his help with the photography and Jennifer Pinard and Julia Burrows for their contributions during the initial stages of this project.
Footnotes
This is scientific contribution number 2076 from the New Hampshire Agricultural Experiment Station.
REFERENCES
- 1.Akkermans, A. D. L., F. Hafeez, W. Roelofsen, A. H. Chaudhary, and R. Baas. 1984. Ultrastructure and nitrogenase activity of Frankia grown in pure culture and in actinorhizae of Alnus, Colletia and Datisca spp., p. 311-319. In C. Vegger and W. E. Newton (ed.), Advances in nitrogen fixation research. Nyhoff/Junk Publishers, The Hague, The Netherlands.
- 2.Baker, D., W. Newcomb, and J. G. Torrey. 1980. Characterization of an ineffective actinorhizal microsymbiont, Frankia sp. EuI1 (Actinomycetales). Can. J. Microbiol. 26:1072-1089. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. R., and W. B. Silvester. 1993. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol. Rev. 57:293-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, N. L., D. A. Rouch, and B. T. O. Lee. 1992. Copper resistance determinants in bacteria. Plasmid 27:41-51. [DOI] [PubMed] [Google Scholar]
- 5.Callaham, D., P. Del Tridici, and J. G. Torrey. 1978. Isolation and cultivation in vitro of the actinomycete causing root nodulation in Comptonia. Science 199:899-902. [DOI] [PubMed] [Google Scholar]
- 6.Cervantes, C., J. Campos-Gracia, S. Devars, F. Gutiérrez-Corona, H. Loza-Tavera, J. C. Torres-Guzman, and R. Moreno-Sánchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25:335-347. [DOI] [PubMed] [Google Scholar]
- 7.Cervantes, C., and F. Gutrierrez-Corona. 1994. Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol. Rev. 14:121-138. [DOI] [PubMed] [Google Scholar]
- 8.Dabbs, E. R., B. Gowan, and S. J. Andersen. 1990. Nocardioform arsenic resistance plasmids and construction of Rhodococcus cloning vectors. Plasmid 23:242-247. [DOI] [PubMed] [Google Scholar]
- 9.Garbisu, C., S. Gonzalez, W.-H. Yang, B. C. Yee, D. L. Carlson, A. Yee, N. R. Smith, R. Otero, B. B. Buchanan, and T. Leighton. 1995. Physiological mechanisms regulating the conversion of selenite to elemental selenium by Bacillus subtilis. Biofactors 5:29-37. [PubMed] [Google Scholar]
- 10.Gilotra, U., and S. Srivastava. 1997. Plasmid-encoded sequestration of copper by Pseudomonas pickettii strain US321. Curr. Microbiol. 34:378-381. [DOI] [PubMed] [Google Scholar]
- 11.Golab, Z., B. Orlowska, M. Glubiak, and K. Olejnik. 1990. Uranium and lead accumulation in cells of Streptomyces sp. Acta Microbiol. Pol. 39:177-188. [PubMed] [Google Scholar]
- 12.Hafeez, F., A. D. L. Akkermans, and A. H. Chaudhary. 1984. Morphology, physiology, and infectivity of two Frankia isolates, An1 and An2 from root nodules of Alnus nitida. Plant Soil 78:45-59. [Google Scholar]
- 13.Kessi, J., M. Ramuz, E. Wehrli, M. Spycher, and R. Bachofen. 1999. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 65:4734-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalonde, M., H. E. Calvert, and S. Pine. 1981. Isolation and use of Frankia strains in actinorhizae formation, p. 296-299. In A. H. Gibson and W. E. Newton (ed.), Current perspectives in nitrogen fixation. Australian Academy of Science, Canberra, Australia.
- 15.Levinson, H. S., I. Mahler, P. Blackwelder, and T. Hood. 1996. Lead resistance and sensitivity in Staphylococcus aureus. FEMS Microbiol. Lett. 145:421-425. [DOI] [PubMed] [Google Scholar]
- 16.Levinson, H. S., and I. Mahler. 1998. Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus. FEMS Microbiol. Lett. 161:135-138. [DOI] [PubMed] [Google Scholar]
- 17.Lumini, E., and M. Bosco. 1996. PCR-restriction fragment length polymorphism identification and host range of single-spore isolates of the flexible Frankia sp. strain UFI 132715. Appl. Environment. Microbiol. 62:3026-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meesters, T. M., S. T. van Genesen, and A. D. L. Akkermans. 1985. Growth, acetylene reduction activity and localization of nitrogenase in relation to vesicle formation in Frankia strains Cc11.7 and Cp1.2. Arch. Microbiol. 143:137-142. [Google Scholar]
- 19.Mergeay, M. 1995. Heavy metal resistances in microbial ecosystems, p. 6.1.7.1-6.1.7.17. In A.D.L. Akkermans, J. D. van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Press, Dordrecht, The Netherlands.
- 20.Mirza, M. S., W. M. Akkermans, and A. D. L. Akkermans. 1994. PCR-amplified 16S rRNA sequence analysis to confirm nodulation of Datisca cannabina L. by the endophyte of Coriaria nepalensis Wall. Plant Soil 160:147-152. [Google Scholar]
- 21.Moore, M. D., and S. Kaplan. 1992. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 174:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullin, B. C., and C. S. An. 1990. The molecular genetics of Frankia, p. 195-214. In C. R. Schwintzer and J. D. Tjepkema (ed.), The biology of Frankia and actinorhizal plants. Academic Press, San Diego, Calif.
- 23.Normand, P., and M. Lalonde. 1986. The genetics of actinorhizal Frankia: a review. Plant Soil 90:429-453. [Google Scholar]
- 24.Silver, S., and L. T. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 25.Simonet, P., P. Normand, A. M. Hirsch, and A. D. L. Akkermans. 1990. The genetics of the Frankia-actinorhizal symbiosis, p. 77-109. In P. M. Gresshoff (ed.), Molecular biology of symbiotic nitrogen fixation. CRC Press, Inc., Boca Raton, Fla.
- 26.Tisa, L., M. McBride, and J. C. Ensign. 1983. Studies of growth and morphology of Frankia strains EAN1pec, EuI1c, CpI1 and ACN1AG. Can. J. Bot. 61:2768-2773. [Google Scholar]
- 27.Tisa, L. S., M. S. Chval, G. D. Krumholz, and J. Richards. 1999. Antibiotic resistance patterns of Frankia strains. Can. J. Bot. 77:1257-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall, L. G. 2000. The actinorhizal symbiosis. J. Plant Growth Regul. 19:167-182. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Z., M. F. Lopez, and J. G. Torrey. 1984. A comparison of cultural characteristics and infectivity of Frankia isolates from root nodules of Casuarina species. Plant Soil 78:79-90. [Google Scholar]