Abstract
Understanding the behavior of Cryptosporidium oocysts in the environment is critical to developing improved watershed management practices for protection of the public from waterborne cryptosporidiosis. Analytical methods of improved specificity and sensitivity are essential to this task. We developed a nested PCR-restriction fragment length polymorphism assay that allows detection of a single oocyst in environmental samples and differentiates the human pathogen Cryptosporidium parvum from other Cryptosporidium species. We tested our method on surface water and animal fecal samples from the Wachusett Reservoir watershed in central Massachusetts. We also directly compared results from our method with those from the immunofluorescence microscopy assay recommended in the Information Collection Rule. Our results suggest that immunofluorescence microscopy may not be a reliable indicator of public health risk for waterborne cryptosporidiosis. Molecular and environmental data identify both wildlife and dairy farms as sources of oocysts in the watershed, implicate times of cold water temperatures as high-risk periods for oocyst contamination of surface waters, and suggest that not all oocysts in the environment pose a threat to public health.
Cryptosporidium parvum is an intracellular protozoan parasite responsible for an acute gastrointestinal and, less frequently, respiratory infection in humans that is self-limiting in immunocompetent people but prolonged and potentially life-threatening for the immunocompromised population (31). Gastrointestinal cryptosporidiosis is characterized by watery diarrhea, abdominal pain, low-grade fever (<39°C), general malaise, weakness, fatigue, loss of appetite, nausea, vomiting, and weight loss (10, 34). Symptomatic infection may last from a few days to a few weeks in immunocompetent individuals, although extreme cases of up to 12 weeks of severe diarrhea have been reported (34). Cryptosporidiosis is particularly serious for immunosuppressed people because no curative treatment currently exists.
The existence of multiple species of Cryptosporidium, including C. parvum, C. muris, C. felis, C. wrairi, and C. andersoni (mammals), C. baileyi and C. meleagridis (birds), C. serpentis (reptiles), and C. nasorum (fish), has been suggested on the basis of oocyst morphology, host specificity, infectivity, and 18S rRNA sequence comparisons (33, 34, 36). There is some uncertainty with respect to the validity of these taxa. For example, C. wrairi appears to be a strain of C. parvum that is isolated from guinea pigs, while C. andersoni is a recently proposed species characterized by C. muris-like oocysts that infect cattle (21). Classifications based on host species may not be appropriate given that C. felis, associated with cryptosporidial infection in cats, was recently isolated from a cow (4). There are now multiple reports of species other than C. parvum infecting humans, particularly immunocompromised people (12, 17, 25, 27, 28, 39). Due to the confusion surrounding the taxonomy of Cryptosporidium, it is difficult to conclusively assess the human public health threat attributable to Cryptosporidium species other than C. parvum.
Numerous outbreaks of waterborne cryptosporidiosis in the United States have occurred over the past 20 years (6, 31) in both rural and urban areas, spanning the nation from Pennsylvania to Oregon. Cryptosporidium species are a threat to water supplies because they are resistant to chlorine disinfection, small (∼5 μm in diameter) and thus difficult to filter, and harbored by many animal species (10). The largest waterborne outbreak in U.S. history occurred in Milwaukee, Wis., in the spring of 1993 and affected an estimated 403,000 people served by the Milwaukee Water Works. The Wisconsin Division of Health found that the outbreak was responsible for the premature deaths of at least 69 individuals, most of whom were human immunodeficiency virus positive. The sources of oocyst contamination, although not identified conclusively, were suspected to include cattle waste, slaughterhouse waste, and human sewage. The combination of severe spring rains and snowmelt runoff that occurred just prior to the outbreak could have carried oocysts from these suspected sources into Lake Michigan and subsequently into the intakes of the Milwaukee Water Works treatment plants. Treatment processes at the South Milwaukee Water Works plant included the following: chlorine and permanganate addition at the raw water intake, polyaluminum chloride coagulation, rapid mixing, flocculation, sedimentation, rapid sand filtration, chlorination, and fluoride addition. Despite such thorough water treatment, the turbidity of the South Milwaukee Water Works plant effluent exceeded the 1993 Environmental Protection Agency (EPA) limit of 1.0 nephelometric turbidity unit(s) (NTU), peaking at 1.7 NTU in late March 1993 (10, 23, 31)
This episode of Cryptosporidium oocysts passing through a water treatment plant bolsters the argument that successful public health measures must include appropriate watershed management. Improved watershed management requires a better understanding of the behavior of Cryptosporidium oocysts in the environment, and this in turn requires improved analytical detection methods. We now report a sensitive and specific nested-PCR-restriction fragment length polymorphism (PCR-RFLP) assay for detection of Cryptosporidium oocysts in environmental samples. This nested PCR targets a 434-bp hypervariable region of the 18S rRNA gene, a multicopy gene (20 copies per oocyst) ideal for species identification. Application to surface water and animal fecal samples from the Wachusett Reservoir watershed in central Massachusetts confirms the method's high degree of sensitivity and specificity and provides new hypotheses regarding the control of Cryptosporidium oocyst contamination in surface waters.
Molecular methods for detection of Cryptosporidium oocysts in wastewater and surface water have been reported (22, 38, 40), and we have extended these studies with the development of a novel assay and its application to the investigation of sources and species of oocysts in a geographic area that has not been previously described. The Wachusett Reservoir, a drinking water source for Boston, Mass., and surrounding cities, has recently been the subject of litigation concerning appropriate measures to protect against waterborne parasites such as C. parvum and Giardia lamblia. Our goal of understanding the sources, species, and seasonal trends of oocyst contamination in watersheds will contribute to the development of better watershed management practices to prevent waterborne outbreaks of cryptosporidiosis in drinking water watersheds.
MATERIALS AND METHODS
Oocysts.
GCH1 C. parvum oocysts were a kind gift of Giovanni Widmer at Tufts University School of Veterinary Medicine in North Grafton, Mass.
Surface water sample selection.
Sampling sites in the Wachusett Reservoir watershed in central Massachusetts (Fig. 1) were chosen to encompass a variety of potential sources of Cryptosporidium contamination. Surface water sites (and their suspected source of contamination) included Stillwater River (wildlife); Quinapoxet River (wildlife); Gates Brook (sewage); and two small, unnamed brooks, designated Brook JF and Brook SF, downgradient from dairy farms (agricultural runoff). Stillwater River and Quinapoxet River were sampled monthly from February 2000 to January 2001, often side-by-side with the Metropolitan District Commission (MDC) of the Commonwealth of Massachusetts. The MDC adhered to the Information Collection Rule (9), using conventional yarn-wound filters and an immunofluorescence microscopy assay (IFA) for oocyst detection. Gates Brook, Brook JF, and Brook SF were sampled periodically, but not as frequently, from March 1999 to January 2001.
FIG. 1.
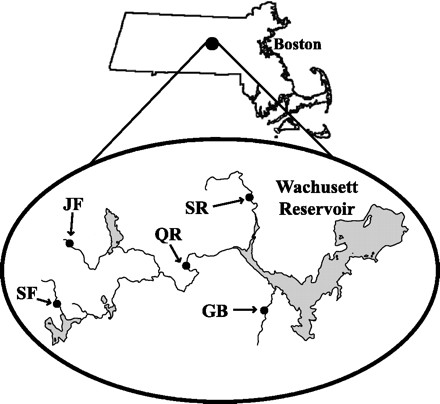
Schematic of the Wachusett Reservoir watershed sampling sites in central Massachusetts. Abbreviations: SR, Stillwater River; QR, Quinapoxet River; GB, Gates Brook; SF, Brook SF; JF, Brook JF. Suspected sources of oocyst contamination include wildlife (SR and QR), sewage (GB), and agricultural runoff from dairy farms (SF and JF).
Sample collection.
Surface waters were filtered through Gelman Envirochek Sampling Capsules (Pall Gelman Sciences, Inc., Ann Arbor, Mich.) at 1 to 2 liters min−1 according to manufacturer's recommendations. During filtration, water temperature was recorded. Filtration continued for 1 h or until the backpressure exceeded the filter rating (30 lb/in2 [psi]), whichever came first. Typically, 40 to 80 liters of water were filtered. Filters were transported to the laboratory on ice, and samples were eluted according to manufacturer's recommendations within 36 h of sample collection. Eluted solids were resuspended in 10 ml of laboratory-grade water (Milli-Q System; Millipore Corp., Bedford, Mass.) for each 0.5 ml of solids, stored at 4°C, and processed within 24 h.
Fecal samples were collected in sterile 50-ml polypropylene tubes and transported to the laboratory on ice. Fecal samples were suspended in 10 ml of laboratory-grade water for each 0.5 ml of solids, stored at 4°C, and processed within 24 h of collection.
Immunomagnetic separation of oocysts.
Oocysts were purified from water and fecal samples by using immunomagnetic separation (IMS) with the Crypto-Scan IMS kit (ImmuCell, Portland, Maine) according to the recommendations of the manufacturer. After being dissociated from magnetic beads, oocysts were transferred to a new microcentrifuge tube and treated with 5 μl of 1 N NaOH to neutralize the pH. The oocysts were pelleted for 2 to 3 min at 16,000 × g, resuspended in 50 μl of laboratory-grade water, and stored at 4°C.
Positive and negative IMS controls were processed with each set of field samples. Positive IMS controls consisted of 9.9 ml of laboratory-grade water and 100 μl of a 104 oocyst ml−1 suspension; negative IMS controls consisted of 10 ml of laboratory-grade water. IMS controls were processed as described above.
Genomic DNA extraction.
Oocysts were lysed by adding 25 μl of IMS product to 475 μl of Tris-EDTA (TE) buffer containing 0.2 g of proteinase K liter−1 and 0.4% sodium dodecyl sulfate and incubating the mixture overnight at 45°C. Positive and negative DNA extraction controls were included for each set of field samples. Positive DNA extraction controls consisted of 25 μl of a suspension of 104 oocysts ml−1 in 475 μl of TE buffer; negative DNA extraction controls consisted of 25 μl of laboratory-grade water in 475 μl of TE buffer. DNA was extracted several times with phenol-chloroform, precipitated with 0.2 M NaCl and 2 volumes of absolute ethanol, and resuspended in 30 μl of TE buffer.
Nested PCR assay.
PCR amplification was performed in a 50-μl volume containing 10 mM Tris-HCl, 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2, 0.015 mM concentrations of each deoxynucleoside triphosphate (Perkin-Elmer, Wellesley, Mass.), 0.2 μM concentrations of each primer, and 2 U of Taq DNA polymerase (Promega Corp., Madison, Wis.). The initial amplification reaction was performed with 15 μl of DNA template, and 3 μl of the initial amplification product was used as a template for the secondary PCR. Positive and negative PCR controls were included with each set of samples. For the initial amplification reaction, positive PCR controls contained 12 μl of laboratory-grade water and 3 μl of genomic C. parvum DNA (at a concentration equivalent to 104 oocysts μl−1); negative PCR controls contained 15 μl of laboratory-grade water. For the secondary amplification reaction, positive PCR controls contained 3 μl of genomic C. parvum DNA (at a concentration equivalent to 104 oocysts μl−1); negative PCR controls contained 3 μl of laboratory-grade water.
Both amplification reactions used forward and reverse oligonucleotide primers that are complementary to Cryptosporidium 18S rRNA gene sequences (Fig. 2). The initial 1,056-bp product was obtained with a forward primer, KLJ1 (5"-CCACATCTAAGGAAGGCAGC-3"), corresponding to nucleotides 389 to 408, and a reverse primer, KLJ2 (5"-ATGGATGCATCAGTGTAGCG-3"), corresponding to nucleotides 1422 to 1441 of C. parvum L16996 in GenBank (3). The final 434-bp product was obtained by using forward and reverse primers CPB-DIAGF and CPB-DIAGR, respectively (16). Cycling conditions consisted of an initial denaturation (5 min at 80°C, followed by 30 s at 98°C), 40 cycles of amplification (denaturation for 30 s at 94°C, annealing for 30 s at 53°C, and extension for 1 min at 72°C), and a final extension (10 min at 72°C). Secondary PCR products were visualized after electrophoresis on a 1.2% agarose gel stained with ethidium bromide.
FIG. 2.
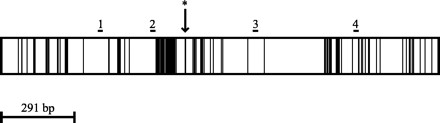
Schematic of the 1,746-bp Cryptosporidium 18S rRNA gene (based on GenBank accession no. L16996). Within the gene, dark areas are regions of sequence variability, and white areas are regions of sequence conservation. Primer binding locations are indicated above the gene (1, KLJ1; 2, CPB-DIAGF; 3, CPB-DIAGR; 4, KLJ2). An asterisk identifies the NdeI digest site.
RFLP analysis.
Digestion of amplified 18S rRNA gene products with NdeI can be used to differentiate most C. parvum isolates from other Cryptosporidium species. The 434-bp final amplicon of most C. parvum isolates (with the exception of GenBank accession numbers AF112570 and AF108860, isolates from a kangaroo and a koala in Australia, respectively, and AF112576, the dog genotype) contains a single NdeI site (Fig. 2), while the amplicons from other Cryptosporidium species (C. muris, C. baileyi, C. serpentis, and C. felis) do not. Restriction digestion was carried out in a 20-μl volume containing 10 μl of secondary PCR product, 20 U of NdeI (New England Biolabs, Beverly, Mass.), 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, and 100 μg of bovine serum albumin ml−1 and then incubated at 37°C for 1 h. Digestion products were visualized after electrophoresis on a 1.2% agarose gel stained with ethidium bromide.
Cloning.
Secondary PCR products from water or fecal samples positive for Cryptosporidium were cloned into the pGEM-T Easy Vector System (Promega) and used to transform XL1-Blue E. coli cells (Stratagene, La Jolla, Calif.). Clones were selected on Luria-Bertani (LB) agar supplemented with 100 μg of ampicillin ml−1 and cultured overnight in LB broth supplemented with 100 μg of ampicillin ml−1. Plasmid DNA was isolated from clones by using the QIAPrep Spin Miniprep Kit (Qiagen, Inc., Valencia, Calif.) and digested with NotI (New England Biolabs) to verify the presence of the secondary PCR amplicon insert. Plasmids with the insert were further digested with NdeI. All digestion products were visualized after electrophoresis on a 1.2% agarose gel stained with ethidium bromide.
Sequencing.
Representative clones of the secondary PCR products were sequenced on an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems, Foster City, Calif.) by using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit with the AmpliTaq DNA Polymerase, FS (PE Applied Biosystems). If multiple NdeI digestion patterns existed among clones from a given sample, at least one clone of each digestion pattern was sequenced. At least two clones for each positive sample were sequenced in any case and confirmed by sequencing both strands. The basic local alignment search tool (BLAST) algorithm was used to compare cloned DNA sequences with GenBank sequences and to determine the species of Cryptosporidium present in the sample (1, 3). Multiple sequence alignments and phylogenetic trees were generated with MacVector 7.0 (Genetics Computer Group, Madison, Wis.) with manual adjustment.
RESULTS
By seeding PCRs with known quantities of oocyst DNA, initial PCR amplification of the 18S rRNA gene was found to detect as few as 500 oocysts; the lower limit of detection of nested PCR was a single oocyst (Fig. 3). This detection limit assay, however, was performed under ideal conditions and did not account for the possible presence of PCR inhibitors in environmental samples. The potential for PCR inhibition was tested by processing two filters side-by-side for a single surface water source: one filter contained the surface water only, and the second filter contained the surface water seeded with 500 C. parvum oocysts. Using one-half of the eluted water pellets for IMS, one-half of the IMS products for DNA extraction, and one-thirtieth of the DNA extract for PCR, the initial PCR of the seeded sample received the DNA equivalent of 4.2 oocysts. After the secondary amplification reactions, no oocysts were detected in the surface water sample alone; oocysts were clearly detected in the spiked surface water sample (Fig. 4).
FIG. 3.
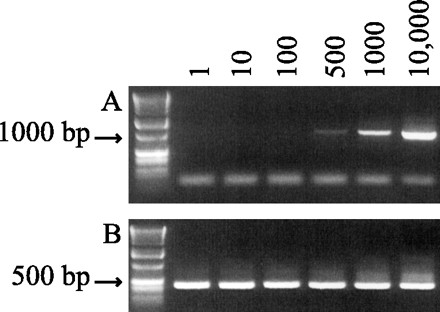
Detection limit of nested PCR assay. (A) Initial PCR products (primers KLJ1/2). (B) Secondary PCR products (primers CPB-DIAGF/R). PCRs were seeded with known quantities of DNA representative of 1 to 10,000 oocysts (as indicated at the top of each lane). Corresponding lanes on gels A and B represent the same seeded sample. The first lanes of gels A and B are molecular weight standards.
FIG. 4.
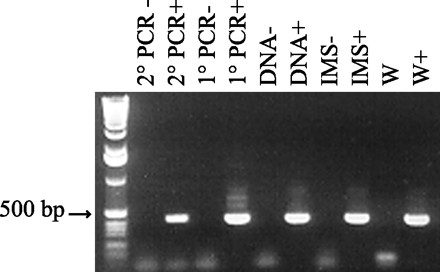
The potential for PCR inhibition was tested by seeding a surface water sample with 500 oocysts. From left to right, the lanes are as follows: molecular weight standard; negative and positive controls for secondary (2°) PCR, respectively; negative and positive controls for initial (1°) PCR, respectively; negative and positive controls for DNA extraction, respectively; negative and positive controls for IMS, respectively; surface water sample (W); seeded surface water sample (W+).
For the year spanning February 2000 to January 2001, 34 surface water samples were collected for Cryptosporidium detection and 5 (14.7%) were determined to be positive by nested PCR. In addition, 44 water samples were collected by the MDC and 5 (11.4%) were found to be positive by IFA. Table 1 includes all of the surface water samples that were determined to be positive for Cryptosporidium by either nested PCR or IFA and two additional samples analyzed in March and July of 1999. Of the seven samples determined to be positive by nested PCR, C. parvum was identified in three samples (samples 2/1/00, 4/4/00, and 11/7/00). Sample 2/1/00 was a mixed population of C. parvum and C. muris, and C. muris appeared to be more prevalent since only one of the 12 clones could be digested with NdeI (the single clone containing an NdeI site was sequenced and identified as C. parvum, and 2 of the remaining clones were identified as C. muris). C. muris and C. baileyi were identified in three and one of the seven positive samples, respectively. One positive sample could not be cloned and sequenced due to insufficient sample quantity.
TABLE 1.
Surface water samples that tested positive for Cryptosporidium spp.
| Sample (date)a | Location | Sample identification | MDCb | Molecular results
|
||
|---|---|---|---|---|---|---|
| Nested PCR | NdeI digestc | Sequence resultsd | ||||
| 3/1/99 | Brook SF | SF | NDe | + | − | 3/C. muris |
| 7/12/99 | Quinapoxet River | QR | + | + | − | 3/C. muris |
| 2/1/00 | Stillwater River | SR | ND | + | 1/+ | 1/C. parvum |
| 11/− | 2/C. muris | |||||
| 2/22/00 | Quinapoxet River | + | − | |||
| 3/7/00 | Quinapoxet River | + | ND | |||
| 4/3/00 | Quinapoxet River | + | ND | |||
| 4/4/00 | Gates Brook | GB | ND | + | 11/− | 5/C. parvum |
| 4/4/00 | Brook JF | ND | + | QNSf | QNSf | |
| 10/23/00 | Stillwater River | + | ND | |||
| 11/7/00 | Quinapoxet River | QR1.5, QR2 | − | + | 6/− | 2/C. parvum |
| 12/5/00 | Stillwater River | SR1.5, SR2 | − | + | 10/− | 6/C. baileyi |
| 12/5/00 | Quinapoxet River | + | − | |||
That is, month/day/year.
Results of MDC samples processed by IFA: +, presumptive positive for C. parvum; −, not found.
1/+, One nested PCR clone was cut with NdeI; 11/−, eleven nested PCR clones were not cut with NdeI. For samples 3/1/99 and 7/12/99, the complete nested-PCR products were not cut with NdeI (the nested-PCR clones were not digested individually).
3/C. muris, the nucleotide sequences of three nested-PCR clones were most closely related to those of C. muris.
ND, not done.
QNS, quantity not sufficient.
Agricultural and wildlife fecal samples were collected in June and August of 2000. Results are summarized in Table 2. Among wildlife samples, C. parvum was found only in fresh deer feces, and C. baileyi was identified in the feces from cormorants alone. No Cryptosporidium spp. were isolated from adult cattle on farm SF or from calves on farm JF. C. baileyi and C. muris were identified in adult cattle and in the manure pit, respectively, on farm JF.
TABLE 2.
Results of fecal sampling
| Sample (date)a | Location | Sample identification | Source | Nested PCR | NdeI digestb | Sequence resultsc |
|---|---|---|---|---|---|---|
| 6/26/00 | Farm SF | Adult cattle | − | |||
| 6/26/00 | Farm JF | Cow | Adult cattle | + | 5/− | 2/C. baileyi |
| Calves | − | |||||
| Manure | Manure pit | + | 11/− | 3/C. muris | ||
| 8/21/00 | Wachusett Reservoir | Geese | − | |||
| Deer (old)d | − | |||||
| Deer | Deer (fresh) | + | 3/− | 3/C. parvum | ||
| Geese and cormorante | − | |||||
| Cormorant | Cormorant | + | 9/− | 3/C. baileyi |
See Table 1, footnote a.
5/−, five nested PCR clones did not digest with NdeI.
2/C. baileyi, the nucleotide sequences of two nested-PCR clones were most closely related to those of C. baileyi.
Dessicated deer feces.
That is, a mixture of geese and cormorant feces.
DISCUSSION
Nested-PCR targeting the variable region of the 18S rRNA gene enabled detection of a single Cryptosporidium oocyst (Fig. 3); this compares favorably to other sensitive PCR-RFLP methods for detection of Cryptosporidium (16, 22). Given a 50% infective dose of 132 oocysts (7), our nested PCR should allow detection of oocysts in environmental samples at and below infectious levels. For all water and fecal samples that tested positive for Cryptosporidium oocysts, nested PCR was necessary for detection (i.e., no signal was detected in any sample after initial PCR amplification). Our findings suggest that single PCR, which has been used for both laboratory and environmental samples (2, 16, 19, 20, 22, 26, 32), may not be sensitive enough for detection of commonly occurring levels of oocyst contamination in the environment.
This Cryptosporidium detection assay offers a high degree of sensitivity and species-level oocyst identification. Although the assay does not provide information about oocyst viability, detection of any C. parvum oocysts in environmental samples from source water watersheds is a warning that precautionary measures should be considered to protect public health. Oocyst viability is influenced by many environmental factors, including temperature, hydration, starvation, predation, and UV exposure (8, 14, 24, 29). The presence of oocysts in the environment, even if nonviable at one time, is an indication that potentially viable oocysts may be present under different environmental conditions in the future.
We were able to detect multiple species of Cryptosporidium oocysts in water and fecal samples, including C. parvum, C. muris, and C. baileyi (Tables 1 and 2). The 434-bp secondary PCR product is ideal for species identification because it spans the most hypervariable region of the 18S rRNA gene but also includes recognizable, conserved anchors (Mitchell L. Sogin, personal communication).
U.S. EPA Method 1622 for Cryptosporidium analysis in water (35) uses IFA for detection of oocysts in environmental samples. Comparison of our results to those obtained by IFA illustrates that IFA may not be a reliable indicator of public health risk (Table 1). First, IFA results are based on visual identification of oocysts and do not classify the Cryptosporidium species. Thus, oocysts identified by IFA must be assumed to be infectious in order to protect public health. By our molecular method, we were able to identify C. muris in a sample presumed to be positive for C. parvum by IFA (sample 7/12/99), illustrating the importance of species-level oocyst detection. A second limitation of IFA is the possibility that sample debris cross-reacting with the fluorescent antibodies may lead to false-positive reports. We believe this is the most likely explanation for samples that were determined to be positive for Cryptosporidium by IFA (samples 2/22/00 and 12/5/00) but negative by our molecular method. We believe our results for these samples are true negatives because we have shown that a single oocyst can be detected under ideal circumstances (Fig. 3) and have run controls that discount the likelihood of PCR inhibitors (Fig. 4). Although we do not routinely run controls for PCR inhibitors, they should be sufficiently removed during filtration and IMS (13, 16, 30). Third, low numbers of oocysts in the environment may go undetected by IFA due to sample dilution and competition of sample debris with fluorescent antibodies. We also identified Cryptosporidium oocysts (C. parvum and C. baileyi [samples 11/7/00 and 12/5/00, respectively]) in water samples that were negative by IFA.
Although some of the differences between IFA and our molecular method may be explained by the random distribution of oocysts in the water (i.e., if the concentration of oocysts in surface water is low, one filter may trap an oocyst while another filter running simultaneously does not), our data suggest that it is possible to incorrectly estimate the public health threat for cryptosporidiosis with conventional IFA analyses. Not all Cryptosporidium species in the environment are C. parvum. In fact, C. baileyi and C. muris were identified more often than C. parvum in the water samples analyzed in the present study (Table 1). Of the wildlife fecal samples analyzed (Table 2), C. parvum oocysts were found in fresh deer stool only. In contrast, C. baileyi was found in fecal samples from cormorants and adult dairy cattle, and C. muris was identified in a dairy farm manure pit. To our knowledge, infection by C. baileyi in cattle has never been described. We speculate that the feed may have been contaminated with C. baileyi by birds on the farm and that the oocysts passed transiently through the cattle (the cattle were passing normal feces). The fact that no C. parvum oocysts were isolated from the dairy farm cattle or the manure pit is especially pertinent since dairy cattle are considered a major source of infectious oocysts. Also relevant is the fact that C. muris (and not C. parvum) was identified in the manure pit on farm JF and in Brook SF (where the suspected source of oocysts is agricultural runoff) in sample 3/1/99. A recent study (21) proposed that the large form of Cryptosporidium (previously thought to be C. muris) infecting the abomasum of cattle is a new species, C. andersoni; however, the lack of 18S rRNA sequence data in GenBank precludes the identification of Cryptosporidium oocysts in our samples as C. andersoni instead of C. muris.
Phylogenetic analysis of the sequence data derived from our water and fecal samples indicate that the oocysts isolated from both wildlife and dairy farm fecal samples are closely related to the oocysts found in surface waters in the Wachusett Reservoir watershed (Fig. 5). The fact that we found a mixed population of oocysts in sample 2/1/00 at Stillwater River (C. parvum and C. muris) suggests that either one source may harbor multiple oocyst species or that multiple sources exist for this site. Because wildlife are abundant in the area, the existence of multiple sources is plausible. C. muris appeared to be more abundant than C. parvum at this site (as indicated by the fact that only 1 of the 12 nested-PCR clones had the C. parvum-like NdeI restriction pattern). Additional studies to determine whether wildlife are a significant source of oocysts pathogenic for people are therefore needed.
FIG. 5.
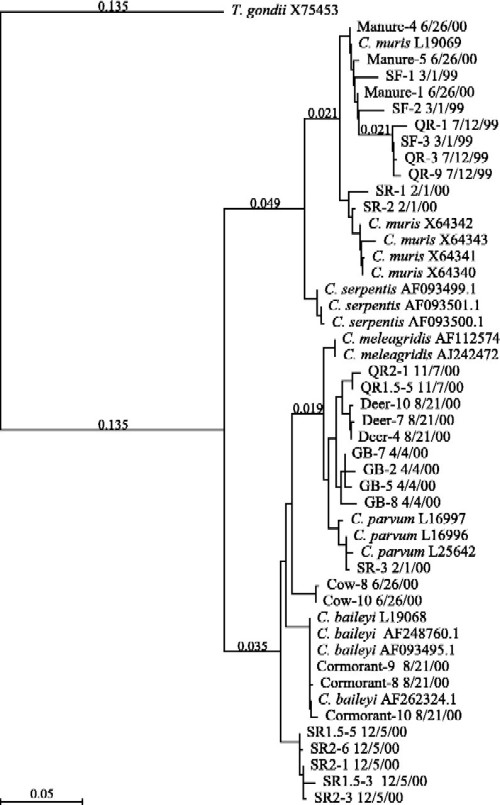
Phylogenetic relationships among field samples and GenBank Cryptosporidium sequences (2, 5, 11, 15, 18, 31-33, 35-37). Phylogeny based upon multiple sequence alignments performed with MacVector 7.0 by using the Tamura-Nei algorithm. A distance of 0.10 indicates a 10% difference between sequences. Field samples are labeled as follows: source-clone no. date sampled (e.g., “Manure-4 6/26/00” denotes clone 4 of manure sampled on 26 June 2000).
Our data also indicate a seasonal pattern in oocyst contamination of surface waters. Water samples positive for oocysts were limited to late fall, winter, and early spring (Table 1). No oocysts were found in water samples between mid-April and mid-October with one exception (sample 7/12/99). High-risk periods for oocyst contamination are often thought to be linked to calving season in late winter and early spring, but the detection of oocysts in late fall and early winter suggests that additional factors are operating. The observed seasonal pattern correlates well with temperature; the maximum water temperature at which positive samples were found during 2000 was 9°C. Given that wildlife and dairy farm fecal samples collected in the summer (when water temperatures were >9°C) were positive for Cryptosporidium oocysts, it appears that oocysts are present in the watershed year round. Although hydrologic factors are often and probably correctly thought to influence oocyst transport to streams, it is also possible that grazers or predators may limit surface water populations of Cryptosporidium in the summer. Possibly other chemical or biotic factors limit oocyst survival in surface waters in warmer temperatures.
The nested-PCR protocol described here can be helpful in the identification of sources and species of oocysts in watersheds, as well as the times of year when surface waters are most susceptible to oocyst contamination. Such information will aid in the development and implementation of the most appropriate watershed management policies and water treatment technologies to protect the public from exposure to C. parvum.
Acknowledgments
We are grateful for materials and technical assistance provided by Giovanni Widmer, Tufts University School of Veterinary Medicine, North Grafton, Mass. We thank the Metropolitan District Commission of the Commonwealth of Massachusetts for their generous help in identifying sampling locations and sharing the results of their IFA studies.
This work was supported by a National Science Foundation Graduate Research Fellowship, a U.S. EPA “Science to Achieve Results” (STAR) Fellowship, and a feasibility project funded by the National Institute for Environmental Health Sciences.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awad-el-Kariem, F. M., D. C. Warhurst, and V. McDonald. 1994. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. J. Parasitol. 109:19-22. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. A., M. S. Boguski, D. J. Lipman, J. Ostell, and B. F. F. Ouellette. 1998. GenBank. Nucleic Acids Res. 26:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornay-Llinares, F. J., A. J. da Silva, I. N. S. Moura, P. Myjak, H. Pietkiewicz, W. Kruminis-Lozowska, T. K. Graczyk, and N. J. Pieniazek. 1999. Identification of Cryptosporidium felis in a cow by morphologic and molecular methods. Appl. Environ. Microbiol. 65:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, J., M. D. Collins, V. McDonald, and D. E. Thompson. 1992. PCR cloning and nucleotide sequence determination of the 18S rRNA genes and internal transcribed spacer 1 of the protozoan parasites Cryptosporidium parvum and Cryptosporidium muris. Biochim. Biophys. Acta 1131:317-320. [DOI] [PubMed]
- 6.Deneen, V. C., P. A. Belle-Isle, C. M. Taylor, L. L. Gabriel, J. B. Bender, J. H. Wicklund, C. W. Hedberg, and M. T. Osterholm. 1998. Outbreak of cryptosporidiosis associated with a water sprinkler fountain--Minnesota, 1997. Morb. Mortal. Wkly. Rep. 47:856-859. [PubMed] [Google Scholar]
- 7.DuPont, H. L., C. L. Chappell, C. R. Sterling, P. C. Okhuysen, J. B. Rose, and W. Jakubowski. 1995. The infectivity of Cryptosporidium parvum in healthy volunteers. N. Engl. J. Med. 332:855-859. [DOI] [PubMed] [Google Scholar]
- 8.Fayer, R., and T. Nerad. 1996. Effects of low temperatures on viability of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 62:1431-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Federal Register. Information collection rule. Fed. Regist. 61:24354-24388.
- 10.Fox, K. R., and D. A. Lytle. 1996. Milwaukee's crypto outbreak: investigation and recommendations. J. Am. Water Works Assoc. 88:87-94. [Google Scholar]
- 11.Guerrant, R. L. 1997. Cryptosporidiosis: an emerging, highly infectious threat. Emerg. Infect. Dis. 57:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyot, K., A. Follet-Dumoulin, E. Lelièvre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallier-Soulier, S., and E. Guillot. 1999. An immunomagnetic separation polymerase chain reaction assay for rapid and ultra-sensitive detection of Cryptosporidium parvum in drinking water. FEMS Microbiol. Lett. 176:285-289. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. C., C. E. Enriquez, I. L. Pepper, T. L. Davis, C. P. Gerba, and J. B. Rose. 1997. Survival of Giardia, Cryptosporidium, poliovirus, and Salmonella in marine waters. Water Sci. Technol. 35:261-268. [Google Scholar]
- 15.Johnson, D. W., N. J. Pieniazek, and J. B. Rose. 1993. DNA probe and PCR detection of Cryptosporidium parvum compared to immunofluorescence assay. Water Sci. Technol. 27:77-84. [Google Scholar]
- 16.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsumata, T., D. Hosea, I. G. Ranuh, S. Uga, T. Yanagi, and S. Kohno. 2000. Short report: possible Cryptosporidium muris infection in humans. Am. J. Trop. Med. Hyg. 62:70-72. [DOI] [PubMed] [Google Scholar]
- 18.Kilani, R. T., and W. M. Wenman. 1994. Geographical variation in 18S rRNA gene sequence of Cryptosporidium parvum. Int. J. Parasitol. 24:303-306. [DOI] [PubMed] [Google Scholar]
- 19.Laxer, M. A., B. K. Timblin, and R. J. Patel. 1991. DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 45:688-694. [DOI] [PubMed] [Google Scholar]
- 20.Leng, X., D. A. Mosier, and R. D. Oberst. 1996. Differentiation of Cryptosporidium parvum, C. muris, and C. baileyi by PCR-RFLP analysis of the 18S rRNA gene. Vet. Parasitol. 62:1-7. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay, D. S., S. J. Upton, D. S. Owens, U. M. Morgan, J. R. Mead, and B. L. Blagburn. 2000. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporidiae) from cattle, Bos taurus. J. Eukaryot. Microbiol. 47:91-95. [DOI] [PubMed] [Google Scholar]
- 22.Lowery, C. J., J. E. Moore, B. C. Millar, D. P. Burke, K. A. J. McCorry, E. Crothers, and J. S. G. Dooley. 2000. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J. Med. Microbiol. 49:779-785. [DOI] [PubMed] [Google Scholar]
- 23.MacKenzie, W. R., N. J. Hoxie, M. E. Proctor, M. S. Gradus, K. A. Blair, D. E. Peterson, J. J. Kazmierczak, D. G. Addiss, K. R. Fox, J. B. Rose, and J. P. Davis. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-167. [DOI] [PubMed] [Google Scholar]
- 24.Medema, G. J., M. Bahar, and F. M. Schets. 1997. Survival of Cryptosporidium parvum, Escherichia coli, faecal enterococci and Clostridium perfringens in river water: influence of temperature and autochthonous microorganisms. Wat. Sci. Technol. 35:249-252. [Google Scholar]
- 25.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel, S. M., and J. Heptinstall. 1997. Species-specific polymerase chain reaction to detect Cryptosporidium parvum and C. muris. Biochem. Soc. Trans. 25:19S. [DOI] [PubMed]
- 27.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterization of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 28.Pedraza-Diaz, S., C. F. Amar, J. McLauchlin, G. L. Nichols, K. M. Cotton, P. Godwin, A. M. Iversen, L. Milne, J. R. Mulla, K. Nye, H. Panigrahl, S. R. Venn, R. Wiggins, M. Williams, and E. R. Youngs. 2001. Cryptosporidium meleagridis from humans: molecular analysis and description of affected patients. J. Infect. 42:243-250. [DOI] [PubMed] [Google Scholar]
- 29.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58:3494-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sluter, S. D., S. Tzipori, and G. Widmer. 1997. Parameters affecting polymerase chain reaction detection of waterborne Cryptosporidium parvum oocysts. Appl. Microbiol. Biotechnol. 48:325-330. [DOI] [PubMed] [Google Scholar]
- 31.Solo-Gabriele, H., and S. Neumeister. 1996. U.S. outbreaks of cryptosporidiosis. J. Am. Water Works Assoc. 88:76-86. [Google Scholar]
- 32.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed] [Google Scholar]
- 33.Sreter, T., G. Kovacs, A. J. Da Silva, N. J. Pieniazek, Z. Szell, M. Dobos-Kovacs, K. Marialigeti, and I. Varga. 2000. Morphologic, host specificity, and molecular characterization of a hungarian Cryptosporidium meleagridis isolate. Appl. Environ. Microbiol. 66:735-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ungar, B. L. P. 1995. Cryptosporidium, p. 2500-2510. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 35.U.S. Environmental Protection Agency. 1999. Method 1622: Cryptosporidium in water by filtration/IMS/FA. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 36.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 40.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]


