Abstract
The phylogenetic diversity of the intestinal bacterial community in pigs was studied by comparative 16S ribosomal DNA (rDNA) sequence analysis. Samples were collected from a total of 24 pigs representing a variety of diets, ages, and herd health status. A library comprising 4,270 cloned 16S rDNA sequences obtained directly by PCR from 52 samples of either the ileum, the cecum, or the colon was constructed. In total, 375 phylotypes were identified using a 97% similarity criterion. Three hundred nine of the phylotypes (83%) had a <97% sequence similarity to any sequences in the database and may represent yet-uncharacterized bacterial genera or species. The phylotypes were affiliated with 13 major phylogenetic lineages. Three hundred four phylotypes (81%) belonged to the low-G+C gram-positive division, and 42 phylotypes (11.2%) were affiliated with the Bacteroides and Prevotella group. Four clusters of phylotypes branching off deeply within the low-G+C gram-positive bacteria and one in the Mycoplasma without any cultured representatives were found. The coverage of all the samples was 97.2%. The relative abundance of the clones approximated a lognormal distribution; however, the phylotypes detected and their abundance varied between two libraries from the same sample. The results document that the intestinal microbial community is very complex and that the majority of the bacterial species colonizing the gastrointestinal tract in pigs have not been characterized.
The microbial ecology of gastrointestinal tract ecosystems is not well understood due to the inadequacy of classical, culture-dependent microbiological methods. Two decades ago, substantial efforts were put into characterizing the intestinal microbiota of pigs by using microbiological methods based on culturing and phenotypic analysis of the isolates (1, 28, 36, 37, 39, 40). These studies showed that the majority of the culturable bacteria are gram-positive, strict anaerobic streptococci, lactobacilli, eubacteria, clostridia, and peptostreptococci, while the gram-negative part of the microbiota is dominated by Bacteroides. Because culture-based methods are very time-consuming, thereby limiting the number of samples that can be processed, no information on the population dynamics or community responses to perturbations was obtained.
Detailed information of the microbial community composition in natural systems can be gained from the phylogenetic analysis of 16S ribosomal DNA (rDNA) sequences obtained directly from samples by PCR amplification, cloning, and sequencing, although this procedure may be biased as well (9, 35, 46, 49, 50). 16S rDNA cloning and sequencing has been applied to analyze the intestinal bacterial community in humans (45, 56) and in a pig (33) and compared to culture-based methods. The results showed that the microbial community is complex and that the bacterial diversity cannot be comprehended by culturing. However, these studies were limited, as only a single individual was sampled, the number of clones analyzed was small relative to the expected diversity, and the phylogenetic analysis was based on partial sequences.
To improve our understanding of the intestinal ecosystem we have initiated the Pig Intestinal Molecular Microbiology Project, which will implement and develop molecular methods for structural and functional analysis of intestinal bacterial communities. This paper presents the analysis of a library of bacterial 16S rDNA sequences cloned from the gastrointestinal (GI) tract microbiotas of 24 pigs. The purpose was to provide an inventory of the phylotypes (bacteria defined only by their 16S rDNA sequence) that can be found in the intestinal tracts of Danish pigs. In the second phase of the project, specific probes for the sequences will be designed and implemented on oligonucleotide arrays (biochips) to facilitate dynamic analysis of the intestinal microbiota.
MATERIALS AND METHODS
Animals and sampling.
Samples were collected from 24 pigs and analyzed. The animals were 12 to 18 weeks of age and weighed 30 to 50 kg, except for 6 animals that were 2, 4, and 6 weeks old. The pigs were fed different diets as listed in Table 1. Two animals were experimentally infected with Salmonella enterica serovar Typhimurium but showed no clinical symptoms.
TABLE 1.
Basic conditions of pigs that were sampled
| Piga | Diet | Sample analyzedh
|
||
|---|---|---|---|---|
| Ileum | Cecum | Colon | ||
| SPF-Ab | Standardf | √ | √ | √ |
| SPF-Bb | Standard | √ | √ | |
| SPF-Cb | Standard | √ | √ | |
| Organicc | Standard | √ | √ | √ |
| s-962d | Standard, no Tylosin | √ | √ | √ |
| s-959d | Standard, 60 ppm Tylosin | √ | √ | √ |
| A-65 | Standard | √ | ||
| A-42 | Standard | √ | ||
| G-55 | Standard, 2% lactic acid | √ | ||
| G-23 | Standard, 2% lactic acid | √ | √ | √ |
| SwA | Standard | √ | √ | √ |
| F-9 | Fermented standard feed | √ | √ | √ |
| B-18 | CPg | √ | √ | |
| E-24 | CP, 12% sugar beet pulp | √ | ||
| 6C | CP, 10% potato starch | √ | ||
| 2W-a,be | Sow's milk | √ | √ | √ |
| 4W-a,be | Sow's milk and dry feed | √i | √ | √ |
| 6W-a,be | Standard | √ | √ | √ |
| 28-Fa | Standard, 1.8% formic acid | √ | ||
| 29-stf | Standard, fine | √ | ||
| 30-stc | Standard, coarse | √ | ||
Unless otherwise stated, the pigs originated from experimental studies at the Danish Veterinary Laboratory and were 12 to 16 weeks of age.
SPF-A, SPF-B, and SPF-C were bought from a specific pathogen-free high-health, commercial herd.
Organically reared pig.
Experimentally challenged with Salmonella enterica serovar Typhimurium, with or without Tylosin as a growth promoter.
2W-a,b, 4W-a,b, and 6W-a,b were six animals bought from a commercial herd and were 2, 4, and 6 weeks old, respectively.
Standard feed based on wheat, barley, soybeans, and fish meal, without antibiotic growth promoters.
CP, cooked rice and animal protein.
Marked samples were analyzed.
The ileal sample from pig 4-Wb was not analyzed.
For sampling, the pigs were sacrificed and 5- to 10-cm sections of the ileum, the cecum, or the top of the spiral colon were tied off and stored at −80°C until further processing.
Extraction and purification of DNA.
DNA was extracted from intestinal luminal contents by a bead-beating method as previously described (21). Briefly, 200 mg of intestinal content was suspended in 600 μl of phosphate-buffered saline, vortexed, and centrifuged 2 min at 200 × g. After transfer to a new tube, the sample was centrifuged at 12,000 × g for 5 min and the pellet was resuspended in 570 μl of TE (10 mM Tris-HCl-1 mM EDTA [pH 8.0]). The bacterial cells were lysed by shaking for 4 min on a minibead beater (Biospec Products Inc., Bartlesville, Okla.) on Hi speed in 2-ml screw-cap tubes to which 350 to 400 mg of 100-μm-diameter Zirconia/Silica beads (Biospec Products Inc.) and 30 μl of 10% sodium dodecyl sulfate had been added. After a brief spin in a microcentrifuge, the samples were transferred to a microcentrifuge tube and the DNA was purified by the CTAB method (6); the DNA was dissolved in 50 μl of TE and stored at −21°C.
16S rDNA amplification and cloning.
The 16S rDNA was amplified using primers S-D-Bact-0008-a-S-20 (5"-AGAGTTTGATCMTGGCTCAG-3" [19]) and S-*-Univ-1492-a-A-19 (5"-GGTTACCTTGTTACGACTT-3" [modified from that described in reference 19]) (oligonucleotide probe nomenclature according to Alm et al. [2]). The primers were purchased from DNA Technology, Aarhus, Denmark. Reaction conditions were as follows: 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 1 mg of bovine serum albumin ml−1, 400 μM concentrations of each deoxynucleoside triphosphate, 1 mM MgCl2, 0.25 μM concentrations of each primer, 2.5 U of Pfu polymerase with exonuclease activity (Stratagene, Cambridge, United Kingdom), and 2 μl (≈30 ng of DNA) of template DNA in a total volume of 50 μl. PCR was performed using a PTC-200 Thermal Cycler (MJ Research, Watertown, Mass.) using the following program: 3 min 15 s of initial denaturation at 94°C followed by 30 cycles of denaturation (45 s at 94°C), annealing (45 s at 53°C), and extension (3 min 30 s at 72°C), with a final extension at 72°C for 10 min. Five reactions were made from each sample, and the PCR products were pooled before further processing. Amplified DNA was verified by electrophoresis of 8-μl aliquots of the pooled PCR products in 1.5% agarose gels. The PCR products were purified on QIAquick PCR purification kit columns (QIAGEN GmbH, Hilden, Germany), redissolved in double-distilled water, and stored at −20°C. Blunt-end PCR products were cloned into linearized pCR-Blunt vectors (Invitrogen, Groningen, The Netherlands), and One Shot TOP10 Competent Escherichia coli organisms (Invitrogen) were transformed using a Zero Blunt PCR Cloning Kit (Invitrogen), according to the manufacturer's instructions. Colonies were grown on low-salt Luria-Bertani (Invitrogen) plates for 18 to 24 h. Ninety-six single colonies from each sample were picked randomly, transferred to SOB-Zeocin (2% tryptone, 0.5% yeast extract, 0.05% NaCl, 2.5 mM KCl, 10 mM MgCl2, 50 μg of Zeocin ml−1) medium (Invitrogen), and grown for 24 h at 37°C either in 15-ml tubes or in 2-ml-well flat-bottom 96-well blocks (QIAGEN GmbH) sealed with AirPore film (QIAGEN GmbH).
Two hundred-microliter stocks of each clone were stored in SOB-glycerol in 2.2-ml Deep Well Plates (Advanced Biotechnologies LTD, Surrey, United Kingdom) sealed with Adhesive Plate Seal (Advanced Biotechnologies LTD) at −80°C.
Plasmid preps were done either in single tubes with the QIAprep Spin Miniprep Kit (QIAGEN GmbH) or in 96-well blocks with a QIAprep 96 Turbo Miniprep Kit (QIAGEN GmbH) using a QIAvac vacuum manifold (QIAGEN GmbH).
Sequencing of rDNA.
The 16S rDNA nucleotide sequences of the inserts were determined by cycle sequencing using the ABI PRISM BigDye Terminator Cycle Sequencing Kit (PE Biosystems, Foster City, Calif.) according to the manufacturer's instruction. Sequences were read with an automatic sequence analyzer (ABI PRISM 377 DNA Sequencer; PE Biosystems). Partial and near-full-length sequences were generated. All clones were sequenced at the 3"-terminal part of the molecule with a single primer, S-D-Bact-0907-a-S-20 (5"-AAACTCAAAGGAATTGACGG-3"). This provided sequences of approximately 450 bp, which were used for a provisional grouping of the sequences (see below). Representative sequences from each group were subjected to near-full-length, bidirectional sequencing of both strands using the following primers: S-D-Bact-0008-a-S-20, S-*-Univ-0519-a-A-18 (5"-GTATTACCGCGGCTGCTG-3"), S-D-Bact-0786-a-A-20 (5"-GACTACCAGGGTATCTAATC-3"), S-D-Bact-0907-a-S-20 (5"-AAACTCAAAGGAATTGACGG-3"), S-D-Bact-1054-a-A-20 (5"-ACGAGCTGACGACAGCCATG-3"), and S-*-Univ-1492-a-A-19.
Sequence analysis.
The sequences were aligned to the Ribosomal Database Project (RDP) small subunit rRNA alignment version 7.1 (26), running locally in the ARB sequence environment (4). The RDP 7.1 alignment comprises 7,008 entries of mostly full-length or partial sequences for the Bacteria domain. The cloned sequences were automatically aligned using the ARB Sequence Editor v2.0, and the alignment was inspected and corrected manually based on known secondary structure when necessary. The aligned sequences were placed on the phylogenetic tree of the RDP alignment version 7.1 using the constrained maximum-parsimony method provided in the ARB software. This method facilitated the addition of the new sequences to the validated and optimized framework tree of the RDP alignment, produced by the maximum-likelihood method (25), without allowing for changes in the overall tree topology (24). Furthermore, with this method any bias introduced by the selection of a subset of sequences a priori, necessary to redraw the trees of the involved phylogenetic groups with other treeing methods, was avoided. Only unambiguous nucleotide positions were included in the analysis. Distance matrices and similarities were calculated according to the Jukes-Cantor model (16). The percentage of coverage (13) was calculated by the formula [1 − (n/N)] × 100, where n is the number of phylotypes represented by one clone and N is the total number of clones.
Initially the partial sequences were analyzed. Single sequences, or clusters of sequences that grouped together on the tree, with less than 97% similarity to any other cloned sequence were identified as provisional phylotypes. At least one representative from each of the provisional phylotypes was selected for bidirectional sequencing of the region from positions 27 to 1492 of the 16S rDNA (E. coli numbering).
All near-full-length sequences were tested for possible chimeric structures. Chimeras were detected by the RDP analysis service Check__Chimera (20) by comparing the phylogenetic positions of the partial and near-full-length sequences from the clones and during the manual inspection of the alignment.
The near-full-length sequences were aligned and drawn on the RDP alignment 7.1 tree as described above, and the similarity to the closest related sequence in the alignment was calculated according to the Jukes-Cantor model. The sequences were assigned to individual operational taxonomic units (OTUs; phylotypes) based on their phylogenetic positions and the 97% sequence similarity criterion.
When clusters of phylotypes did not group with any sequences in the database, the phylogenetic positions of these clusters were tested in neighbor-joining trees with 1,000 times resampling to determine the significance of the clusters from bootstrap values with the ARB software. For the neighbor-joining trees, datasets were selected to cover the phylogenetic diversity in the taxonomic group which the unknown phylotypes were affiliated with.
All dendrograms in this paper present near-full-length sequences added to the tree provided with the RDP alignment 7.1 using the maximum-parsimony method in ARB without changing the original topology of the tree. However, for clarity and to minimize the sizes of the figures, replicate sequences of individual species were removed from the figures. A complete phylogenetic tree of the RDP alignment 7.1 with all detected phylotypes added is available upon request from the corresponding author.
Nucleotide sequence accession numbers.
The near-full-length 16S rDNA sequences have been deposited in the GenBank database under accession numbers AF371468 to AF371949.
RESULTS
A total of 3.5 Mb of 16S rDNA was sequenced. Fifty-two samples from the 24 animals were analyzed (Table 2). From each sample, 96 clones were picked randomly. The average cloning efficiency was 86%, and in total 4,270 clones contained an insert that could be sequenced.
TABLE 2.
Diversity and coverage of phylotypes from the porcine GI tract in 24 pigs
| No. of samples | No. of clones | No. of phylotypes | Coverage (%)a | Hb | |
|---|---|---|---|---|---|
| Ileum | 13 | 968 | 86 | 96.2 | 4.65 |
| Cecum | 16 | 1,262 | 231 | 94.1 | 6.78 |
| Colon | 23 | 2,040 | 298 | 95.8 | 6.90 |
| Total | 52 | 4,270 | 375 | 97.8 | 6.88 |
According to Good (13).
The Shannon-Wiener index, H, is defined as − Σ pi log2 pi, where pi is the decimal fraction of individuals (clones) of the ith species (phylotype).
The partial sequencing with the S-D-Bact-907-a-S-20 primer provided approximately 450 bp at the 3"-terminal part of the 16S rDNA. This part of the 16S rDNA, comprising the variable regions V6, V7, V8, and V9 (30), was used for the provisional grouping of the sequences.
One representative of each of the preliminary groups was selected for sequencing of the region encompassed by primers S-D-Bact-0008-a-S-20 to S-*-Univ-1492-a-A-19 of the 16S rDNA, and the phylogenetic analysis was done on these near-full-length 16S rDNA sequences.
Sixty-five of the near-full-length sequences (11.9%) were identified as possible chimeras and were rejected from further analysis, because the Check__Chimera program suggested a chimera, and/or a significant difference between the phylogenetic positioning of the partial and the near-full-length sequence from a clone was seen, and/or the similarity between a clone sequence and a database entry abruptly shifted from high similarity in one part of the sequence to a lower similarity in the remaining part of the sequence.
Based on the 97% sequence similarity criterion, 375 operational taxonomic units were found. Each of these represents a phylotype and may be a representative of a bacterial species. The number and distribution of phylotypes and hence the bacterial diversity varied among the three sampling locations. The Shannon-Wiener index (55) for the ileum was 4.65, increasing to 6.78 and 6.90 in the cecum and colon, respectively (Table 2).
The overall mean similarity between the 375 phylotypes and their closest related known bacterial species for which the 16S rRNA sequences are available in the RDP alignment 7.1 was 92.6%. Sixty-six OTUs (17.6%) had a similarity of 97% or higher to any sequence in the RDP, i.e., 309 of the phylotypes were putative representatives of hitherto unknown or unsequenced bacterial genera or species.
Ninety-four phylotypes were detected only once, and the percentage of coverage was 97.8% in all samples from the 24 pigs analyzed, i.e., 2.2% of the clones in a new library produced from those pigs would represent previously undetected phylotypes. The coverage in the individual parts of the GI tract was slightly lower (Table 2).
The phylotypes were assigned to 13 major phylogenetic lineages (Table 3). Three hundred four (81%) of the phylotypes belonged to the low-G+C gram-positive division.
TABLE 3.
Major phylogenetic lineages to which the phylotypes from the porcine GI tracts were affiliated
| Phylogenetic groupa | RDP reg. no. | No. of phylotypes detected | Similarity (%)b |
|---|---|---|---|
| Eubacterium and relatives | 2.30.4 | 125 | 93.0 |
| Clostridium and relatives | 2.30.9 | 109 | 92.2 |
| Bacillus-Lactobacillus-Strepto-coccus subdivision | 2.30.7 | 46 | 96.7 |
| Flexibacter-Cytophaga-Bacteroides group | 2.15 | 42 | 87.5 |
| Proteobacteria | 2.28 | 20 | 94.8 |
| Sporomusa and relatives | 2.30.3 | 15 | 94.7 |
| Mycoplasma and relatives | 2.30.8 | 8 | 78.6 |
| High-G+C bacteria | 2.30.1 | 4 | 93.5 |
| Spirochetes and relatives | 2.27 | 2 | 86.4 |
| Clostridium purinolyticum group | 2.30.5 | 1 | 94.4 |
| Planctomyces and relatives | 2.20 | 1 | 86.0 |
| Flexistipes sinusarabici assemblage | 2.14 | 1 | 85.9 |
| Anaerobaculum thermoterrenum group | 2.11 | 1 | 84.3 |
Phylogenetic grouping according to the RDP.
Mean similarity of all the phylotypes affiliated with that group to the most closely related sequences in the RDP alignment version 7.1.
Clostridium and relatives (RDP reg. no. 2.30.9).
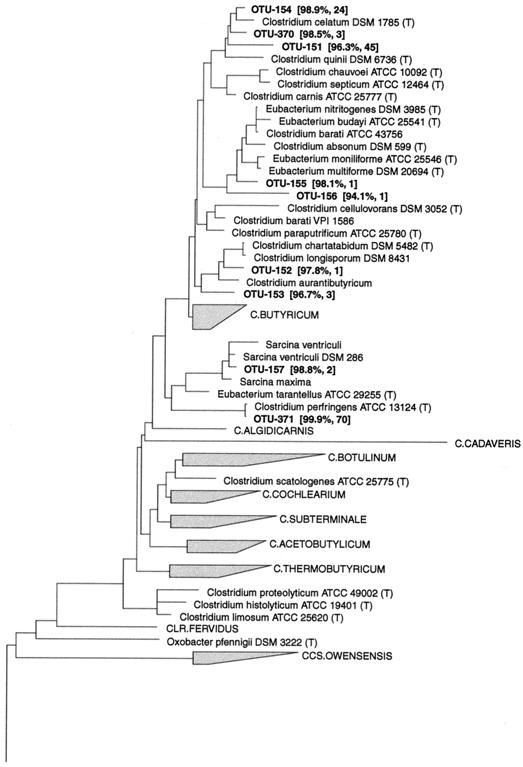
Figures 1 and 2 show the phylogenetic inferences among the OTUs affiliated with the Clostridium botulinum group (RDP registration [reg.] no. 2.30.9.2) and the Clostridium leptum subgroup (RDP reg. no. 2.30.9.1.3).
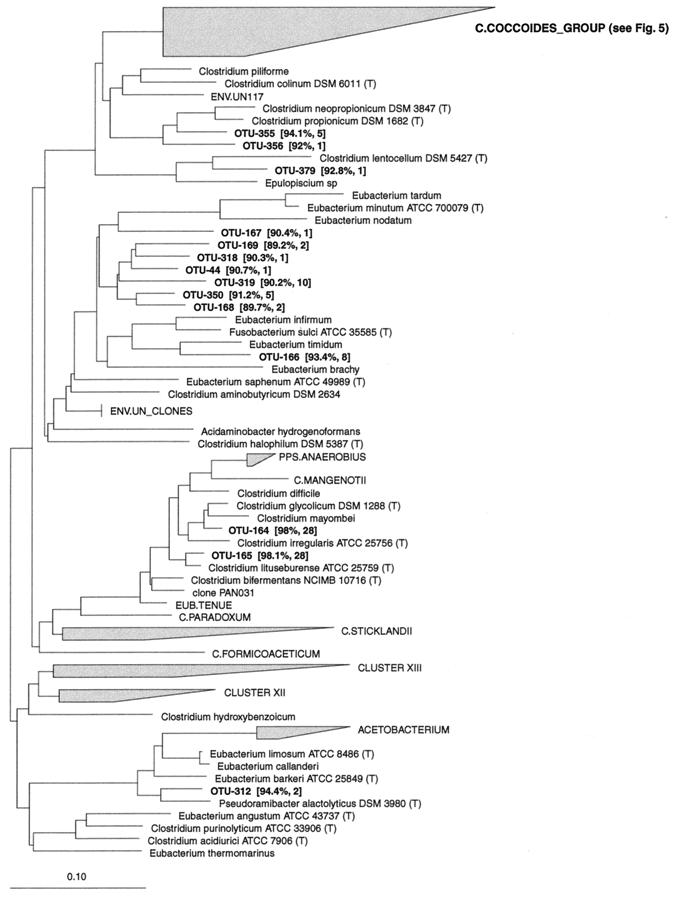
FIG. 1.
Dendrogram showing the phylogenetic affiliation of OTUs in the Clostridium botulinum group (RDP reg. no. 2.30.9.2) and four deep-branching clusters of OTUs from the porcine GI tract. Near-full-length 16S rDNA sequences were aligned to the Ribosomal Database Project small subunit rRNA alignment version 7.1 and added to the matching phylogenetic tree without changing the topology of the tree by using the constrained maximum-parsimony method provided in the ARB software. 16S rDNA sequences from the bovine rumen (superscript a [47] and b [54]) and the GI tract of a pig (superscript c [33]) were added to this tree. The scale bar represents a 10% estimated sequence divergence. For each OTU the similarity to the closest related known bacteria and the number of clones belonging to that phylotype are indicated. The most closely related bacteria in the database and the mean similarity for the deep-branching clusters were as follows: CLUSTER__B, Acetivibrio cellulolyticus (84.1%); CLUSTER__C, Acetivibrio cellulolyticus (86%); CLUSTER__D, Eubacterium plautii (84.6%); CLUSTER__E, STR.16SX-1 (81.7%).
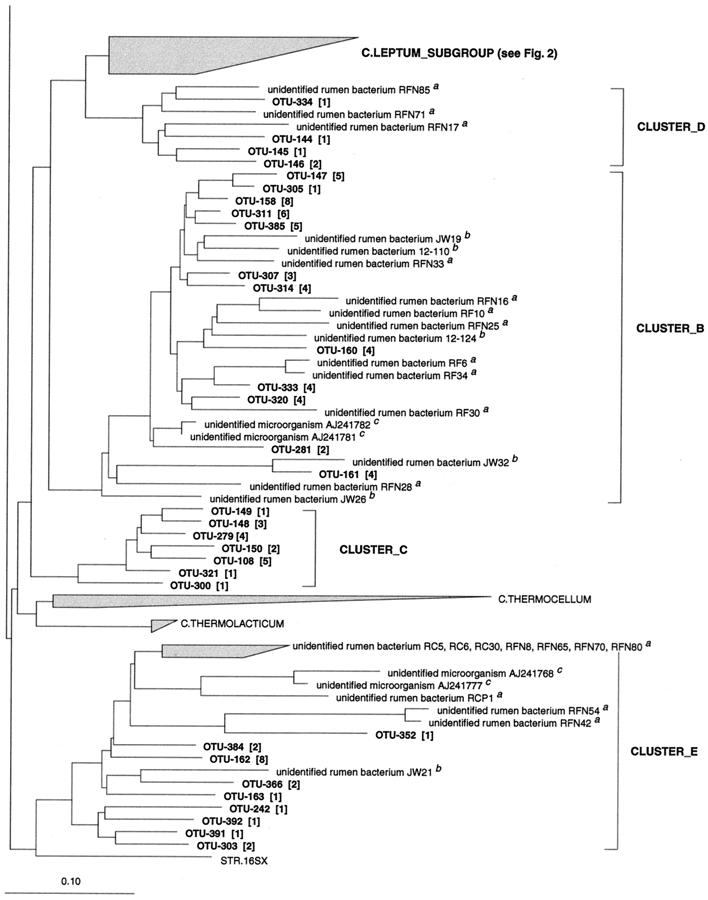
FIG. 2.
Dendrogram showing the phylogenetic affiliation of OTUs from the porcine GI tract in the Clostridium leptum subgroup (RDP reg. no. 2.30.9.1.3). See the legend to Fig. 1 for explanation.
Nine OTUs belonged to the Clostridium botulinum group (Fig. 1). They were related to known bacterial species isolated from intestines or rumen. Seven OTUs were found in the Clostridium barati subgroup (RDP reg. no. 2.30.9.2.11). Clostridium celatum was represented by three phylotypes with high similarity. They were abundant among the clones (72 clones together) and were detected in 14 pigs at all sampling sites. C. barati and Clostridium longisporum were each represented by two phylotypes found at low frequencies.
Two OTUs were affiliated with the Clostridium perfringens subgroup (RDP reg. no. 2.30.9.2.11.3) and were related to Sarcina maxima and C. perfringens. Seventy clones belonged to the latter phylotype; however, the distribution was biased because they were found only in the 2-week-old pigs. They were detected in the ilea, ceca, and colons of those animals.
Four large clusters of phylotypes that had no cultured representatives were found close to the Clostridium leptum subgroup (Fig. 1). These clusters branched off deeply in the tree, suggesting that they were higher-order taxonomic structures. CLUSTER__B, CLUSTER__C, and CLUSTER__D were positioned between the Clostridium leptum subgroup (RDP reg. no. 2.30.9.1.3) and the Clostridium thermolacticum subgroup (RDP reg. no. 2.30.9.1.2), with bootstrap values of 97 to 99%; whereas CLUSTER__E was located near the STR.16SX subgroup (RDP reg. no. 2.30.9.1.4). The bootstrap value was slightly lower (85%) for this cluster.
CLUSTER__B consisted of 12 OTUs with similarities among one another from 81 to 96.9%. These OTUs clustered together with published phylotypes from the bovine rumen (54, 47) and from the large intestine of a pig (33), with low sequence similarity. All but one OTU were detected more than once. CLUSTER__C had seven phylotypes with low intracluster sequence similarity (86.5 to 94.6%). None of the published sequences from the bovine rumen or the pig GI tract clone libraries were affiliated with this cluster. The four OTUs in CLUSTER__D grouped together with sequences from three unidentified rumen bacteria and had mutual similarities from 85.8 to 90.7%. CLUSTER__E consisted of nine phylotypes with similarities from 78.1 to 91.8%. They clustered with sequences from both the bovine rumen and the pig GI tract with low sequence similarity.
Sixty-eight phylotypes were affiliated with the Clostridium leptum subgroup (RDP reg. no. 2.30.9.1.3), (Fig. 2). The OTUs grouped into 15 clusters of which 11 included previously cultured representatives. Six OTUs clustered around the intestinal bacterium Fusobacterium prausnitzii with sequence similarity values from 89.9 to 98.9%. The phylotypes in this cluster were among the most abundant and were found in the large intestines, but not in the ilea, in 23 out of 24 pigs. Other previously cultured intestinal bacteria in the Clostridium leptum subgroup, to which OTUs were related, included Eubacterium siraeum (two OTUs), Ruminococcus callidus (three OTUs), Ruminococcus flavefaciens (one OTU), Ruminococcus albus (three OTUs), Ruminococcus bromii (three OTUs), C. leptum (two OTUs), and Eubacterium desmolans (three OTUs). Twelve OTUs were most closely related to the Sporobacter genus, which has a single cultured representative, Sporobacter termitidis, isolated from the intestine of a wood-feeding termite. The similarities between these OTUs and S. termitidis were low (87.6 to 93.1%); however, the 12 OTUs comprised a cluster of abundant phylotypes, with 203 clones found in the large intestines of 20 pigs. They were not found in the ilea. Three clusters of OTUs had no cultured representatives but were loosely associated with the Ruminococcus genus. The sequence similarity to known species was in the range of 84.4 to 92% (mean = 89.1%), indicating that these clusters may represent new taxonomic entities above the species level. The cluster comprising OTU-116, OTU-117, OTU-121, and OTU-122 had a moderate bootstrap value of 77%, whereas the remaining clusters had values of less than 50%. The phylotypes were not found frequently; however, 7 of 14 OTUs were detected more than once.
Five OTUs formed a cluster with sequence similarity values from 88.4 to 91.5% to Eubacterium plautii. In addition, four phylotypes were related to this species at low similarity. These phylotypes were detected in the large intestines but not in the ilea.
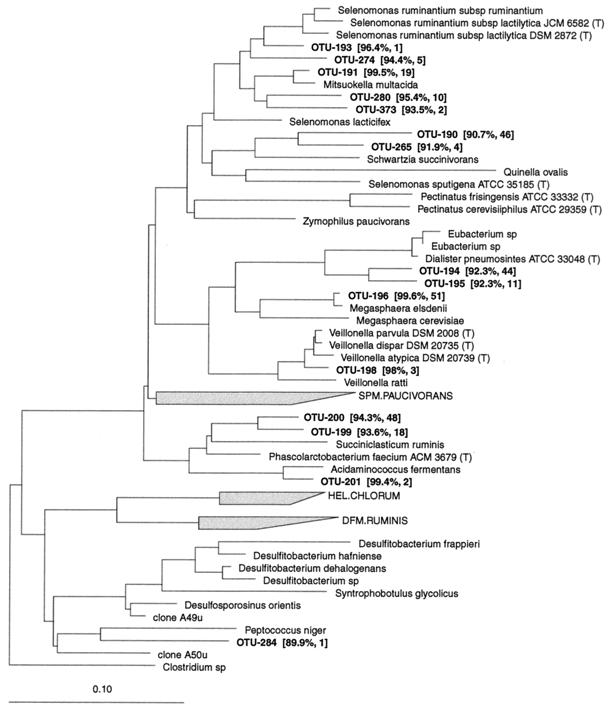
Sporomusa and relatives (RDP reg. no. 2.30.3).
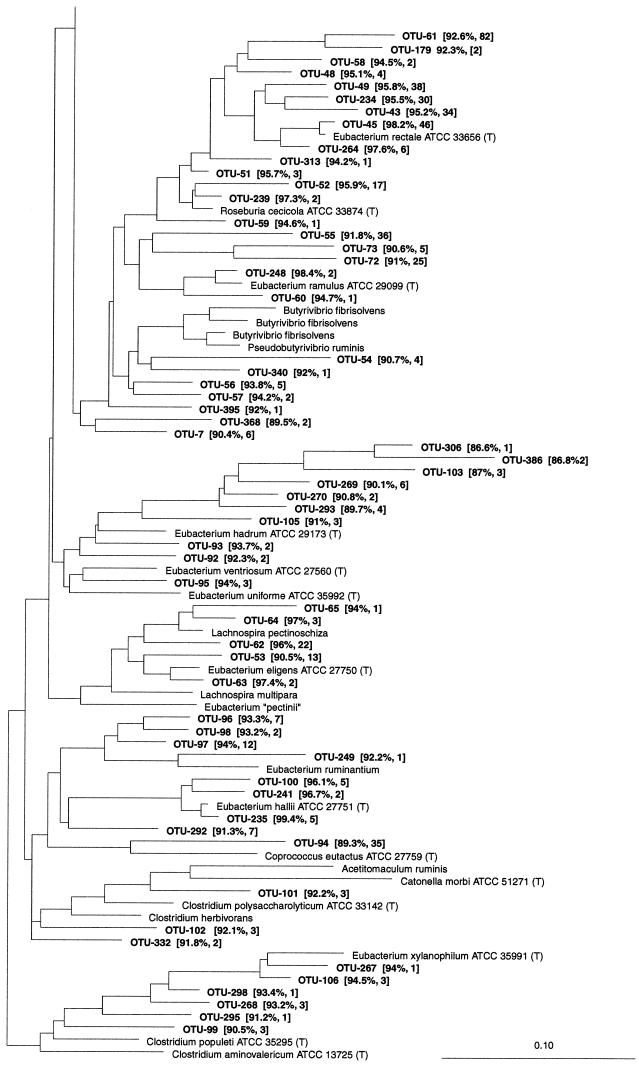
Most of the phylotypes affiliated with this group were related to previously cultured intestinal or rumen bacteria (Fig. 3). Seven OTUs belonged to the Selenomonas ruminantium subgroup (RDP reg. no. 2.30.3.1.1). S. ruminantium was the closest relative to two phylotypes, with a modest sequence similarity. Mitsuokella multacida was represented by three phylotypes, Schwartzia succinivorans by two. In the Veillonella parvula subgroup (RDP reg. no. 2.30.3.1.2), OTU-194 and OTU-195 were found among 55 of the clones. The closest relative was Dialister pneumosintes; however, the similarity was only 92.3%, suggesting that these phylotypes may represent a new genus. In this subgroup Megasphaera elsdenii and Veillonella dispar were represented by a single OTU each with high similarity. One phylotype related to Acidaminococcus fermentans with a high sequence similarity was found, while two phylotypes were related to Phascolarctobacterium faecium with modest similarity. The P. faecium-related phylotypes were detected in the large intestines of 15 pigs. A single OTU was found with Peptococcus niger as the closest relative, but at a sequence similarity level of only 89.9%.
FIG. 3.
Dendrogram showing the phylogenetic affiliation of OTUs from the porcine GI tract in the Sporomusa and relatives group (RDP reg. no. 2.30.3). See the legend to Fig. 1 for explanation.
Clostridium purinolyticum group (RDP reg. no. 2.30.5).
OTU-312 was affiliated with the C. purinolyticum group (Fig. 4). The closest related known bacterium was a human clinical isolate of Pseudoramibacter alactolyticus (94.4% sequence similarity). OTU-312 was detected in two clones, both from the ileum of one animal (SwA).
FIG. 4.
Dendrogram showing the phylogenetic affiliation of OTUs from the porcine GI tract in the Eubacterium and relatives group (RDP reg. no. 2.30.4) and in the C. purinolyticum group (RDP reg. no. 2.30.5). See the legend to Fig. 1 for explanation.
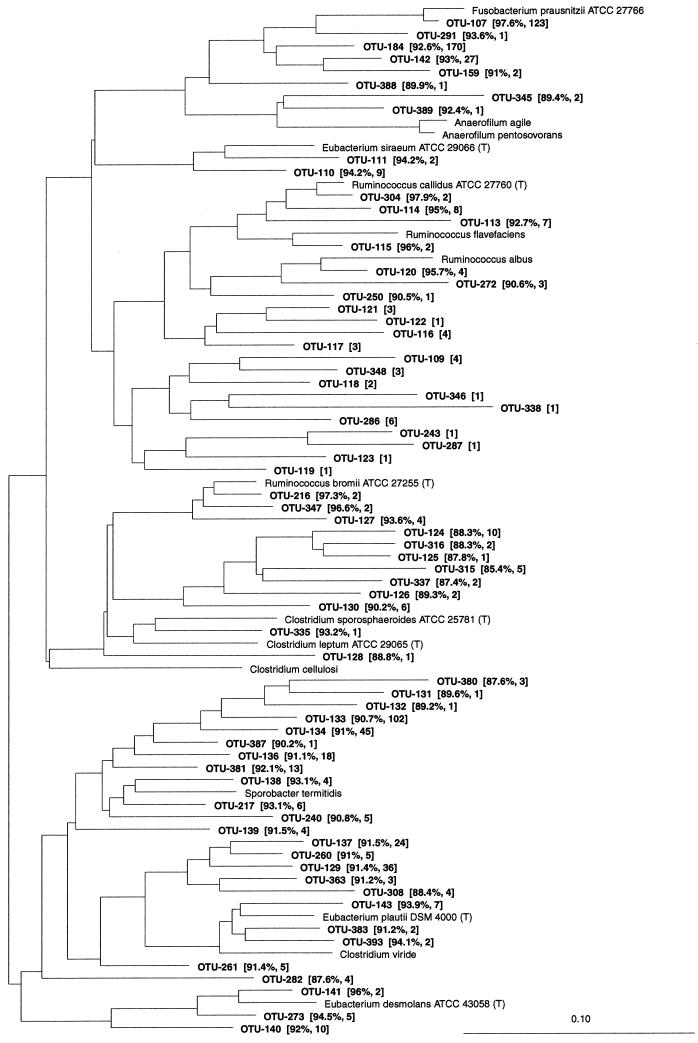
Eubacterium and relatives (RDP reg. no. 2.30.4).
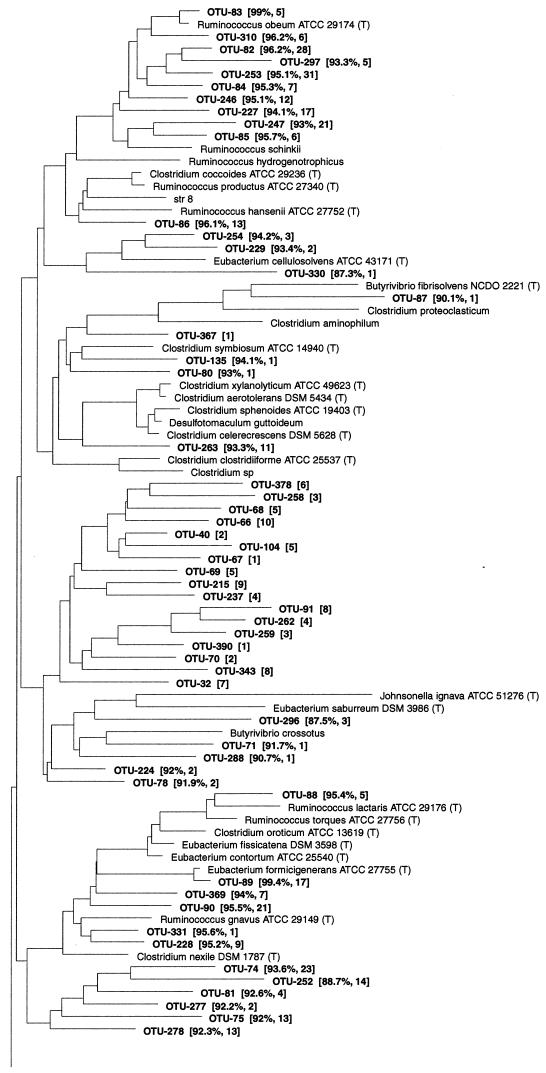
One hundred twenty-five OTUs were affiliated with this phylogenetic group (Fig. 4 and 5). One hundred twelve of these belonged to the Clostridium coccoides group (RDP reg. no. 2.30.4.1) (Fig. 5). Only nine phylotypes in this group had sequence similarity values of 97% or more to any known bacteria. However, clusters of three or more OTUs often formed around species previously cultured from intestines or the rumen. These included Ruminococcus obeum (8 OTUs), Eubacterium cellulosolvens (3 OTUs), Butyrivibrio crossotus (5 OTUs), Ruminococus gnavus (4 OTUs), Eubacterium rectale (11 OTUs), Roseburia cecicola (3 OTUs), Eubacterium ramulus (5 OTUs), Butyrivibrio fibrisolvens and Pseudobutyrivibrio ruminis (7 OTUs), Eubacterium hadrum (9 OTUs), Lachnospira pectinoschiza (3 OTUs), Eubacterium ruminantium (4 OTUs), Eubacterium hallii (4 OTUs), and Eubacterium xylanophilum (5 OTUs). Two clusters that were only distantly related to cultured species were found. In the Clostridium xylanolyticum subgroup (RDP reg. no. 2.30.4.1.1), 17 OTUs formed a cluster with two likely subclusters of sequences having low similarity to any known species (C. xylanolyticum; mean sequence similarity = 91.2%). However, resampling of these phylotypes revealed bootstrap values of less than 50%, and their phylogenetic positions are uncertain. Except for OTU-390, these phylotypes were all found more than once and were detected in the large intestines of 20 pigs (a total of 83 clones). In the Ruminococcus gnavus subgroup (RDP reg. no. 2.30.4.1.4), another cluster of OTUs without any cultured representatives was found. The 69 clones in this cluster grouped into six phylotypes that were 92.2% (mean value) similar to Clostridium nexile. However, the bootstrap value was only 25%, and the phylogenetic positions of these phylotypes are uncertain.
FIG. 5.
Dendrogram showing the phylogenetic affiliation of OTUs from the porcine GI tract in the Clostridium coccoides group (RDP reg. no. 2.30.4.1). See the legend to Fig. 1 for explanation.
Besides the phylotypes found in clusters, 18 phylotypes related to known species were found. Two OTUs were related to Ruminococcus schinkii, two to Clostridium symbiosum, two to Clostridium aerotolerans, two to Eubacterium eligens, and two to Clostridium herbivorans. One OTU (each) was related to C. coccoides, Butyrivibrio fibrisolvens NCDO 2221 (T), Ruminococcus lactaris, Eubacterium formicigenerans, Eubacterium ventriosum, Coprococcus eutactus, Clostridium polysaccharolyticum, and Clostridium populeti.
Several of the phylotypes in this group were among the most abundant clones from the large intestines of the pigs. The OTUs related to E. rectale were found in 20 of the 24 pigs analyzed, those related to R. obeum were found in 19 pigs, E. ramulus-related phylotypes were found in 18 pigs, B. fibrisolvens-related phylotypes were found in 14 pigs, and Eubacterium hadrum- and Ruminococcus schinkii-related phylotypes were each found in 13 pigs.
In the remaining groups of the Eubacterium and relatives, two OTUs were most closely related to Clostridium propionicum, isolated from black mud. The sequence similarity was low, and the phylotypes were found only in the ceca of the 4- and 6-week-old animals. OTU-379, which was detected in the cecum of only one 2-week-old pig, was related to another environmental ecotype, Clostridium lentocellum, with a low sequence similarity.
A cluster of seven OTUs with a bootstrap value of 32% were affiliated with the Eubacterium group (RDP reg. no. 2.30.4.4). The closest relative was Clostridium aminobutyricum (mean sequence similarity = 90.2%), which was isolated from a swamp. OTU-166 was related to Eubacterium timidum, which was isolated from the subgingival region of a human mouth. OTU-164 and OTU-165 had high similarities to Clostridium glycolicum and Clostridium lituseburense, respectively, which were isolated from mud. The two phylotypes were both represented by 28 clones and were detected in the ilea, ceca, or colons of 8 and 12 pigs, respectively.
Bacillus-Lactobacillus-Streptococcus subdivision (RDP reg. no. 2.30.7).
The 46 phylotypes affiliated with this group generally had a higher sequence similarity to known species than any of the other phylogenetic groups, and they did not form clusters of phylotypes around the cultured species. The mean sequence similarity of all phylotypes to their closest related known species was 92.6%, whereas the similarity of the phylotypes within the Bacillus-Lactobacillus-Streptococcus subdivision to known species was 96.9%.
The most abundant phylotype was OTU-180, with 337 clones found in the ilea or the large intestines of 13 pigs. OTU-180 had a high similarity to Streptococcus alactolyticus (99.7%). Other cultured Streptococci represented by phylotypes were S. dysgalactiae (OTU-372, 99.8% similarity, 1 clone), S. hyointestinalis (OTU-221, 99.8% similarity, 2 clones), S. gallolyticus (OTU-358, 99.8% similarity, 34 clones), S. suis (OTU-374, 99.4%, 4 clones). These species have previously been isolated from the pig GI tract. OTU-360, represented by nine clones, was most closely related to Streptococcus saliva (96.5% similarity).
Phylotypes affiliated with the Enterococci were detected in low numbers among the clones. They had high sequence similarity to Enterococcus faecium (OTU-178, 99.7%) and E. hirae (OTU-328, 100% similarity, 1 clone).
Twenty-four OTUs belonged to the Lactobacilli (RDP reg. no. 2.30.7.17), and 22 known species of Lactobacilli were represented by the phylotypes. Three of the phylotypes in the Lactobacilli were among the most abundant clones. They had high sequence similarity to Lactobacillus amylovorus (OTU-171, 99.2% similarity, 191 clones), Lactobacillus johnsonii (OTU-170, 99.8% similarity, 138 clones), and Lactobacillus reuteri (OTU-173, 99.5% similarity, 91 clones). L. johnsonii and L. reuteri have been isolated from the human GI tract, whereas, L. amylovorus was isolated from cattle corn silage. The phylotypes were found in the ilea or large intestines in half of the pigs analyzed. Forty-eight clones found in the ileum and cecum of a pig fed fermented standard feed (F-9), represented a phylotype related to Lactobacillus sharpeae (OTU-208, 94.9% similarity, 45 clones), which was isolated from municipal sewage.
Other abundant representatives of Lactobacillus included Lactobacillus vaginalis (OTU-47, 98.8% similarity, 50 clones), L. mucosae (OTU-174, 98% similarity, 23 clones), L. plantarum (OTU-271, 97.1% similarity, 23 clones), L. salivarius subsp. salivarius (OTU-364, 99.7% similarity, 15 clones), previously isolated from animals or humans. Less abundant representatives among the clones of bacterial species of human or animal origin were L. brevis (OTU-289, 97.6% similarity, 2 clones), L. agilis (OTU-351, 99.7% similarity, 3 clones), L. ruminis (OTU-329, 99.9% similarity, 1 clone), and L. murinus (OTU-175, 99.6% similarity, 1 clone). Phylotypes related to Lactobacillus with known habitats in human foodstuffs included L. delbrueckii, which was represented by three OTUs (OTU-172, 98.7% similarity, 1 clone; OTU-353, 95.7% similarity, 3 clones; OTU-50, 96.3% similarity, 16 clones), L. pontis (OTU-173, 99.5% similarity, 18 clones), L. farciminis (98.5% similarity, 19 clones), L. alimentarius (OTU-285, 93.4% similarity, 16 clones), L. panis (OTU-341, 99.2% similarity, 9 clones; OTU-354, 95% similarity, 8 clones), and L. collinoides (OTU-302. 97%, 1 clone). Two phylotypes were most closely related to Pediococcus parvulus (OTU-290, 98.9% similarity, 19 clones; OTU-309, 97.3% similarity, 7 clones), while Weissella confusus was represented by one phylotype (OTU-77, 99.9% similarity, 3 clones). Both species have been isolated from silage.
Bacterial species belonging to other genera in the Bacillus-Lactobacillus-Streptococcus subdivision represented by the phylotypes with low similarity included Gemella haemolysans (OTU-176, 87.1% similarity, 102 clones; OTU-255, 94.9% similarity, 1 clone), Abiotrophia adiacens (OTU-177, 88.8% similarity, 1 clone), and Staphylococcus epidermidis (OTU-181, 99.9% similarity, 1 clone).
Mycoplasma and relatives (RDP reg. no. 2.30.8).
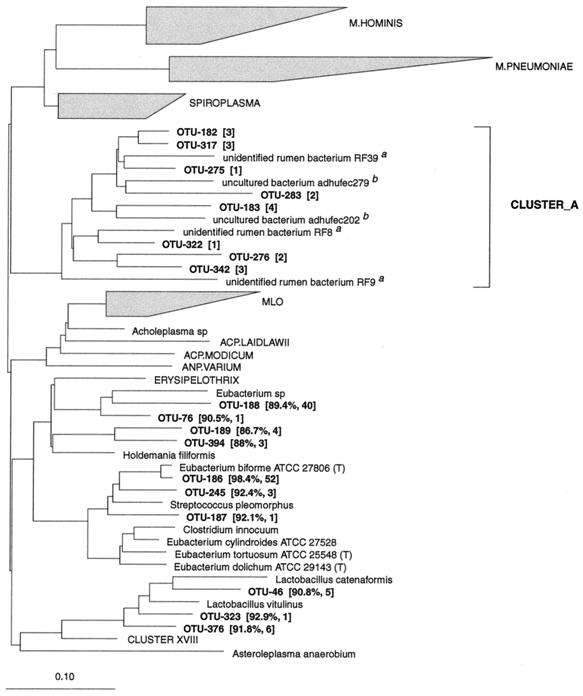
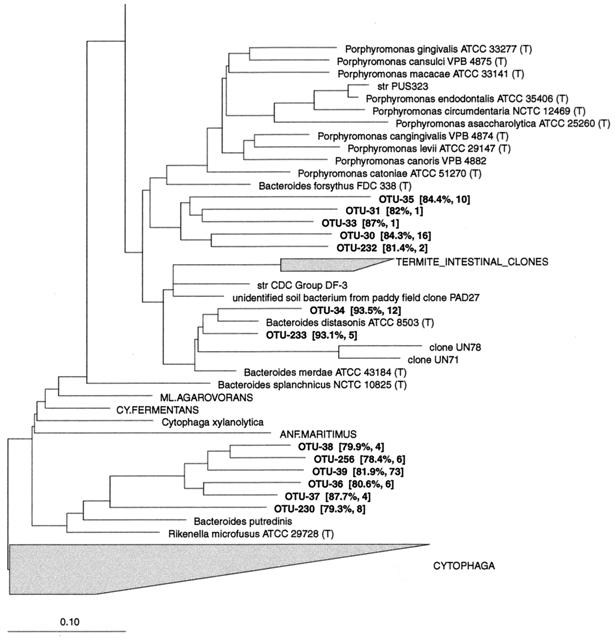
Eight phylotypes affiliated with this group were found in a deep-branching cluster (CLUSTER__A), with low sequence similarity to any known bacteria (Fig. 6). Five sequences from 16S rDNA clone libraries derived from the bovine rumen (47) and from human feces (45) clustered with these phylotypes. CLUSTER__A was positioned near the Spiroplasma group (RDP reg. no. 2.30.8.3), in the Mycoplasma and relatives group (RDP reg. no. 2.30.8), with a 90% bootstrap value. The intracluster similarities ranged from 75.6 to 96.4%. These phylotypes were not found often, but six of them were detected more than once.
FIG. 6.
Dendrogram showing the phylogenetic affiliation of OTUs from the porcine GI tract in the Mycoplasma and relatives (RDP reg. no. 2.30.8). 16S rDNA sequences from the bovine rumen (superscript a, from Tajima et al. [47]) and feces from an adult human (superscript b, from Suau et al. [45]) were added to this tree. The most closely related bacterium in the database to the phylotypes in CLUSTER__A was Mesoplasma seiffertii (81.5%) affiliated with the spiroplasmas. See the legend to Fig. 1 for explanation.
In the Eubacterium cylindroides subgroup (RDP reg. no. 2.30.8.2.9), OTU-186 was closely related to Eubacterium biforme and was detected in 12 pigs. Forty clones belonged to a phylotype (OTU-188) most closely related to a Eubacterium sp. The remaining phylotypes in this group had low similarities to known species.
Flexibacter-Cytophaga-Bacteroides (RDP reg. no. 2.15).
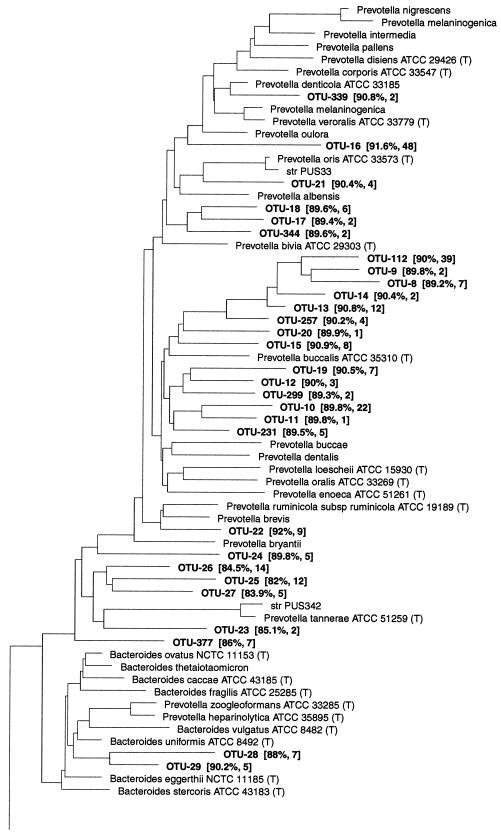
This was the most diverse group of gram-negative bacteria found, with 42 phylotypes affiliated with the Bacteroides group (RDP reg. no. 2.15.1.2) (Fig. 7). Three hundred ninety-three clones (9.2%) belonged to this group. They were found in the large intestine only, except OTU-16 and OTU-112, which were each detected by a single clone in the ileum of pig A-42. None of the phylotypes in this group had sequence similarity above 97% to known bacteria, and the mean similarity to isolated bacteria for which the 16S rRNA is available in the RDP was 87.5%.
FIG. 7.
Dendrogram showing the phylogenetic affiliation of OTUs from the porcine GI tract in the Bacteroides group (RDP reg. no. 2.15.1.2). See the legend to Fig. 1 for explanation.
Six OTUs belonged to the Prevotella nigrescens subgroup (RDP reg. no. 2.15.1.2.13). They were most closely related to Prevotella denticola, Prevotella veroralis, Prevotella oris, and Prevotella albensis. In the Prevotella buccae subgroup (RDP reg. no. 2.15.1.2.12), 14 OTUs formed two clusters that were not affiliated with any known species. The known bacterial species with the highest sequence similarity was Prevotella loescheii, a member of the human oral flora. The 115 clones comprising these phylotypes were found in 17 of the pigs analyzed. Two OTUs were related with low sequence similarity to the bovine rumen isolate Prevotella ruminicola and to Prevotella bryantii, respectively. Four phylotypes were found in the Prevotella tannerae subgroup (RDP reg. no. 2.15.1.2.9), and two phylotypes were found in the Bacteroides fragilis subgroup (RDP reg. no. 2.15.1.2.8), with Bacteroides uniformis as the closest related known species. In the Porphyromonas macacae subgroup (RDP reg. no. 2.15.1.2.7), five OTUs clustered together, with similarities in the range of 81.4 to 87%, to the closest related known species, Bacteroides merdae, previously isolated from human feces. Two OTUs were most closely related to Bacteroides distasonis. Six OTUs were affiliated with the Rikenella microfusus subgroup (RDP reg. no. 2.15.1.2.1). The closest related known bacterium was an isolate of Bacteroides putredinis from human feces; but the sequence similarity was only 78.4 to 87.7%.
Phylogenetic groups with few phylotypes.
Twenty OTUs were affiliated with the Proteobacteria (RDP reg. no. 2.28). In the gamma subdivision, OTU-1 was related to E. coli with 99% sequence similarity. Nine clones of OTU-226 were closely related (99.7%) to Pantoea agglomerans. In the Haemophilus-Pasteurella group (RDP reg. no. 2.28.3.26), the phylotypes were closely related to a Haemophilus sp. (99%), Actinobacillus minor (99.1%), Actinobacillus porcinus (98.8%), and Pasteurella aerogenes (99.7%). They were all found in low numbers. OTU-2 was represented by five clones and was distantly related to Thiocapsa roseopersicina (90.7%). In the Oceanospirillum group (RDP reg. no. 2.28.3.11), the two clones of OTU-203 were closely related to the marine isolate, Halomonas aquamarina (99% sequence similarity). Five OTUs belonged to the beta subgroup of the Proteobacteria. They were related to Roseateles depolymerans (97.3% sequence similarity), Oxalobacter formigenes (97.8% sequence similarity), Sutterella wadsworthensis (90.5 and 91.7% sequence similarity), and Gallionella ferruginea (86.6% sequence similarity). Two phylotypes were found in the alpha subgroup. They were related to Blastobacter denitrificans (99.8% sequence similarity), isolated from freshwater, and the marine Oceanospirillum pusillum (85.5% sequence similarity). In the delta subdivision two phylotypes were related to Desulfomonas pigra (86.4 and 94.1%), and in the epsilon subdivision, two OTUs were related with high sequence similarity to Helicobacter rodentium (99.5%) and a Campylobacter sp. (97.4%), while OTU-349 was distantly related to Pelobacter propionicus (85.4% sequence similarity).
One phylotype, represented by three clones, was affiliated with the Planctomyces and relatives (RDP reg. no. 2.20). The most closely related known bacterium was the aquatic Pirellula marina; however, the sequence similarity was only 86%.
Two phylotypes belonged to the spirochetes and relatives (RDP reg. no. 2.27). They were related with low sequence similarity to Treponema bryantii (88.1%) and a Spirochaeta sp. (84.7%).
In the group of high-G+C bacteria (RDP reg. no. 2.30.1) four phylotypes were found. They were related to Sanguibacter inulinus (99.8% sequence similarity), Bifidobacterium asteroides (93.5%), Eggerthella lenta (89.7% sequence similarity), and Atopobium parvulum (91.1% sequence similarity). These phylotypes were found infrequently.
OTU-266 was affiliated with the Flexistipes sinusarabici assemblage (RDP reg. no. 2.14), with Geovibrio ferrireducens as the closest relative (85.9% sequence similarity), while OTU-359 was most closely related to Dethiosulfovibrio peptidovorans (84.3% sequence similarity) in the Anaerobaculum thermoterrenum group (RDP reg. no. 2.11). Both phylotypes were detected only once among the clones.
Abundance of clones.
The relative abundance of the clones approximated a lognormal distribution. Thus, there were few numerically dominant species and few very rare species and many species of intermediate abundance. Nine phylotypes each constituted 2% or more of the clones: OTU-180 (7.9%, S. alactolyticus-like); OTU-1 (4.8%, E. coli-like); OTU-171 (4.5%, L. amylovorus-like); OTU-184 (4%, F. prausnitzii-like); OTU-170 (3.2%, L. johnsonii-like); OTU-107 (2.9%, F. prausnitzii-like); OTU-133 (2.4%, S. termitidis-like); OTU-176 (2.4%, G. haemolysans-like); OTU-173 (2.1%, L. reuteri-like).
No consistent relations between the abundance of individual phylotypes and the differences between animals with respect to diet, animal age, or herd health were found.
Comparison of two clone libraries from the same sample.
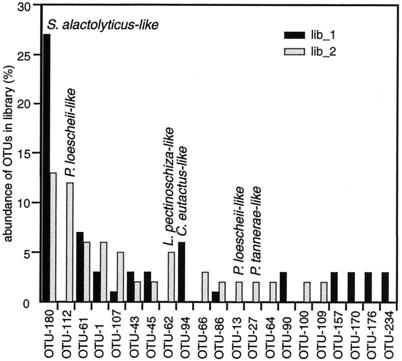
Two clone libraries were made from the colon sample of one pig (SPF-A), i.e., from one DNA extraction, but separate PCR amplifications and the abundances of phylotypes in the two libraries were compared. The coverage values of the two libraries were similar (library 1 [lib_1] coverage = 64.2%; lib_2 coverage = 67.4%). The relative abundance of the OTUs varied between the two samples (Fig. 8). In the first library, no phylotypes affiliated with the Bacteroides group were found, whereas in the second library they constituted 22.1% of the clones. In lib_1, OTU-180 (S. alactolyticus-like) was found in 26.9% of the clones. In lib_2, OTU-180 constituted only 12.8% of the clones. In lib_1, OTU-62 (L. pectinoschiza-like) was detected in 5% of the clones, whereas the C. eutactus-like OTU-94 was not found. In lib_2 this situation was reversed.
FIG. 8.
The relative abundance of phylotypes in two 16S rDNA clone libraries from one DNA extraction sampled from the colon of pig SPF-A. Only phylotypes representing >1% of the clones in each library are shown. Twenty-four phylotypes in lib_1 and 43 phylotypes in lib_2 were found. The names of the most closely related known bacterial species are indicated for some of the abundant OTUs.
DISCUSSION
The clone library presented in this paper is, to our knowledge, the most comprehensive collection of 16S rDNA sequences cloned from a single type of ecosystem. The overall coverage was 97.8%; thus, the clones comprised most of the phylotypes present in the 52 samples. Recently, Paster et al. (32) reported on a large clone library of bacteria associated with human subgingival plaque. The overall conclusions in their report and in our study regarding the complexity and diversity of the two ecosystems agree.
The pigs were selected to cover a variety of diets, animal ages, and herd health conditions because these parameters are known to influence the bacterial community structure in the porcine GI tract (10, 21, 28, 37, 48).
Only the luminal contents of the GI tract were analyzed. Differences between luminal and mucosa-associated microbiota have been indicated (33, 39, 41). However, due to the normal mucus excretion, epithelial turnover, and peristaltic movements in the GI tract, we assume that the mucosa-associated microbiota is a subset of the luminal microbiota and that this fraction was roughly represented among our clones.
A 10:1 ratio of PCR products to the cloning vector is required by the cloning procedure. To obtain sufficient rDNA for the cloning, we used 30 PCR cycles and pooled the PCR products from five individual reactions. Reports in the literature have indicated that amplification through many cycles may decrease the observed diversity (54, 56), although there is no consensus on this issue in samples from complex ecosystems (46, 58). However, we did not intend to characterize the diversity of individual samples by the cloning and sequencing procedure, but to comprehend as much as possible the bacterial diversity in the GI tracts of Danish pigs through analysis of a number of representative animals.
The frequency of formation of chimeric molecules increases by the number of amplification cycles and may be as high as 32% after 30 cycles, dependent on template sequence similarity and PCR conditions (50, 51), whereas typical frequencies are in the range of 5 to 10% (49). We found, using stringent criteria, that 11.9% of the near-full-length sequences were possible chimeras; however, it cannot be excluded that some chimeras may have eluded all three methods of detection.
Two striking features of the PCR-derived 16S rDNA sequences obtained in this study were the large proportion of phylotypes with sequence similarity of less than 97% to any known species and the clustering of phylotypes around a single cultured bacterial species.
Only 66 of the OTUs had sequence similarity of 97% or more to any previously cultured species for which the 16S rRNA sequence is available, i.e., only 17% of the phylotypes likely represented known bacterial species. Large fractions of uncharacterized phylotypes in 16S rDNA libraries have been found since the introduction of the rRNA approach to microbial ecology (8, 11, 44, 53), and this has provided the now generally accepted view that bacterial diversity in natural systems extends far beyond what can currently be perceived from culturing methods (15, 31, 43).
The phylophenetic concept of species for prokaryotes currently in use by taxonomists is based on a combined genomic, phenotypic, and phylogenetic characterization of bacterial isolates, and the comparative analysis of 16S rDNA sequences alone is insufficient for establishing the borders of a prokaryotic species (38). However, the phylogenetic characterization of an organism requires only the sequence of one or more genes, e.g., the 16S rDNA (31). The 97% sequence similarity used as a criterion for assigning 16S rDNA sequences to individual phylotypes and to assign phylotypes to known species is based on the empirical observation that bacteria sharing less than 97.5% sequence similarity unlikely will be identified as different species in the phylophenetic species concept (42). The criterion is justified for comparisons of full 16S rDNA sequences only, because of the inherent structure of conserved and variable regions of the rRNA molecule. Therefore, and because correct phylogenetic relationships of sequences without any close relatives can be determined only from full 16S rDNA sequences (15, 22), the phylogenetic analysis presented in this paper was based on the near-full-length sequences. It is important to note that our estimate of 375 phylotypes found in the pig GI tract is not conclusive but varies according to the conditions set for the analysis. It is a conservative value which does not discriminate closely related genera such as Salmonella and Escherichia. Employing, for instance, a 97.5 or 98% sequence similarity criterion would substantially increase the number of phylotypes defined.
The comparison of two libraries from one sample showed that 16S rDNA clone libraries may fail to integrate significant parts of the bacterial community (in this study, the dominating gram-negative bacteria in lib_1). Distortion of clone libraries is attributed to differential PCR amplification caused by various factors (49) but may also be due to the random variation of picking a relatively small number of clones (96) to represent a highly complex system. This limitation of the method may be the reason that the distribution of phylotypes could not be related to specific conditions of the animals. Quantitative hybridization methods could provide this kind of information.
In their classic paper, Moore and Holdeman (27) indicated that the number of different kinds of bacteria in the human GI tract exceeds 400 to 500 species. This was a statistical estimate based on the 113 different kinds of bacteria actually detected by culturing in that study. From the morphological and functional resemblance of the human and porcine GI tract, it is reasonable to assume that the complexity of the pig microbiota would resemble that of the human GI tract. However, it is difficult to correlate our finding of 375 phylotypes in the pig GI tract to Moore and Holdeman's estimate, beyond the fact that they are in the same range, due to the problems of ascribing a certain value of 16S rRNA sequence similarity to the taxonomic definition of a species. Most of our phylotypes with a sequence similarity greater than 97% to known bacterial species were related to species previously isolated from the human GI tract or the rumen. This finding merely reflects that microbiologists have focused far more on these two habitats than on the porcine GI tract.
What do the phylotypes with low similarity to any sequence in the database represent? Their phylogenetic inferences can be estimated from the dendrograms, but the extrapolation of functional properties from well-characterized cultured strains to distantly related phylotypes may not be justified. Part of the low-similarity phylotypes undoubtedly represents novel species and higher taxonomic entities. Other phylotypes may already have been cultured and characterized, but their 16S rRNA sequence is not available in the databases. Thus, Ramsak et al. (34) have shown that for some of the large clusters of Prevotella and Bacteroides detected by molecular methods, cultured strains are indeed available. This view is also supported by our finding of a high overall sequence similarity of the phylotypes affiliated with the lactic acid bacteria, a group that has received much attention throughout the history of microbiology. In contrast, all the Prevotella- and Bacteroides-affiliated phylotypes had low similarity to cultured strains. None of the Prevotella sequences in the database are from intestinal isolates.
Five clusters of phylotypes that branched off deeply in the tree were found. They were supported by high bootstrap values and had intracluster sequence similarities as low as 75.6% (in CLUSTER__A). In comparison, the interdivisional 16S rRNA sequence difference in the Bacteria domain is typically 20 to 25% (15). The deep-branching clusters probably represent diverse taxa above the species level. Since no representatives of the clusters have been cultured (or at least sequenced), their phenotype cannot be predicted. In four of the clusters our phylotypes grouped with sequences from clone libraries derived from bovine rumen and from the GI tract of a human individual, suggesting that these taxa are indeed associated with the GI ecosystem.
We often found clusters of phylotypes around a single cultured species. This is a common feature of environmental clone libraries (3). The use of in situ hybridization has shown that it is a true reflection of the diversity in certain complex ecosystems (5, 12, 23). In the human intestine five coexisting populations of bacteria with minor phenotypic differences, all identified as E. rectale, were found by culturing (27), and based on DNA hybridizations Hudman and Gregg (14) concluded that the diversity of rumen bacteria (B. fibrisolvens, B. ruminicola) may be far greater than indicated by the current taxonomy. It is thus likely that the clusters of phylotypes represent the true diversity of the GI tract. A possible source of error that would produce bush-like phylogenies is intraspecies variation in the rRNA operon sequences, and it has been suggested that sequences from single operons cannot adequately represent taxa (7). A few examples of intraspecies rRNA operon sequence variation greater than 3% have been reported (29, 52, 57); however, a compilation of data from the genome sequences indicates that such high heterogeneity is exceptional (17).
Our results confirm previous observations that the Bacteroides and Prevotella group constitutes the largest fraction of gram-negative bacteria in the porcine GI tract. This has been documented both by the use of culturing methods (28, 36, 37, 39) and by molecular approaches (21, 33).
The majority of the phylotypes affiliated with the Prevotella in our study belonged to the nonruminal Prevotella supercluster as defined by Ramsak et al. (34). However, the sequence similarity to the cultured species of this group was low, and none of the phylotypes represented known, cultured species for which the 16S rRNA sequence is available in the RDP 7.1 alignment. Most of the nonruminal prevotellas are of human oral origin, and these cultured strains are likely poor representatives of the intestinal bacterial community. The clustering of most of the prevotellas with the nonruminal Prevotella species is in agreement with the findings from a human GI tract 16S rDNA clone library (45) but is in contrast to the clustering of prevotellas in the bovine rumen. A supercluster of very diverse phylotypes related to P. ruminicola was found in the bovine rumen clone libraries (47, 54), whereas we found only a single OTU in this cluster.
The Prevotella phylotypes were detected only in the cecum and colon, except for two clones which came from the ileum of one pig. Apparently the ecotype of this bacterial group is restricted to the conditions in the large intestines. The sensitivity of the prevotellas to bile, which inhibits their growth (18), may be the reason for their absence in the small intestines.
In agreement with what is known from culturing, our data show that the low-G+C gram-positive bacteria dominate the pig GI tract. However, the diversity of this group, indicated by the abundant bush-like clustering of phylotypes around known species found in this study, has never been realized by culturing, and it raises an intriguing question: what do these clusters represent in terms of metabolic functioning?
A large fraction of the low-G+C gram-positive phylotypes were affiliated with the Clostridium coccoides group and the Clostridium leptum subgroup, which contains many of the classic intestinal bacteria. The Sporomusa group also includes known intestinal and rumen bacteria, and phylotypes closely related to these strains were found. Phylotypes within the Clostridium botulinum group all had similarities greater than 97% to cultured intestinal or ruminal strains, whereas most phylotypes affiliated with the Eubacterium group had low sequence similarity to known bacteria. The known ecotype of the latter group is mainly environmental, and the strains most closely related to our phylotypes were isolated from mud and sediments.
The lactic acid bacteria and in particular the Lactobacilli included some of the most abundant phylotypes. This is in agreement with the findings from experiments using culturing methods with healthy pigs (37), but it differs from the situation in the human GI tract (27, 45). OTU-180, related to S. alactolyticus, was the most abundant phylotype, but the distribution of OTU-180 varied substantially between samples, and even between two libraries from the same sample. S. alactolyticus is a predominant member of the pig colon microbiota and may account for more than 50% of the cultured strains (37). As in the human GI tract (45, 56), F. prausnitzii-like phylotypes were abundant and formed a cluster of related OTUs. Clusters of phylotypes related to S. termitidis have been found both in the human GI tract (45) and in the bovine rumen (47) and were abundant among our clones. Surprisingly, OTU-1 related highly to E. coli and was the second most abundant phylotype, but this may be an error caused by the E. coli-based cloning system.
Nevertheless, the distribution of the clones was characterized by a large fraction of phylotypes with intermediate abundance and only few dominating or rare phylotypes. The lognormal distribution of the relative clone abundance suggests a complex structure of the bacterial community in the pig GI tract, where the extent of the resource space occupied by each phylotype is determined by a large number of independent variables affecting the relative success of one phylotype in competition with the other phylotypes (55).
This study has provided an inventory of phylotypes in the GI tracts of a collection of Danish pigs. The results document a hitherto unknown bacterial diversity and indicate that the majority of intestinal bacteria are uncharacterized. To validate the data, and to investigate the distribution and abundance of the phylotypes in individual samples, we are in the process of designing specific oligonucleotide probes for the phylotypes and to implement the probes in oligonucleotide arrays. These will facilitate the analysis of a large number of samples to address questions of how the intestinal bacterial community responds to perturbations such as different diets and the use of antibiotics, probiotics, etc.
Acknowledgments
We are indebted to Niels Larsen for his thoughtful encouragement throughout this project and for his generous providing of Perl-scripts that made it so much easier to maintain the data. We thank H. Rex Gaskins for critical review of the manuscript.
This work was supported by the Research Secretariat of the Danish Ministry of Food, Agriculture and Fisheries (project number Alt-98-1).
REFERENCES
- 1.Allison, M. J., I. M. Robinson, J. A. Bucklin, and G. D. Booth. 1979. Comparison of bacterial populations of the pig cecum and colon based upon enumeration with specific energy sources. Appl. Environ. Microbiol. 37:1142-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. 2000. Who is out there? Microbial aspects of biodiversity. Syst. Appl. Microbiol. 23:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R., and W. Ludwig. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555-565. [DOI] [PubMed] [Google Scholar]
- 5.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K.-H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1988. Preparation and analysis of DNA, p. 2.4.1-2.4.5. In F. M. Ausubel (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 7.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 8.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaskins, H. R. 2001. Intestinal bacteria and their influence on swine growth, p. 585-608. In A. J. Lewis and L. L. Southern (ed.), Swine nutrition. CRC Press, Boca Raton, Fla.
- 11.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 12.Glöckner, F. O., H.-D. Babenzien, J. Wulf, and R. Amann. 1999. Phylogeny and diversity of Achromatium oxaliferum. Syst. Appl. Microbiol. 22:28-38. [DOI] [PubMed] [Google Scholar]
- 13.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrica 40:237-264. [Google Scholar]
- 14.Hudman. J. F., and K. Gregg. 1989. Genetic diversity among strains of bacteria from the rumen. Curr. Microbiol. 19:313-318. [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 17.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause, D. O., and J. B. Russell. 1996. How many ruminal bacteria are there? J. Dairy Sci. 79:1467-1475. [DOI] [PubMed] [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 20.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The ribosomal database project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Møller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig, W., and K.-H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, W. E. C., L. V. H. Moore, E. P. Cato, T. D. Wilkins, and E. T. Kornegay. 1987. Effect of high-fiber and high-oil diets on the fecal flora of swine. Appl. Environ. Microbiol. 53:1638-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mylvaganam, S., and P. P. Dennis. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neefs, J.-M., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace, N. R. 1996. New perspective on the natural microbial world: molecular microbial ecology. ASM News 62:463-470. [Google Scholar]
- 32.Paster. B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pryde, S., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsak, A., M. Peterka, K. Tajima, J. C. Martin, J. Wood, M. E. A. Johnston, R. I. Aminov, H. J. Flint, and G. Avgustin. 2000. Unravelling the genetic diversity of ruminal bacteria belonging to the CFB phylum. FEMS Microbiol. Ecol. 33:69-79. [DOI] [PubMed] [Google Scholar]
- 35.Reysenback, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, I. M., M. J. Allison, and J. A. Bucklin. 1981. Characterization of the cecal bacteria of normal pigs. Appl. Environ. Microbiol. 41:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson, I. M., S. C. Whipp, J. A. Bucklin, and M. J. Allison. 1984. Characterization of predominant bacteria from the colons of normal and dysenteric pigs. Appl. Environ. Microbiol. 48:964-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 39.Russel, E. G. 1979. Types and distribution of anaerobic bacteria in the large intestine of pigs. Appl. Environ. Microbiol. 37:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salanitro, J. P., I. G. Blake, and P. A. Muirhead. 1977. Isolation and identification of fecal bacteria from adult swine. Appl. Environ. Microbiol. 33:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Optimization of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-180. [DOI] [PubMed] [Google Scholar]
- 42.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 43.Stahl, D. A. 1995. Application of phylogenetically based hybridization probes to microbial ecology. Mol. Ecol. 4:535-542. [Google Scholar]
- 44.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1985. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl. Environ. Microbiol. 49:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tajima, K., R. I. Aminov, T. Nagamine, K. Ogata, M. Nakamura, H. Matsui, and Y. Benno. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 29:159-169. [Google Scholar]
- 48.Varel, V. H., I. M. Robinson, and H.-J. G. Jung. 1987. Influence of dietary fiber on xylanolytic and cellulytic bacteria of adult pigs. Appl. Environ. Microbiol. 53:22-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Wang, G. C.-Y., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 51.Wang, G. C.-Y., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Y., Z. Zhang, and R. Narendrakumar. 1997. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 54.Whitford, M. C., R. J. Forster, C. E. Beard, J. Gong, and R. M. Teather. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153-163. [DOI] [PubMed] [Google Scholar]
- 55.Whittaker, R. H. 1975. Communities and ecosystems, p. 82-104. MacMillan Publishing Co., Inc., New York, N.Y.
- 56.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the Actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoetendal, E. G., A. D. L. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]