Abstract
The presence of Yersinia ruckeri in a French fish farm was investigated. Y. ruckeri was isolated mainly from algae and sediment samples rather than from water. Twenty-two Y. ruckeri isolates were obtained, and three strains were distinguished by enterobacterial repetitive intergenic consensus PCR amplification. These strains were able to adhere to solid supports. This characteristic was correlated with flagellum-mediated motility. Killing experiments showed that sessile cells were more resistant to oxolinic acid than their planktonic counterparts. Our results demonstrate that surface colonization of fish farm tanks by Y. ruckeri biofilms is a potential source of recurrent infection for extended periods of time.
Yersiniosis or enteric redmouth disease (ERM), caused by Yersinia ruckeri, is a serious infectious disease in the rainbow trout farming industry that causes economic problems in many countries (3). ERM is characterized by the presence of congestive or hemorrhagic zones in various tissues and organs, particularly around the mouth and in the intestines. Its mode of transmission has been related to wild or farmed carrier fish and other putative vectors, such as aquatic invertebrates and birds (27). The pathogen has been isolated from the feces of carrier fish 2 months after an ERM outbreak (18). Therefore, the ability of Y. ruckeri to survive and remain infective in the aquatic environment must be considered a major determinant in the spread of the disease (19). It is now accepted that bacterial biofilms are prevalent on most wet surfaces in nature (7). Very few (if any) studies on the ability of Y. ruckeri to form biofilms have been performed, however.
In this study, we investigated the presence of Y. ruckeri in a rearing tank on a French fish farm during routine monthly sampling for 1 year. Y. ruckeri was recovered from water, algae, and sediment samples. The genetic diversity among Y. ruckeri environmental isolates was studied by using enterobacterial repetitive intergenic consensus PCR (ERIC-PCR). The ability of the majority strain to adhere to wood supports was then investigated. The motility of environmental strains was compared to that of a reference strain. The resistance of fixed cells to oxolinic acid (an antibiotic used routinely to treat yersiniosis) was also compared to that of their free-floating counterparts.
MATERIALS AND METHODS
Sampling.
Water, sediment, and alga samples were collected monthly from the outdoor rearing tank of a French fish farm from October 1999 to September 2000. This farm is located at the source of the Cailly River, an oligotrophic river that flows into the Seine River in Rouen, France. Water samples were collected aseptically in sterile glass bottles at a depth of 10 cm. Temperature and pH were determined with hand-held probes. During the investigation period, the pH and temperature of the water were uniform (pH 7.5 and 11°C, respectively). Surficial sediments in the tank were collected with a dredge. Algae located on tank walls were removed with a scraper. Three rainbow trout (Oncorhynchus mykiss) were sampled each month at the same time. Samples were immediately placed in a 4°C cooling box and were processed within 1 h after collection.
Reference strains.
Y. ruckeri ATCC 29473, Escherichia coli K-12 strain CIP 54-117, and Hafnia alvei ATCC 13337 were purchased from the Pasteur Institute Collection. F. Esnault (Trouw-France S.A., Fontaine-Les-Vervins, France) kindly provided two Y. ruckeri strains (strains YR55 and YR80) that were freshly isolated from moribund trout kidneys. One clinical strain of Yersinia enterocolitica was provided by the Microbiology Laboratory of Charles Nicolle Hospital in Rouen, France.
Enumeration of Y. ruckeri and total flora.
Water samples (0.25, 0.1, and 0.05 ml) were plated onto SW-DCN medium (SW medium [21, 23] supplemented with 1.2 mM sodium deoxycholate,7.2 μM cefsulodin, and 3.9 μM novobiocin). After filtration under a vacuum with cellulose nitrate membrane filters (pore size, 0.22 μm; Millipore), alga and sediment samples were weighed and resuspended in 20 ml of 0.1 M phosphate buffer (pH 7). The suspensions were homogenized by mixing them with a vortex mixer for 1 min, sonicated in a water bath (Deltasonic, Meaux, France) for 4 min at 50 W and 4°C, and vortex mixed again for 1 min. After centrifugation at 4 × g for 5 min, decimal dilutions of the supernatants were plated onto SW-DCN medium. Simultaneously, the numbers of total cultivable flora in water, alga, and sediment samples were determined by plating 0.05-ml portions of suitable decimal dilutions of samples onto Trypticase soya agar (TSA) (Difco) plates. The plates were incubated for 48 h at 25°C.
Kidneys were removed aseptically from fish. The organs were weighed and blended individually with 10 ml of 0.1 M phosphate buffer (pH 7). The presence of Y. ruckeri in the tissue homogenates was determined by plating 25 μl of each homogenate onto SW-DCN medium. The plates were incubated at 25°C for 4 days. Isolates suspected of being Y. ruckeri (isolates that produced green colonies with a zone of Tween 80 hydrolysis) were identified presumptively by whole-cell seroagglutination by using an antiserum provided by CNEVA (Brest, France).
PCR amplification.
Two forward random primers, YER1 (5"-ACGAATCAGGCTGTTACCG-3") and YER2 (5"-TGCCTGTGCCAATGTTGGC-3"), previously described by Argenton et al. (2), were used for PCR assays. One colony of a suspected Y. ruckeri isolate was resuspended in 200 μl of sterile distilled water. Amplification was performed with a reaction mixture containing 1 μl of the bacterial suspension, each deoxynucleoside triphosphate (Eurogentec, Seraing, Belgium) at a concentration of 200 μM, each primer (Genset SA, Paris, France) at a concentration of 1 μM, 5 μl of 10× Taq buffer [750 mM Tris-HCl (pH 8.8), 200 mM (NH4)2SO4, 0.1% (vol/vol) Tween 20], 1.25 U of Taq polymerase (Eurogentec), and 2.5 mM MgCl2. The final volume was adjusted to 50 μl with sterile distilled water. The PCR system 9700 thermocycler (Applied Biosystems) used for thermal amplification was programmed as follows: (i) an initial denaturation step consisting of 94°C for 5 min; (ii) 30 reaction cycles consisting of 94°C for 45 s, 60°C for 1 min, and 72°C for 1 min; and (iii) a final extension step consisting of 72°C for 5 min. PCR-generated products (10 μl of each amplification mixture) were detected by electrophoresis by using 1.5% agarose gels.
ERIC-PCR.
The oligonucleotide ERIC primers ERIC1R (5"-ATGTAAGCTCCTGGGGATTCAC-3") and ERIC2 (5"-AAGTAAGTGACTGGGGTGAGCG-3") were designed as described elsewhere (22). One colony of a Y. ruckeri isolate was resuspended in 200 μl of sterile distilled water. Amplification was performed with the reaction mixture described above except that 2 U of Taq polymerase and 1.5 mM MgCl2 were used. The thermocycler was programmed as follows: (i) an initial denaturation step consisting of 95°C for 5 min; (ii) 35 reaction cycles consisting of 94°C for 45 s, 52°C for 1 min, and 72°C for 8 min; and (iii) a final extension step consisting of 72°C for 15 min. PCR-generated products were detected as described above.
Two strains were considered different when their profiles differed in at least one band.
Determination of API 20E profiles.
Y. ruckeri strains were subjected to the API 20E test and the supplementary test to determine xylose-fermenting ability in order to distinguish Y. ruckeri from H. alvei (11).
Growth conditions.
Bacterial strains were maintained as glycerol stocks and stored at −80°C. Cultures were grown in 50-ml Erlenmeyer flasks containing 15 ml of Columbia medium (Difco). The flasks were incubated for 18 h at 25°C in a gyratory water bath shaker at 115 rpm. After collection of bacteria by centrifugation, each pellet was washed and resuspended in sterile distilled water. The size of the cell population was estimated by measuring the optical density at 550 nm and comparing it to a calibration curve for plate counts.
Bacterial adhesion to solid supports.
The ability of Y. ruckeri strains to initiate biofilm formation was tested by using wood supports (length, 5.5 cm; width, 3.5 cm; thickness, 5 mm), for which Y. ruckeri exhibits a strong affinity (L. Coquet, P. Cosette, G.-A. Junter, E. Beucher, J.-M. Saiter, and T. Jouenne, Colloids Surfaces B. Biointerfaces, submitted for publication). Each sterile support, suspended at the end of a glass rod, was immersed in 300 ml of diluted (10%, vol/vol) Columbia medium in an Erlenmeyer flask. The flasks were inoculated with an 18-h-old culture of Y. ruckeri (initial cell population density, 106 CFU ml−1) and incubated at 25°C with gyratory agitation at 115 rpm for at least 2 weeks. The medium was replaced every 3 days. Five supports were used for each experiment.
The supports were periodically (every 3 days) removed from the flasks and rinsed with sterile distilled water to remove weakly fixed organisms. Adherent cells were recovered by sonication of each support in 20 ml of sterile 0.1 M phosphate buffer (pH 7) precooled at 4°C. The resulting cell suspension was vortex mixed and then serially diluted in decimal steps. Aliquots (25 μl) of each dilution were spread on TSA plates. The plates were incubated for 48 h at 25°C. The numbers of bacteria were expressed as log CFU per square centimeter of support.
Antibiotic and MIC determination.
Oxolinic acid was purchased from Sigma-Aldrich. The MIC was determined by the macrodilution method (14) by using Mueller-Hinton broth (Difco) and an inoculum containing 106 CFU ml−1. A value of 1.5 μg ml−1 was obtained for the Y. ruckeri strain tested (strain PBM1).
Susceptibility tests.
Wood supports were incubated for 6 days in 10% (vol/vol) Columbia broth as described above and then were transferred into 300 ml of Columbia broth supplemented with 285 μg of oxolinic acid ml−1 (i.e., 190 times the MIC). Every 2 h, one of the supports was removed, rinsed, and then placed in 20 ml of sterile 0.1 M phosphate buffer (pH 7). Viable sessile cells were enumerated as described above. Bactericidal assays were also performed with suspensions of Y. ruckeri. The initial cell concentration (107 CFU ml−1) was chosen based on the number of sessile bacteria and the volume of culture medium used for bactericidal assays on biofilms. Three independent experiments were performed.
Motility assays.
Bacteria were stab inoculated with a needle into the bottoms of Columbia agar plates (0.3% agar; average depth, 3 mm). The plates were incubated at 25°C for 24 h, and the migration zones were measured (15).
Preparation and analysis of outer membrane extracts.
Crude outer membrane extracts were prepared from bacterial pellets by using the spheroplast procedure of Mizuno and Kageyama (13). Briefly, bacteria were harvested by centrifugation for 15 min at 3,500 × g and washed twice with 20% (wt/vol) sucrose. Cells (ca. 1.5 g, wet weight) were suspended in a digestion solution containing 28 ml of 2 M sucrose, 10 ml of 0.1 M Tris-HCl (pH 7.8 at 25°C), 0.8 ml of 1% (wt/vol) Na-EDTA (pH 7.0), and 1.8 ml of 0.5% lysozyme. The mixture was incubated for 1 h at 30°C in the presence of DNase and RNase (each at 3 μg ml−1; Sigma). Spheroplasts were collected by centrifugation for 20 min at 10,000 × g. Outer membranes were then pelleted by centrifugation at 150,000 × g for 1 h at 4°C, resuspended in 500 μl of sterile distilled water, and dialyzed overnight against 1 mM Na-EDTA by using a dialysis membrane with a molecular mass cutoff of 25 kDa. The amount of proteins in a dialyzed sample was measured by the Bio-Rad protein assay. The protein patterns were analyzed by two-dimensional gel electrophoresis (2-DE). Two hundred micrograms of protein was added to isoelectric focusing buffer (final volume, 400 μl) consisting of 5 M urea, 2 M thiourea, 33 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2 mM tributyl phosphine, 10 mM dithiothreitol, 2% (vol/vol) carrier ampholytes (pH 3.5 to 10; Sigma), and 0.4% (wt/vol) bromophenol blue. The first-dimension gel separation was carried out with Immobiline Dry Strips NL (pH 3 to 10; Pharmacia). The second-dimension separation was obtained by performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 12.5% (wt/vol) polyacrylamide resolving gel (width, 16 cm; length, 20 cm; thickness, 0.75 mm). After migration, the proteins were visualized by silver nitrate staining.
N-terminal amino acid sequence analysis.
After 2-DE, proteins were electrotransferred onto polyvinylidene difluoride membranes (Millipore). Protein spots were visualized by staining the membranes with 0.1% (wt/vol) Coomassie brilliant blue G-250 (Sigma), and spots of interest were cut out. The N-terminal sequences of proteins were determined by introducing the blots into an Applied Biosystems 492 automated protein sequencer. The sequences obtained were compared to sequences in public protein sequence databases by using the BLASTP algorithm (1).
RESULTS
Isolation of Y. ruckeri and bacterial counting.
A total of 147 suspected Y. ruckeri isolates were obtained from the different environmental samples. No isolates suspected of being Y. ruckeri were obtained from fish kidney homogenates.
Twenty-two of the isolates (including 16 isolates obtained from algae or sediments) displayed positive seroagglutination and produced a strong PCR amplification product (data not shown). Amplification revealed a band at approximately 550 bp, in agreement with the expected size (2). Control experiments performed with H. alvei ATCC 13337 and a clinical strain of Y. enterocolitica yielded no PCR product, which confirmed the specificity of the YER1 and YER2 primers (2).
Twenty of the Y. ruckeri isolates had the same API 20E numeric profile (5105500), while two isolates had slightly different API 20E profiles (5107500 and 5104100). These profiles were similar to those of the reference strains (5104100 for strain ATCC 29473 and 5107100 for strains YR55 and YR80). The observed biochemical differences affected reactions (the Voges-Proskauer, gelatin hydrolysis, and sorbitol fermentation reactions) that were previously described as variable in Y. ruckeri (8).
Enumeration of cultivable cells on TSA showed that the size of the bacterial population remained roughly constant in water, algae, and sediments during the investigation period (Table 1). A total bacterial flora was encountered mainly in sediments and algae. This was particularly true for some very high concentrations of Y. ruckeri cells in alga samples. Some seasonal variations in the number of Y. ruckeri cells were observed with water and sediment samples, and no bacteria were recovered in the winter. In contrast, the number of Y. ruckeri cells in alga samples seemed to be independent of the season.
TABLE 1.
Bacterial enumeration and sampling
| Sampling date (day/mo/yr) | Water samples
|
Sediment samples
|
Alga samples
|
|||
|---|---|---|---|---|---|---|
| Total bacteria (103 cells ml−1) | Y. ruckeri (cells ml−1)a | Total bacteria (106 cells g−1) | Y. ruckeri (103 cells g−1) | Total bacteria (106 cells g−1) | Y. ruckeri (103 cells g−1) | |
| 18/10/99 | 2.5 | 18 | 2.0 | BDTb | 1.1 | 11.0 |
| 15/11/99 | 4.3 | 4 | 3.8 | 0.8 | 6.9 | BDT |
| 20/12/99 | 6.5 | BDT | 1.8 | 0.9 | 4.5 | BDT |
| 21/01/00 | 1.4 | BDT | 1.8 | BDT | 4.9 | 10.5 |
| 25/02/00 | 2.4 | BDT | 6.2 | BDT | 1.5 | BDT |
| 30/03/00 | 3.9 | BDT | 6.5 | BDT | 3.0 | 218.1 |
| 28/04/00 | 4.5 | BDT | 1.4 | BDT | 4.0 | 2.2 |
| 31/05/00 | 4.4 | BDT | 1.5 | BDT | 1.2 | BDT |
| 27/06/00 | 4.8 | 20 | 1.3 | 2.2 | 3.8 | 2.3 |
| 21/07/00 | 2.2 | 4 | 9.0 | 1.7 | 7.8 | 21.0 |
| 25/08/00 | 6.2 | 10 | 3.2 | 4.3 | 3.4 | BDT |
| 26/09/00 | 4.0 | 10 | 5.2 | BDT | 3.2 | 1.8 |
Y. ruckeri was identified by PCR.
BDT, below detection threshold (5 × 102 cells g−1 or 4 cells ml−1).
ERIC-PCR-based differentiation of isolates.
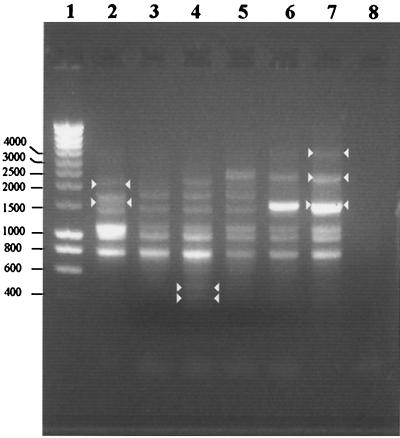
The ERIC-PCR analysis resulted in three different patterns for the 22 Y. ruckeri isolates (data not shown). One of these patterns (the strain PBM1 pattern) was obtained with the majority of the isolates (20 isolates). The other two patterns were obtained for the two isolates identified on the basis of their API 20E numeric profiles (5107500 and 5104100) (see above). These two isolates were designated strains PBM2 and PBM3, respectively. The fingerprints of strains PBM1, PBM2, and PBM3 are shown in Fig. 1. Three fragments (721, 912, and 1,015 bp) were found in all strains. Two bands, one at 1,268 and one at 1,562 bp, were produced by PBM1, PBM2, PBM3, and Y. ruckeri ATCC 29473 but not by Y. ruckeri YR55 and YR80. A band at 1,426 bp was produced only by PBM1, while a band at 1,962 bp was produced by PBM1 and PBM3. ERIC-PCR products that were smaller than 600 bp were obtained only with PBM3. Strains YR55 and YR80 produced bands at 1,384 and 3,439 bp that were not produced by the other strains. A PCR product at 2,104 bp allowed us to distinguish reference strains ATCC 29473, YR55, and YR80 from environmental strains. Surprisingly, analysis of the amplification products generated from Y. ruckeri YR55 and YR80 revealed a single profile.
FIG. 1.
DNA fingerprints of PMB1, PMB2, and PMB3 generated by ERIC-PCR amplification. Whole bacteria were used as templates. Lane 1, standard markers; lane 2, PMB1; lane 3, PMB2; lane 4, PMB3; lane 5, Y. ruckeri ATCC 29473; lane 6, Y. ruckeri YR80; lane 7, Y. ruckeri YR55; lane 8, control without DNA. Bands whose presence or absence was specific for one strain are indicated by arrowheads.
Adhesion of ATCC 29473 and PBM1.
The ability of PBM1 to adhere to wood supports was clearly greater than that of ATCC 29473 (Table 2). After incubation for 2 weeks, the number of adherent bacteria per square centimeter of support was approximately 100 times higher for PBM1 than for the reference strain.
TABLE 2.
Adhesion of Y. ruckeri ATCC 29473 and PBM1 to wood supports
| Length of incubation (days) | 106 cultivable cells cm−2
|
|
|---|---|---|
| ATCC 29473 | PBM1 | |
| 0 | 0 | 0 |
| 3 | 16 ± 5a | 62 ± 24 |
| 6 | 28 ± 15 | 131 ± 42 |
| 9 | 12 ± 3 | 683 ± 111 |
| 13 | 39 ± 28 | 703 ± 49 |
| 15 | 21.0 ± 0.2 | 2,750 ± 65 |
Mean ± standard deviation (n = 3).
Motility assays.
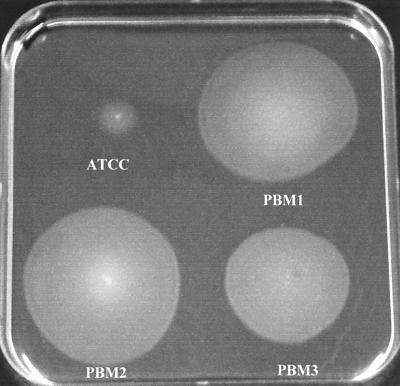
PBM1, PBM2, and PBM3 exhibited aberrant flagellum-mediated motility (Fig. 2). The diameters of the migration zones produced by the environmental strains were three to four times greater than the diameter of the migration zone produced by the reference strain (13, 49, 46, and 39 mm for ATCC 29473, PBM1, PBM2, and PBM3, respectively). The growth rates of the four strains were similar (0.0115 min−1), however.
FIG. 2.
Macroscopic assay for flagellum-mediated motility. An agar (0.3%, wt/vol) plate was stab inoculated with a needle in the bottom of the plate and was incubated for 24 h at 25°C. Migration of cells from the point of inoculation (a turbid zone) indicates that a strain has flagellum-mediated motility.
Outer membrane protein patterns and N-terminal sequencing.
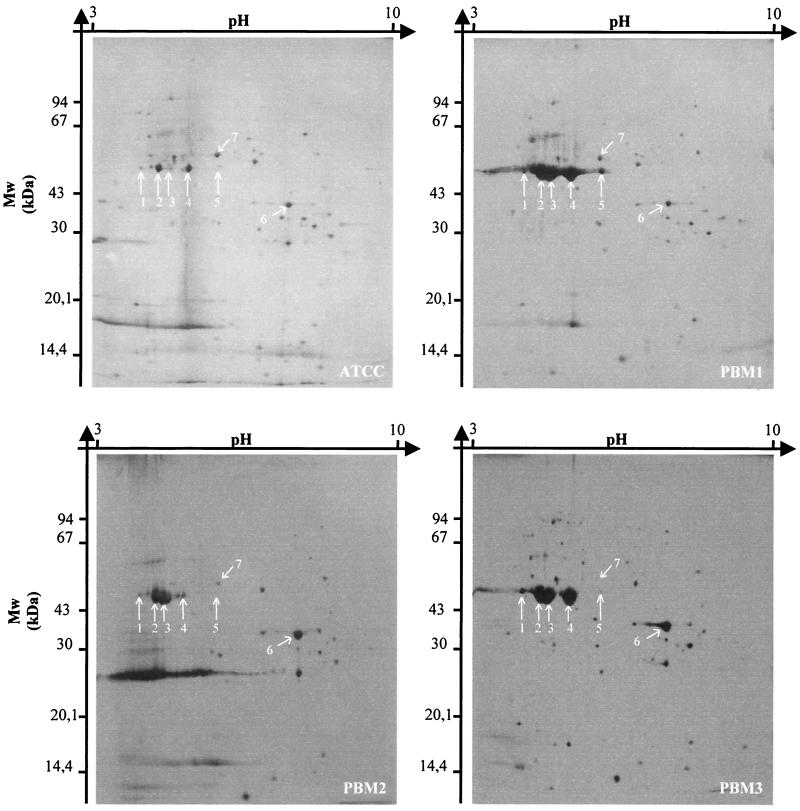
After 2-DE of outer membrane crude extracts, gel analysis revealed that PBM1 but not ATCC 29473 accumulated five proteins (proteins 1 to 5) (Fig. 3). Four of these proteins had the same apparent molecular mass (50 kDa), but the pI values were different (pI 4.8, 5.1, 5.6, and 6.3 for proteins 2 to 5, respectively). These proteins had the same N-terminal amino acid sequence (A-V-I-N-T-N-S-L-S-L-L-T). This sequence exhibited 92% identity with that of flagellins 1 and 2 of Yersinia pestis. The fifth protein (protein 1) had an apparent molecular mass of 52 kDa and a pI of 4.3. Its sequence (F-S-Q-A-V-S-G-L) exhibited 100% identity with the N-terminal sequence of the flagellar hook protein of E. coli. A comparison of the 2-DE patterns of the outer membrane proteins of two of the reference strains revealed that YR55 accumulated proteins 1, 2, 4, and 5 but strain ATCC 29473 did not (data not shown). Protein 6 was commonly accumulated by YR55, PBM2, and PBM3 but not by the other strains. The N-terminal sequence of this protein (A-P-K-D-N-T-X-Y-T-G-G-K-L) exhibited 92% identity with the N-terminal sequence of β-barrel membrane protein OmpA of Yersinia pseudotuberculosis.
FIG. 3.
2-DE of outer membrane proteins of Y. ruckeri strains. Proteins were visualized by silver staining. For identification of the spots, see the text. Mw, molecular mass.
Only proteins 1, 2, and 3 were overexpressed by PBM2 compared with strain ATCC 29473, whereas only proteins 3 and 4 were strongly accumulated by PBM3. On the other hand, protein 5 was not expressed by PBM2 and PBM3. Protein 7 was not expressed by PBM3. The N-terminal amino acid sequence of this protein (E-N-L-L-Q-V-Y-K-Q-A-A-E) exhibited 92% identity with the N-terminal amino acid sequence of the HasABC exporter outer membrane component (a TolC analogue) of Serratia marcescens.
Susceptibility test.
The susceptibility test was carried out with PBM1, which was the majority strain in the fish farm tank. Biofilm-forming cells displayed a high level of resistance to oxolinic acid compared with their free-floating counterparts. Exposure for 10 h to 285 μg of oxolinic acid ml−1 resulted in a 5-log decrease in the size of the population in a free cell culture, whereas a decrease of 2 log units was observed for the sessile cell population subjected to the same antibiotic treatment (Table 3).
TABLE 3.
Oxolinic acid efficacy against planktonic and sessile PBM1 cells
| Length of exposure (h) | Planktonic cells
|
Sessile cells
|
|||
|---|---|---|---|---|---|
| 104 cultivable cells ml−1
|
106 cultivable cells cm−2
|
||||
| Controla | With oxolinic acid (285 μg ml−1) | Controla | With oxolinic acid (285 μg ml−1) | ||
| 0 | 960 ± 210b | 815 ± 255 | 62 ± 25 | 54 ± 24 | |
| 2 | 1,540 ± 530 | 0.5 ± 0.1 | 100 ± 49 | 23 ± 16 | |
| 4 | 2,840 ± 640 | 0.01 ± 0.05 | 45 ± 33 | 7 ± 4 | |
| 6 | 4,380 ± 1,360 | 0.05 ± 0.01 | 68 ± 31 | 0.5 ± 0.2 | |
| 8 | 5,230 ± 2,640 | 0.02 ± 0.01 | 72 ± 42 | 0.5 ± 0.1 | |
| 10 | 9,980 ± 2,030 | 0.01 ± 0.01 | 56 ± 32 | 0.3 ± 0.1 | |
Without antibiotic.
Mean ± standard deviation (n = 3).
DISCUSSION
Y. ruckeri is readily isolated from kidney samples from clinically infected fish by using routine bacteriological media, but isolation from carrier fish is more difficult. In fact, such fish are difficult to recognize because they show no pathology and asymptomatic infection can be detected only in the lower intestine (20). Bush and Lingg (5) demonstrated that up to 25% of the fish in a rainbow trout population could carry Y. ruckeri in their lower intestines. In addition, periodic shedding of the organism in the feces is a source of infection for other fish (18). The finding that two reference strains (YR55 and YR80) that were originally isolated from the Somme River and the Meuse River (two distinct French rivers that are about 200 km apart) are in fact the same organism supports the hypothesis that the organism is transported by birds (27), through channels, or by fish exchange between farmers.
The ability of Y. ruckeri to survive in the aquatic environment must be considered a major determinant in the spread of ERM. Few studies on the persistence of this pathogen in natural waters have been performed, however. Furthermore, no one has addressed the biofilm problem, although it is now well recognized that most bacteria in aquatic environments are associated with surfaces rather than in the planktonic state (6). Wedemeyer and Nelson (26) found that the number of cultivable bacterial fish pathogens was constant for at least 1 month in untreated lake water. Using microcosms, Romalde et al. (19) found that Y. ruckeri persisted longer in sediments than in environmental waters. They also demonstrated that Y. ruckeri might survive in a dormant state under starvation conditions. Our data show that Y. ruckeri may actually grow on surfaces in an aquatic environment. The absence of this phenotype in strain ATCC 29473 might be a consequence of genetic divergence, long-term cultivation, and storage, factors that can result in genetic diversity (10).
The biofilm-forming ability of Y. ruckeri probably reflects adaptation of the bacterium to the detrimental oligotrophic conditions that prevail in the aquatic system; the sessile mode of growth is the sine qua non of bacterial survival in this hostile environment (7). The overexpression of proteins implied in the flagellum structure is part of the phenotypic features of bacteria that display high adhesion efficiency (15). Genetic screening analyses of biofilm-defective mutants have shown that the initial interaction with the surface is accelerated by force-generating organelles, such as pili and flagella (24). Once contact with the surface is made, bacteria use either flagella or pili to move along the surface until other bacteria are encountered and microcolonies are formed or enlarged (15, 16, 25). Our observations agree with those of Rhodes and Kator (17), who reported that there was an increase in the density of microflagella in Salmonella spp. during exposure to estuarine water.
TolC is the outer membrane component of the type I secretory pathway of gram-negative bacteria (4). Underexpression of a TolC analogue protein by PBM3 might indicate that this strain exhibits multidrug susceptibility.
Biofilm bacteria are known to be resistant to a number of adverse conditions (7, 9). The persistence of Y. ruckeri cells in alga samples during the investigation period and the seasonal variations in the number of Y. ruckeri cells in water might reflect this resistance. However, the prolonged seasonal absence of Y. ruckeri in sediments is not explained by this hypothesis. Since biofilms are notoriously difficult to eradicate and are a source of many recalcitrant infections (12), we hypothesized that surface-adherent Y. ruckeri cells were less susceptible to antibiotics than planktonic cells. This hypothesis was confirmed since sessile PBM1 cells displayed strong resistance to oxolinic acid compared with their planktonic counterparts.
In conclusion, this study revealed the tendency of Y. ruckeri to use the sessile state to persist in the natural aquatic environment. A question that arises from our observations is whether this propensity to initiate interactions with a solid surface reflects natural selection in a hostile environment or stable genetic modifications induced by the sessile mode of growth. We favor the first hypothesis since most genetic studies of single-species biofilms have revealed modifications only at the gene transcription level compared with planktonic cells (24).
Our results obviously show that surface colonization in fish farm tanks by Y. ruckeri biofilms may be a source of recurrent infections for extended periods of time. Further studies will address the virulence of the environmental strains in order to investigate the possible interaction between pathogenicity and biofilm-forming properties.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argenton, F., S. De Mas, C. Malocco, L. Dalla Valle, G. Giogetti, and L. Colombo. 1996. Use of random DNA amplification to generate specific molecular probes for hybridization tests and PCR-based diagnosis of Yersinia ruckeri. Dis. Aquat. Org. 24:121-127. [Google Scholar]
- 3.Austin, B., and D. A. Austin. 1993. Bacterial fish pathogens: diseases in farmed and wild fish, 2nd ed., p. 208-216. Ellis Horwood, Chichester, United Kingdom.
- 4.Buchanan, S. K. 2001. Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem. Sci. 26:3-6. [DOI] [PubMed] [Google Scholar]
- 5.Bush, R. A., and A. J. Lingg. 1975. Establishment of an asymptomatic carrier state infection of enteric redmouth disease in rainbow trout (Salmo gairdneri). J. Fish. Res. Board Can. 32:2429-2432. [Google Scholar]
- 6.Costerton, J. W., and H. M. Lappin-Scott. 1995. Introduction to microbial biofilms, p. 1-11. In H. M. Lappin-Scott and J. W. Costerton (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 7.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 8.Davies, R. L., and G. N. Frerichs. 1989. Morphological and biochemical differences among isolates of Yersinia ruckeri obtained from wide geographical areas. J. Fish Dis. 12:357-365. [Google Scholar]
- 9.Gilbert, P., and M. R. W. Brown. 1995. Mechanisms of the protection of bacterial biofilms from antimicrobial agents, p. 118-130. In H. M. Lappin-Scott and J. W. Costerton (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 10.Korona, R. 1996. Genetic divergence and fitness convergence under uniform selection in experimental populations of bacteria. Genetics 143:637-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Minor, L., and C. Richard. 1993. Yersinia, p. 129-135. In L. Le Minor and C. Richard (ed.), Méthodes de laboratoire pour l'identification des entérobactéries. Institut Pasteur, Paris, France.
- 12.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno, T., and M. Kageyama. 1978. Separation and characterization of the outer membrane proteins of Pseudomonas aeruginosa. J. Biochem. 84:179-181. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1987. Methods for determining bactericidal activity of antimicrobial agents. Document M26-P. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 15.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 16.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes, M. W., and H. Kator. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers, C. J. 1992. Development of a selective-differential medium for the isolation of Yersinia ruckeri and its application in epidemiological studies. J. Fish Dis. 15:243-254. [Google Scholar]
- 19.Romalde, J. L., J. L. Barja, B. Magarinos, and A. E. Toranzo. 1994. Starvation-survival processes of the bacterial fish pathogen Yersinia ruckeri. Syst. Appl. Microbiol. 17:161-168. [Google Scholar]
- 20.Rucker, R. R. 1966. Redmouth disease of rainbow trout (Salmo gairdneri). Bull. Off. Int. Epizoot. 65:825-830. [PubMed] [Google Scholar]
- 21.Shotts, E. B. 1991. Selective isolation methods for fish pathogens. J. Appl. Bacteriol. Symp. Suppl. 70:75S-80S. [PubMed] [Google Scholar]
- 22.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waltman, W. W., and E. B. Shotts. 1984. A medium for the isolation and differentiation of Yersinia ruckeri. Can. J. Fish. Aquat. Sci. 41:804-806. [Google Scholar]
- 24.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wedemeyer, G. A., and N. C. Nelson. 1977. Survival of two bacterial fish pathogens (Aeromonas salmonicida and the enteric redmouth bacterium) in ozonated, chloronated, and untreated waters. J. Fish. Res. Board Can. 34:429-432. [Google Scholar]
- 27.Willumsen, B. 1989. Birds and wild fish as potential vectors of Yersinia ruckeri. J. Fish Dis. 12:275-277. [Google Scholar]