Abstract
The bacteria on the surface of a farmhouse smear-ripened cheese at four stages of ripening (4, 16, 23, and 37 days) from inoculated (i.e., deliberately inoculated with Brevibacterium linens BL2) and noninoculated (not deliberately inoculated with B. linens BL2) cheese were investigated. The results show that, contrary to accepted belief, B. linens is not a significant member of the surface flora of smear cheese and no microbial succession of species occurred during the ripening of the cheeses. Of 400 isolates made, 390 were lactate-utilizing coryneforms and 10 were coagulase-negative Staphylococcus spp. A detailed analysis of the coryneforms was undertaken using phenotypic analysis, molecular fingerprinting, chemotaxonomic techniques, and 16S rRNA gene sequencing. DNA banding profiles (ramdom amplified polymorphic DNA [RAPD]-PCR) of all the coryneform isolates showed large numbers of clusters. However, pulsed-field gel electrophoresis (PFGE) of the isolates from the cheeses showed that all isolates within a cluster and in many contiguous clusters were the same. The inoculated and noninoculated cheeses were dominated by single clones of novel species of Corynebacterium casei (50.2% of isolates), Corynebacterium mooreparkense (26% of isolates), and Microbacterium gubbeenense (12.8% of isolates). In addition, five of the isolates from the inoculated cheese were Corynebacterium flavescens. Thirty-seven strains were not identified but many had similar PFGE patterns, indicating that they were the same species. C. mooreparkense and C. casei grew at pH values below 4.9 in the presence of 8% NaCl, while M. gubbeenense did not grow below pH 5.8 in the presence of 5 to 10% NaCl. B. linens BL2 was not recovered from the inoculated cheese because it was inhibited by all the Staphylococcus isolates and many of the coryneforms. It was concluded that within a particular batch of cheese there was significant bacterial diversity in the microflora on the surface.
The most significant period of cheese production is probably the ripening process, during which starter and nonstarter bacteria, chymosin, and the indigenous milk enzymes develop the organoleptic and textural properties of the cheese. This is particularly true for surface-ripened cheese, due to the variety and abundance of microorganisms on its surface. Surface-ripened cheeses can be divided into mold ripened, e.g., Camembert and Brie, and bacteria ripened, e.g., Limburger, Tilsit, Brick, and Münster. The latter cheeses are sometimes called washed-rind cheeses because of the extensive washing given to the surfaces of these cheeses during ripening. They are also called smear cheeses, because of the glistening appearance of the surface, or red-smear cheeses, due to the production of a red carotinoid by Brevibacterium linens, which is thought to be the most common bacterium occurring on the surface of the cheese. For this reason, B. linens is often deliberately inoculated onto the surface of the cheese during the early days of ripening, either as a commercial preparation or as the so-called “old-young” smearing method, in which young cheeses are washed with a brine suspension of microorganisms from the surface of mature cheese. The latter system effects the transfer of desirable microorganisms necessary for the ripening process to the young cheeses, but it can also effect the transfer of undesirable microorganisms, such as Listeria monocytogenes and Staphylococcus aureus (25). Based on these considerations, the microflora of smeared and nonsmeared cheeses should be quite different.
Various studies have shown that the ripening of bacterial smear-ripened cheese is characterized by a succession of ill-defined microbial communities on the surface of the cheese. Surface ripening begins with the growth of yeasts, which metabolize the lactic acid produced by the starter to CO2 and H2O, increase the pH on the cheese surface from 5.0 to >6.0 (3, 6, 7, 12, 19, 25), and produce growth factors for B. linens (17). Both factors promote growth of a gram-positive, catalase-positive, salt-tolerant bacterial microflora composed primarily of Micrococcaeae and coryneform bacteria (5, 14).
It is generally believed that B. linens is the dominant bacterium on red-smear cheese although recent results have shown a large number of several species of other genera, particularly Arthrobacter, Brevibacterium, Corynebacterium, Microbacterium, and Rhodococcus (5, 6, 22, 25). These are all coryneform bacteria, which are difficult to identify by classical phenotypic analyses. Usually, the identification of coryneforms should include determination of the types of peptidoglycan, menaquinones, and fatty acids in the cell wall. Some of these analyses are labor-intensive, which effectively means that only small numbers of isolates can be analyzed.
It is not clear if a progression of different bacteria occurs on the surface of the cheese during ripening since a systematic study of isolates from smear cheese does not appear to have been undertaken. In addition, the contribution that B. linens makes to the surface bacterial flora when it is deliberately inoculated onto the cheese surface has not been studied.
The aim of this study was to identify the bacteria on the surface of a red-smear cheese at four different times during ripening using a polyphasic approach, including phenotypic, chemotaxonomic, and genotypic analyses. A comparison between cheeses, some of which had been deliberately inoculated with B. linens and some of which were not, was also undertaken.
MATERIALS AND METHODS
Cheese manufacture.
Gubbeen cheese was used in this study. It is a washed-rind, surface-ripened cheese made on a milk-producing farm from pasteurized milk by a proprietary method using a mesophilic starter culture, and it is similar to Tilsit, Reblochon, Limburger, and other surface-ripened cheeses. The curd is washed during manufacture and, after brining, the cheese is smeared with a saline suspension of B. linens BL2 (Chr. Hansen Laboratories). During ripening, the cheeses are also turned and washed with dilute saline at regular intervals. The cheeses are flat and cylindrical, weigh from 200 to 500 g, and have a moisture level of ∼45% and a pH of ∼5.0 1 day after manufacture. Thirty cheeses from the same production batch were used in the analysis. Some of the cheeses were not smeared with B. linens; these cheeses were termed “noninoculated” to distinguish them from the normal inoculated cheese.
Microbiological examination.
Cheeses were sampled after salting but before smearing (3 days), after smearing (4 days), and after ripening at 15 to 20°C at a relative humidity of ∼90% for 9, 16, 23, 30, 37, and 44 days. A separate cheese from the same batch was used at each sampling time. The surface of each cheese was marked with the rim of a sterile plastic petri dish (90-mm diameter), and a layer (∼90 mm in diameter and 3 mm in depth) was removed with a sterile knife, placed into a sterile stomacher bag, and weighed and sufficient sterile 2% (wt/vol) trisodium citrate solution was added to yield a 1:10 dilution. The resulting suspension was macerated in a stomacher for 4 min.
The bacteria were enumerated by surface plating on Milk Plate Count Agar (MPCA; Oxoid) containing 5 g of NaCl per 100 ml (MPCA5). Yeast development on the agar was suppressed by aseptically spreading 100 μl of a suspension of pimaricin (20 mg/ml) on the surface of each plate before use (8, 20). Plates were incubated at 30°C for 12 days. Yeast counts were determined on yeast-glucose-chloramphenical agar containing 10 μg of bromophenol blue/liter after incubation at 25°C for 5 days (18).
Phenotypic characterization.
Fifty colonies were selected from countable MPCA5 plates at 4, 16, 23, and 37 days from both the inoculated and noninoculated cheese and were purified by restreaking. Cell morphology was determined on mineral base E, yeast extract, glucose agar (4) after incubation at 30°C for 12 h and 1, 3, and 7 days. Strains which were rod-shaped organisms in young cultures and cocci in older cultures were considered to undergo a rod/coccus transformation. Cultures were also Gram stained and tested for the presence of catalase.
All isolates were tested for 53 biochemical characteristics as described previously (21, 22, 23). These included the utilization of organic acids, amino acids, sugars, and alcohols as growth substrates and the abilities to produce acid from several sugars, to grow in the presence of 10% salt, to reduce nitrate, and to hydrolyze starch, gelatin, and casein. Lipase activity was determined using API ZYM kits (API Biomerieux, Marcy l'Etoile, France) which were read after 24 h of incubation at 30°C (1). For casein hydrolysis, 100 ml of 10% (wt/vol) reconstituted skimmed milk autoclaved at 110°C for 10 min and 900 ml of MPCA autoclaved at 121°C for 15 min were tempered to 45°C and mixed prior to pouring. Each strain was streaked and incubated at 30°C for 4 or 10 days. Proteolysis was determined by flooding the plates with 12% (wt/vol) HgCl2 in 20% (vol/vol) HCl. A clear zone around the colonies indicated the hydrolysis of casein.
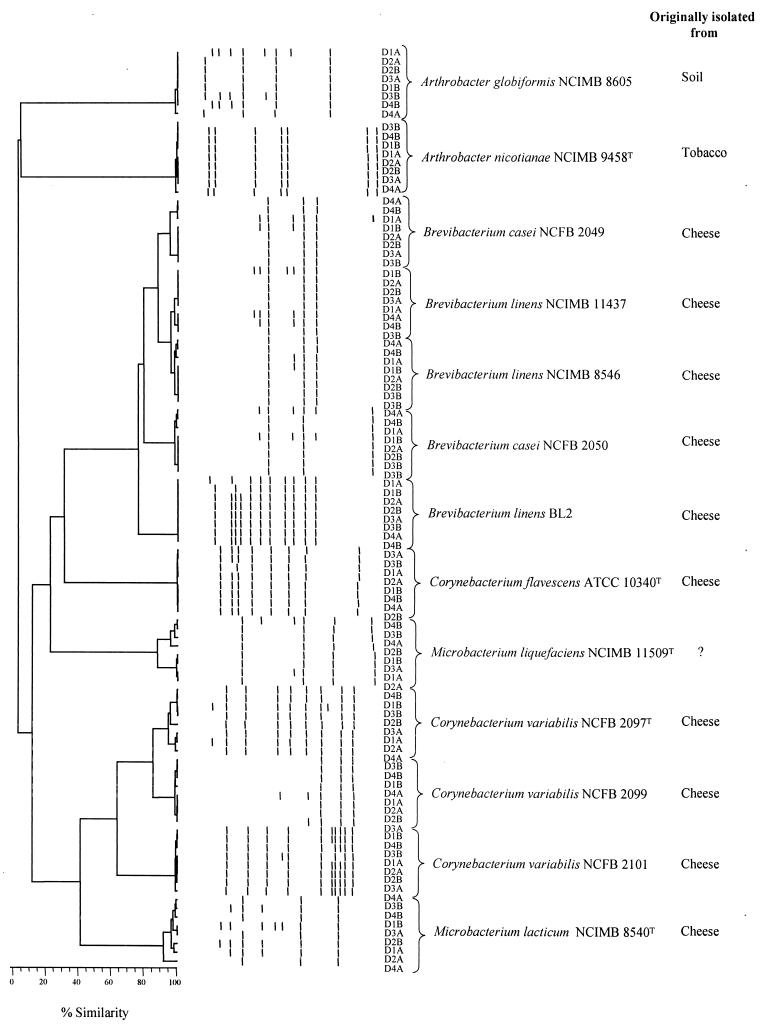
The results were compared with a database of the same tests on 557 strains of Arthrobacter, Brevibacterium, Corynebacterium, Microbacterium, and Rhodococcus by using the computer programs CORYNE and CLUSUM (24). Several collection strains of Brevibacterium, Corynebacterium, and Microbacterium were used as positive controls. These are indicated on Fig. 3. Duplicate analyses of all tests were carried out on a small number (∼20) of strains on the day they were tested and also at regular intervals (5 to 10 days) throughout the study. Production of methanethiol was determined as described previously (1).
FIG. 3.
Reproducibility of the RAPD-PCR profiles of several culture collection strains of coryneform bacteria, most of which have been isolated from cheese. D1, D2, etc. refer to the day on which the PCRs were performed and A and B refer to duplicate PCRs of that strain.
Growth on pH-NaCl gradient plates.
Two-dimensional pH-NaCl gradient plates were prepared by a modification of that described by Wimpenny and Waters (26). Salt and pH gradients were poured at right angles to each other, using four layers (15 ml of each medium) in 10-cm2 petri dishes. Layer 1 medium contained 10 g of peptone, 5 g of yeast extract, 5 g of malt extract, 5 g of Casamino Acids, and 15 g of bacteriological agar no. 1 per liter. The pH was adjusted to 7.2 and autoclaved at 121°C for 15 min (10). To 15 ml of this medium, 220 μl of 1 M dl-lactic acid was added. The medium in layer 2 was the same as that in layer 1 except that 440 μl of 1 M NaOH was added per 15 ml of medium. Layer 3 contained 10 g of peptone, 5 g of yeast extract, 5 g of malt extract, 5 g of Casamino Acids, 15 g of bacteriological agar no. 1, and 320 g of NaCl per liter. Layer 4 contained the same medium as layer 1 without the addition of acid or alkali. All media were tempered to 75°C before the lactic acid or NaOH was added. Plates were poured as follows. One edge of the plate was supported on a 3-mm-thick piece of wood, and layer 1 was poured and allowed to solidify as a wedge shape. After solidification, the plate was placed on a flat surface and layer 2 was poured. Once solidified, the plate was turned through 90°, one side was elevated (3 mm), and the salt layer (layer 3) was poured. Layer 4 was poured after placing the plate on a flat surface. The plates were held for 24 h at 30°C to allow the acid and salt to equilibrate vertically by diffusion. Plates were poured on 5 successive days, and the pH and salt were measured to establish the reproducibility of both salt and pH gradients. pH was measured by excising strips of agar at 1-cm intervals across the pH gradient, melting them in a microwave oven, allowing them to cool to room temperature, and measuring the pH. For the measurement of salt, pieces of agar were excised at 1-cm intervals along the salt gradient, 60 ml of distilled water was added, and the mixture was melted by heating in a microwave oven. The NaCl was measured potentiometrically by titration with 0.1 N AgNO3. The gradient plates were inoculated with biomass from MPCA5 plates resuspended in 1.5 ml of sterile water to a turbidity of McFarland standard 3. One milliliter was then spread aseptically over the surface of the plate and incubated at 30°C for 3 days. One uninoculated plate was incubated with each test series to determine pH and salt gradients. Growth was photographed and mapped with the aid of a template etched at 1-cm intervals in both dimensions. The limits of growth were recorded as x and y coordinates.
Extraction of genomic DNA.
Strains were cultivated on MPCA5 for 5 days at 30°C, and the DNA was extracted as described previously (1).
DNA amplification with random primer.
The randomly amplified polymorphic DNA (RAPD)-PCR was carried out in 50-μl reaction volumes in a Perkin-Elmer 4800 (Applied Biosystems) PCR instrument. Each reaction mixture (50 μl) contained 5 μl of 10× NH4 buffer, 5 μl of 50 mM MgCl2, 1 μl of deoxynucleotide triphosphates (dNTPs) mastermix containing 12.5 μmol of each dNTP, 2.5 μl of a 20 μM stock of primer (M13 forward primer [5"-GTA AAA CGA CGG CCA GT-3"]), 1.0 μl of chromosomal DNA, 0.25 μl of Taq polymerase (5 U/μl) (Bioline), and sufficient sterile water to make the reaction mixture 50 μl. DNA from strains which exhibited a coryneform morphology was amplified as follows. The Taq polymerase was added after a 6-min hot start at 95°C, followed by 34 amplification cycles of 1 min at 94°C, an annealing temperature of 37°C for 1 min, and an extension step of 72°C for 1 min. PCR products (7 μl) were separated on a 1.5% (wt/vol) agarose gel (Promega Corp.) containing ethidium bromide at 100 V for 2 h, and the DNA was detected by UV transillumination. RAPD patterns on Polaroid photographs were digitized with a Scanjet 4P scanner and Deskscan II version 2.7 software (Hewlett-Packard, Dublin, Ireland). Patterns were normalized and processed using GelCompar, version 4.0 (Applied Maths, Kortijk, Belgium); strains were grouped using cluster analysis by the unweighted pair group method (UPGMA) algorithm (16).
Pulsed-field gel electrophoresis (PFGE) analysis.
Biomass was scraped from Funke agar (10) plates, washed twice in 10 mM Tris-HCl (pH 7.6) containing 1 M NaCl, and resuspended in 200 μl of the same solution. The suspensions were then heated to 37°C for 15 min, mixed with an equal volume of 2% low-melting-point agarose (Bio-Rad Laboratories) in 0.125 M EDTA, pH 7.6, and left to solidify in molds (Bio-Rad). Cells were lysed in situ in agarose blocks by gentle shaking for 18 h at 37°C in EC buffer (1 M NaCl-6 mM Tris-HCl-100 mM EDTA-1% [wt/vol] sarkosyl, pH 7.6) containing 10 mg of lysozyme/ml. The blocks were then shaken in proteinase K (0.5 mg/ml in 0.5 M EDTA, 1% sarkosyl, pH 8.0) for 24 h at 55°C and twice in 1 mM phenylmethylsulfonyl fluoride (PMSF) in 10 mM Tris-HCl containing 1 mM EDTA, pH 8.0, and then stored in 10 mM Tris-HCl containing 100 mM EDTA, pH 8.0, at 4°C until required. Slices (1 to 2 mm) were cut from the agarose blocks with sterile coverslips and washed three times at room temperature with gentle shaking in 10 mM Tris-HCl-0.1 mM EDTA, pH 8.0 (Tris-HCl-to-EDTA ratio, 10/0.1). Slices were washed once at 4°C for 30 min with restriction buffer (New England Biolabs) and incubated for 4 h with 20 U of SpeI (New England Biolabs) in a total volume of 100 μl. Following digestion, slices were loaded into wells of a 1% PFGE grade agarose gel (Bio-Rad) and the gel was run in 0.5× Tris-borate buffer using a CHEF-DRIII PFGE apparatus and cooling module (Bio-Rad) at 1 V (6 V · cm−1) for 16 h at 14°C, with the pulse ramped from 1 to 20 s. Gels were stained with ethidium bromide (0.5 μg/ml) in water, destained in water, and photographed using Polaroid type 667 film.
Antimicrobial activity.
One colony from an MPCA5 plate was resuspended in 500 μl of sterile water, and 5 μl of the resultant suspension was spotted onto the same agar and incubated at 30°C overnight. The plates were then overlaid with 5 ml of the appropriate sloppy agar (7.5 g of agar/liter) containing the target (indicator) strain (∼105 CFU/ml) and incubated at the temperature appropriate for growth of the target strain for 2 to 3 days; the diameter of the inhibition zone was measured. The target organisms used were Propionibacterium freundenreichii LMG (Laboratorium voor Microbiologie, University of Ghent, Ghent, Belgium) 16424T, Enterococcus faecalis DPC (Dairy Products Research Centre, Teagasc, Fermoy, Ireland) 3546, and Listeria innocua LMG 11386 and LMG 11387 and B. linens BL1 and BL2 (Chr. Hansen's Laboratory, Little Island, Cork, Ireland).
Chemical analysis of cheese.
Moisture and salt were determined by standard methods (15). pH was determined by placing the electrodes directly into grated cheese.
RESULTS
Microbiology of cheese surface.
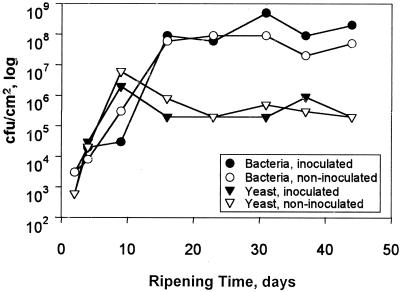
A comparison of the bacterial and yeast counts on the surface of the inoculated and noninoculated cheeses is shown in Fig. 1. The initial numbers of bacteria and yeast ranged from ∼102 to 103 CFU/cm2 and increased to a maximum of ∼108 CFU of bacteria/cm2 after 30 days and ∼106 CFU of yeasts/cm2 after 9 days, after which the numbers of both decreased slightly. Generally, there was little difference between the bacterial and yeast counts of the inoculated and noninoculated cheeses except at the end of ripening, when a ∼0.5 log difference was observed between the bacterial counts. This difference was not considered to be significant.
FIG. 1.
Development of bacteria and yeast on the surface of the inoculated and noninoculated cheese during ripening.
Composition of cheese surface.
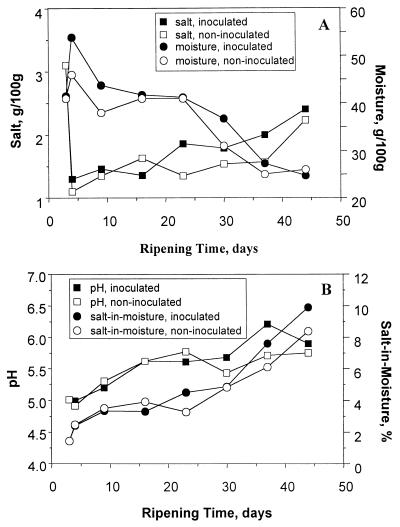
The moisture content of the surface of both cheeses increased between the third and fourth day of ripening, after which it decreased steadily to ∼25 g/100 g (Fig. 2A). The salt content was very high on day 3, probably because the salt had not diffused into the cheese, and decreased rapidly between days 3 and 4, after which it increased gradually to a final level of 2 to 2.5 g/100 g. There was little difference in the percent salt-in-moisture between the inoculated and noninoculated cheese. Moreover, the percent salt-in-moisture gradually increased in both cheeses during ripening; this coincided with the drying out of the cheese surface (Fig. 2B). The pH of both cheeses was 5.0 at day 3 and increased to 5.7 to 6.0 at the end of ripening (Fig. 2B); the increase was greater in the inoculated than in the noninoculated cheese.
FIG. 2.
(A) Salt and moisture levels in the surface layer of the inoculated and noninoculated cheese during ripening. (B) Salt-in-moisture levels and pH in the surface layers of the inoculated and noninoculated cheese during ripening.
Characterization of isolates.
From each cheese, 50 isolates were made at 4, 16, 23, and 37 days, yielding a total of 400 isolates. All of the isolates fell into two groups, gram-positive, catalase-positive cocci (10 isolates) and gram-positive, catalase-positive, irregularly shaped rods (390 isolates). All the cocci were coagulase-negative, facultative anerobes and were, therefore, Staphylococcus species. No effort was made to identify them to species level. The rod-shaped organisms were considered to be coryneforms.
Seiler and Braatz (24) developed two computer programs, CORYNE and CLUSUM, to identify coryneform bacteria, based on 53 biochemical tests on 557 different strains of Arthrobacter, Brevibacterium, Corynebacterium, and Microbacterium spp. The same tests were performed on the 390 coryneforms isolated in the present study, but none of them could be identified with any of the strains in the database. The smallest number of phenotypic differences found between the isolates and those in the database was 5.
Development of RAPD methodology.
The usefulness of RAPD-PCR, using M13 forward as primer, to distinguish between several strains of corynebacteria, most of which had been originally isolated from cheese, was investigated. DNA was extracted from independently grown duplicate cultures and PCR was performed in duplicate on each strain on 4 successive days. The results are shown in Fig. 3. Species of Arthrobacter, Corynebacterium, and Microbacterium clustered very poorly, showing little relationship with each other, while some strains of B. linens and Brevibacterium casei clustered at 75% similarity. However, within a strain, the reproducibility was >96%, except for Microbacterium lacticum NCIMB 8540T and Microbacterium liquefaciens NCIMB 11509T, where it was ∼90%. Because of the high reproducibility of the technique within a species, it was felt that it could be used to cluster monoclonal isolates and that, at 70% similarity, different genera of coryneform bacteria would remain distinct and identifiable from each other.
RAPD analysis of coryneforms from inoculated cheese.
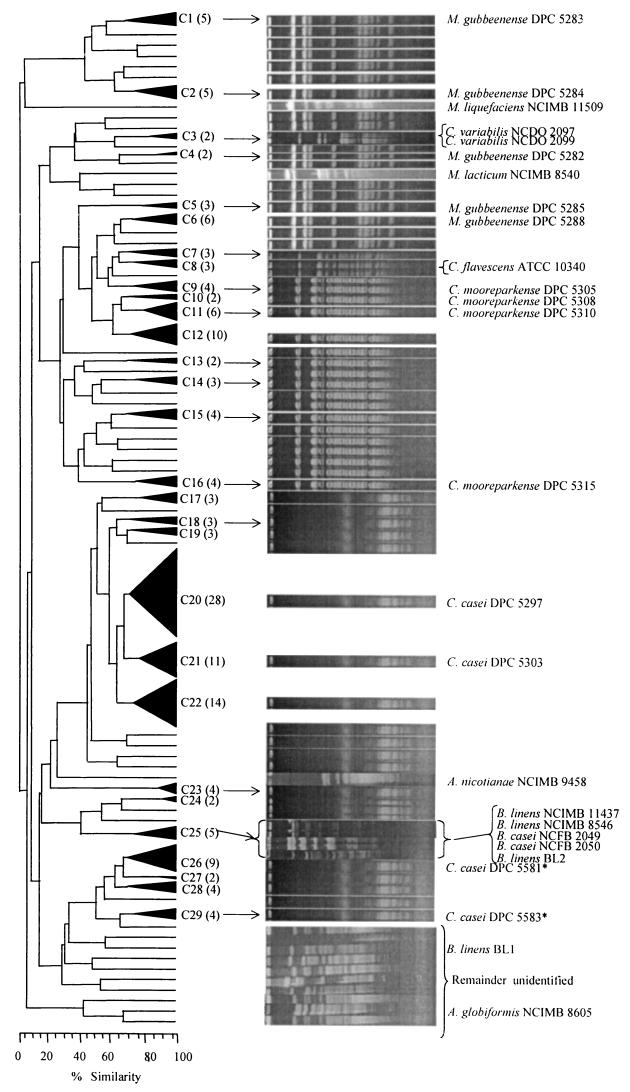
Analysis of the RAPD dendrograms of the 195 coryneform isolates from the inoculated cheese resulted in 29 different clusters and 39 strains, which did not cluster (Fig. 4). None of the cheese isolates clustered with the culture collection strains except DPC 5584 and DPC 5585, which clustered with Corynebacterium flavescens ATCC 10340T in cluster 8. B. casei NCFB 2049 and NCFB 2050 and B. linens BL2, NCIMB 8546, and NCIMB 11437 were found in cluster 25, but this did not include any cheese isolate. Corynebacterium variabilis NCFB 2097T and NCFB 2099 were found in cluster 3, which also did not include any cheese isolate. The other collection strains, A. globiformis NCIMB 8605, A. nicotianae NCIMB 9458T, M. liquefaciens NCIMB 11509T, B. linens BL1, and M. lacticum NCIMB 8540T, did not cluster.
FIG. 4.
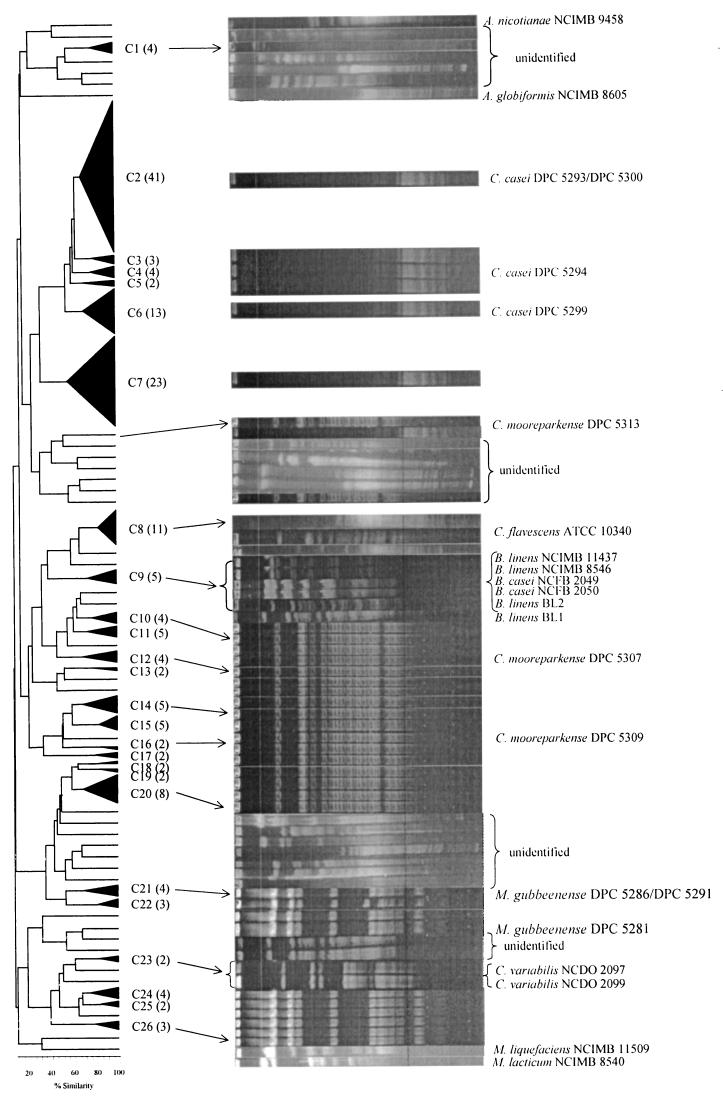
Comparison of RAPD-PCR results and PFGE band patterns of isolates from the inoculated cheese. The cluster numbers (C1 to C29) are shown and the number of strains in each cluster is in parentheses. All isolates within a cluster had the same PFGE pattern and, for clarity, only one example of the band pattern in each cluster is shown. Strains which were previously identified by chemotaxonomic analyses and 16S rRNA sequencing (1, 2) are also indicated, while strains with one asterisk were identified in this study by chemotaxonomic analyses and 16S rRNA sequencing.
PFGE of isolates from inoculated cheese.
SpeI restriction digests of the chromosomal DNA of all isolates from the inoculated cheese were examined by PFGE. The results showed that all isolates within a cluster had identical band patterns (data not shown), except for cluster 25, which contained culture collection strains of B. casei and B. linens. A comparison of the band patterns of one isolate from within a cluster and all the unclustered isolates, with the RAPD dendrogram, is shown in Fig 4. All the isolates in clusters 1, 2, 4, 5, and 6 and the unclustered isolates between these clusters had identical band patterns (Fig. 4). Two culture collection strains, M. liquefaciens NCIMB 11509T and M. lacticum NCIMB 8540T, were also found unclustered in this group, but they had very different banding patterns from the others.
Eleven of the isolates had been previously identified by chemotaxonomic and 16S rRNA sequencing as new species of Microbacterium gubbeenense, Corynebacterium casei, and Corynebacterium mooreparkense (1, 2). The relevant cluster in which these isolates were found is also shown in Fig. 4. The M. gubbeenense isolates were found in clusters 1, 2, 4, 5, and 6. Their PFGE band patterns were identical and were also similar to those of the unclustered isolates between clusters 1 and 6 and to the other isolates in clusters 1, 2, 4, 5, and 6. Therefore, all the isolates in these clusters and the unclustered isolates between clusters 1 and 6 were considered to be M. gubbeenense. Cluster 3 comprised C. variabilis NCDO 2097 and NCDO 2099. Their band patterns were identical but quite different from those of M. gubbeenense.
Similar arguments were used to identify the isolates in clusters 9 to 16, the unclustered isolates between clusters 9 and 16, the isolates in clusters 17 to 29, and the unclustered isolates between clusters 17 and 29 as C. mooreparkense and C. casei, respectively. C. mooreparkense was found in clusters 9, 10, 11, and 16. All the isolates in clusters 9 to 16, as well as the unclustered isolates between these clusters, had band patterns identical to those of C. mooreparkense and hence all these isolates were considered to be this species. Similarly, all the unclustered isolates between clusters 17 and 29, except Arthrobacter nicotinanae ATCC 9458T, as well as all the isolates in clusters 17 to 29, except cluster 25, had patterns which were identical to C. casei and were thus considered to be C. casei.
To confirm the reliability of the PFGE fingerprinting technique to identify strains, the chemotaxonomic analyses and 16S rRNA sequencing used previously (1) were applied to other isolates, four from cluster 20 (DPC 5577, DPC 5578, DPC 5579, and DPC 5580), two from cluster 26 (DPC 5581 and DPC 5582), and one from cluster 29 (DPC 5583). All of these were identified as C. casei sp. nov. The position of two of the latter three isolates in the RAPD dendrogram is also indicated in Fig. 4.
The only cluster which contained both cheese isolates and a culture collection strain was cluster 8, which contained C. flavescens ATCC 10340T. The five cheese isolates in clusters 7 and 8 had similar banding patterns as C. flavescens ATCC 10340T and are therefore considered to be C. flavescens (Fig. 5). Like C. flavescens ATCC 10340T, all five cheese isolates produced yellow colonies.
FIG. 5.
Comparison of RAPD-PCR results and PFGE band patterns of isolates from the noninoculated cheese. The cluster numbers (C1 to C26) are shown and the number of strains in each cluster is in parentheses. All isolates within a cluster had the same PFGE pattern and, for clarity, only one example of the pattern in each cluster is shown. Strains which were previously identified by chemotaxonomic analyses and 16S rRNA sequencing (1, 2) are also indicated.
The PFGE band patterns of C. variabilis NCDO 2097T and NCDO 2099 (cluster 3) were identical, as were those of B. casei NCFB 2049 and NCFB 2050 (cluster 25), confirming the reliability of the PFGE technique for species identification. B. linens was an exception to this. Two of the four strains of B. linens, NCIMB 11437 and NCIMB 8546 (cluster 25), had identical band patterns while B. linens BL2 (cluster 25) and B. linens BL1 (unclustered) had different band patterns from these and from each other. This is not surprising as, based on DNA:DNA hybridization data, B. linens is a mixture of two different species (9); the present data suggest that it might be a mixture of three different species.
The eight unclustered cheese isolates at the end of the RAPD dendrogram were not identified further, but four of them appeared to have identical band patterns.
PFGE of isolates from noninoculated cheese.
RAPD and PFGE analyses were also carried out on the isolates from the noninoculated cheese (Fig. 5). The RAPD analysis divided the 195 coryneform isolates from this cheese into 26 clusters and 47 isolates that did not cluster. None of the isolates clustered with any of the collection strains. Cluster 9 contained B. linens NCIMB 8546, NCIMB 11437, and BL1 and B. casei NCFB 2049 and NCFB 2050, while cluster 23 contained C. variabilis NCFB 2097T and NCFB 2099. Neither of these clusters contained a cheese isolate and all other clusters contained only cheese isolates. The other collection strains, Arthrobacter globiformis NCIMB 8605, A. nicotianae NCIMB 9458T, C. flavescens ATCC 10340T, M. liquefaciens NCIMB 11509T, B. linens BL1, and M. lacticum NCIMB 8540T, did not cluster.
The PFGE band patterns of all isolates within a cluster were identical, except those in cluster 9, which contained B. linens and B. casei. All the isolates in clusters 2 to 8 had identical PFGE band patterns. Four of these isolates, DPC 5293 and DPC 5300 (cluster 2), DPC 5294 (cluster 4), and DPC 5299 (cluster 6) were previously identified by chemotaxonomic analysis and 16S rRNA sequencing as C. casei (1). Therefore, all the strains in these clusters and the unclustered strains between these clusters were considered to be C. casei.
All the isolates in clusters 10 to 20 had identical PFGE patterns as those previously identified as C. mooreparkense DPC 5307 (cluster 11) and C. mooreparkense DPC 5313 (unclustered) and so are considered to be C. mooreparkense (1). Similarly, the isolates in clusters 21, 22, and 24 to 26 had PFGE patterns similar to those previously identified as M. gubbeenense DPC 5286 and DPC 5291 (cluster 21) and M. gubbeenense DPC 5281 (unclustered) (2). Cluster 23 contained C. variabilis NCDO 2097 and NCDO 2099.
The four isolates in cluster 1 and three unclustered isolates around it, together with seven isolates between cluster 20 and cluster 21 and two above cluster 23 were not identified but had different band patterns from C. casei, C. mooreparkense, and M. gubbeenense. Two of them, on either side of cluster 1, two of the unidentified five in the middle of Fig. 5, and two above cluster 21 appear to have similar band patterns, indicating that they were identical isolates, but all the other unidentified isolates had different band patterns.
Progression of species during ripening.
The number of strains of each species isolated at the different times from the inoculated and noninoculated cheeses during ripening are summarized in Table 1. The coagulase-negative staphylococci were mainly found early in ripening and at very low numbers compared with the coryneforms. In both cheeses, C. casei was the dominant species, followed by C. mooreparkense; generally, M. gubbeenense was isolated more frequently late in ripening. C. flavescens was isolated only from the inoculated cheese after 16 days of ripening. With the exception of this organism, the same species were found in both the inoculated and noninoculated cheese at each time point examined. A small number of strains, particularly in the noninoculated cheese, were not identified.
TABLE 1.
Number of strains of different species found in inoculated and noninoculated cheeses at different times during ripening
| Organism | No. of strains found after ripening (days) of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Inoculated cheese
|
Noninoculated cheese
|
||||||||
| 4 | 16 | 23 | 37 | 4 | 16 | 23 | 37 | ||
| Coagulase-negative staphylococci | 1 | 2 | 2 | 5 | |||||
| C. casei | 35 | 24 | 18 | 28 | 24 | 19 | 25 | 23 | |
| C. mooreparkense | 4 | 11 | 23 | 6 | 14 | 16 | 14 | 14 | |
| M. gubbeenense | 8 | 5 | 7 | 13 | 1 | 5 | 11 | ||
| C. flavescens | 5 | ||||||||
| Unidentified coryneforms | 2 | 3 | 2 | 1 | 7 | 14 | 6 | 2 | |
Although the inoculated cheese had been smeared several times during ripening with B. linens BL2, this strain was never isolated at any time point from either cheese (Fig. 4 and 5). This was not due to the inability of the strain to grow on the isolation medium. However, the 10 strains of Staphylococcus isolated from the cheese, 2 strains of C. casei, and strains of C. mooreparkense inhibited B. linens BL2 and B. linens BL1 but not L. innocua LMG 11387 and LMG 13586, P. freudenreichii 16424, or E. faecalis DPC 3546 (Table 2). Several strains of M. gubbeenense had no effect on these indicator strains.
TABLE 2.
Inhibition of several indicator strains by different isolates from the inoculated and uninoculated cheesesa
| Producer strainsb | Inhibition of indicator strains
|
|
|---|---|---|
| B. linens BL1 | B. linens BL2 | |
| Staphylococcus 1 | + | + |
| Staphylococcus 2 | + | + |
| Staphylococcus 3 | + | + |
| Staphylococcus 4 | + | + |
| Staphylococcus 5 | + | + |
| Staphylococcus 6 | + | + |
| Staphylococcus 7 | + | + |
| Staphylococcus 8 | + | + |
| Staphylococcus 9 | + | + |
| Staphylococcus 10 | + | + |
| C. casei DPC 5293 | − | − |
| C. casei DPC 5294 | − | − |
| C. casei DPC 5297 | − | − |
| C. casei DPC 5299 | − | + |
| C. casei DPC 5300 | − | + |
| C. casei DPC 5303 | − | − |
| C. mooreparkense DPC 5305 | − | − |
| C. mooreparkense DPC 5307 | − | + |
| C. mooreparkense DPC 5308 | − | + |
| C. mooreparkense DPC 5309 | − | + |
| C. mooreparkense DPC 5310 | − | + |
| C. mooreparkense DPC 5313 | − | + |
| C. mooreparkense DPC 5315 | − | − |
M. gubbeenense DPC 5291, DPC 5282, DPC 5283, DPC 5284, DPC 5285, DPC 5286, DPC 5288, DPC 5289, and DPC 5291 did not produce any inhibitor for any of the indicator strains tested.
No strain inhibited L. innocua LMG 11386, L. innocua LMG 11387, P. freudenreichii LMG 16424, or E. faecalis DPC 3546.
Ranges of pH and NaCl allowing growth of isolates.
Two-dimensional pH and NaCl gradient plates were used to evaluate the growth response of C. casei, C. mooreparkense, and M. gubbeenense to pH and NaCl. Preliminary studies indicated that the pH and salt gradients in different batches of plates were stable and reproducible and showed no significant change with time over a 4-day incubation at 30°C (data not shown).
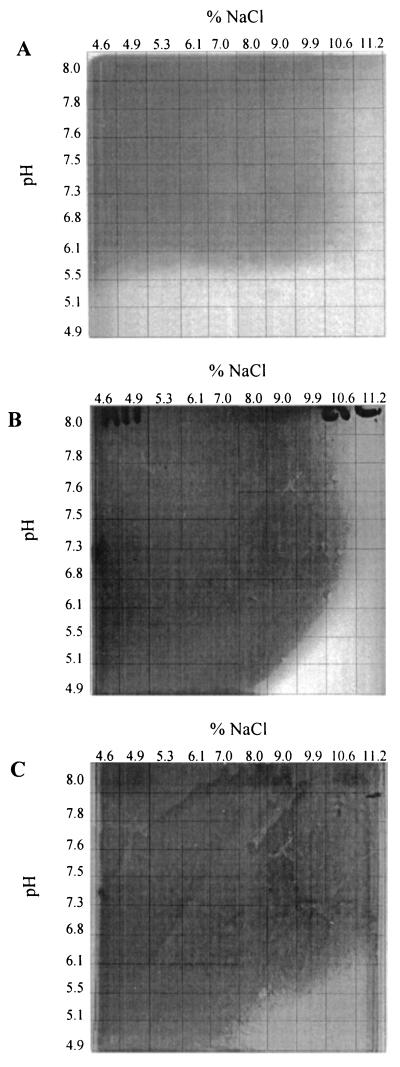
The growth profiles of C. casei, C. mooreparkense, and M. gubbeenense at different pH values and salt concentrations are shown in Fig. 6. C. casei and C. mooreparkense grew at pH values of <4.9 in the presence of 7 and 8% NaCl, respectively, which are well within the ranges of pH and salt concentrations found in the cheese (Fig. 2). In contrast, M. gubbeenense did not grow below pH 5.8 but did grow in the presence of up to 10% NaCl at this pH. The pH and NaCl tolerances of several strains of each species were examined, and all strains yielded the same results (data not shown).
FIG. 6.
Growth of M. gubbeenense DPC 5283 (A), C. mooreparkense DPC 5308 (B), and C. casei DPC 5583 (C) on 2-dimensional pH and salt gradient plates. The dark areas indicate where growth occurred and the light areas indicate where growth did not occur.
Technologically important properties.
All the isolates of C. casei, C. mooreparkense and M. gubbeenense, which were identified using chemotaxonomic methods and 16S rRNA sequencing (1, 2), utilized lactate but produced only small amounts of proteinase. Only C. mooreparkense produced lipase. All of the isolates of these three species, except M. gubbeenense DPC 5286, DPC 5289, and DPC 5288 and C. casei DPC 5297, DPC 5299, and DPC 5300, also produced methanethiol from methionine.
DISCUSSION
In this study, classical and molecular approaches were used to identify 400 isolates from the surface of inoculated and noninoculated smear-ripened cheeses at four different time points during ripening. The bacterial flora was dominated by corynebacteria (390 isolates); the remaining 10 isolates were staphylococci. All the latter were coagulase negative but were not identified further. The coagulase-negative staphylococci in a number of French cheeses have been identified as Staphylococcus equorum, Staphylococcus vitulus, and Staphylococcus xylosus (11). It proved impossible to identify the 390 corynebacteria by classical phenotypic tests despite the fact that a battery of 53 biochemical tests were carried out on each isolate and the availability of a database of 557 strains of coryneforms (23, 24). Therefore, a polyphasic approach to their identification, involving chemotaxonomic RAPD and PFGE, was used. This showed that, except for five isolates which were identified as C. flavescens, most (89%) of the 390 isolates from the inoculated and noninoculated cheese were either C. casei (196 [50.2%] isolates), C. mooreparkense (102 [26%] isolates) or M. gubbeenense (50 [12%] isolates), all of which are new species (1, 2). Thirty-seven (9.3%) isolates were not identified. Isolates from two other batches of cheese are currently been identified to determine if the new species also dominate other batches of this cheese. The fact that most of the isolates were new species, which were not present in the database, explains why the initial phenotypic analysis failed to identify any isolate. It is probable that other bacteria were also present in lower numbers but were not isolated by the techniques used.
The surface of this cheese is deliberately smeared with B. linens BL2 at the beginning of ripening. However, this strain was not recovered from the surface at any time point during ripening. This result was surprising and is most likely due to the fact that all the staphylococcal and many of the coryneform isolates inhibited the growth of B. linens BL2 (Table 2). This inhibition may be due to bacteriocin production, but this aspect was not investigated further. Many bacteriocins are plasmid encoded and easily lost on subculture, which may explain why only some isolates showed inhibition. In reality, the most likely inhibitors are the staphylococci since they grow during the early period of ripening (Table 1).
Inoculating the surface of smear-cheese and other fermented food products with microorganisms from a previous batch, so-called “backslopping,” is a common practice in some fermented foods but is not recommended because of the possibility of transferring pathogens from the older to the younger product (25). In this regard, deliberate smearing of the cheese surface with C. casei, C. moorparkense, and M. gubbeenense as a surface starter culture could be useful. Before this can be done, however, a system of culturing and storing the organisms would need to be developed so as to maximize their survival.
Within each new species, the PFGE patterns of each isolate were virtually identical, implying that the isolates were single clones of that species. This result is not surprising when one considers the way the cheese is handled during ripening. During the early stages, the surface of the cheese is washed several times with dilute saline, which spreads the most rapidly growing microcolonies on the cheese surface, allowing them to develop more uniformly. Thus, the more rapidly growing bacteria will dominate and it is likely that only one or a few strains of each species would be present. In the present study, only one strain of each of the species was found. To our knowledge, there is no other study with which we can compare our data. Many of the 37 unidentified isolates (Table 1) had similar PFGE patterns (Fig. 4 and 5), indicating they were the same strain. Only the dominant bacteria on the surface were isolated, and it would be interesting to determine if other bacteria are present in lower numbers.
The RAPD technique was very reproducible and yielded high similarity values, with several bacteria isolated initially from cheese. Therefore, we felt that it should be useful in grouping large numbers of the same clone. The PFGE analysis of isolates within a cluster showed that this did happen, but it also showed that many isolates in adjacent RAPD clusters had identical band patterns (Fig. 5 and 6). We have no explanation for this result. It does imply that the PFGE technique appears to be much better than the RAPD technique for clarifying the relationships of different isolates from smear-ripened cheese to each other. Despite this, the RAPD technique did bring monoclonal isolates together since all isolates from several contiguous clusters clustered together. The RAPD technique appears to be too discriminating and probably relies on very subtle differences in banding patterns and their intensities to cluster the strains.
There was no significant difference between the bacterial counts on the surfaces of the inoculated and noninoculated cheese. This confirms that the contribution of B. linens BL2 to the smear was of little consequence. In recent report, Kollöffel et al. (13) showed that cell and colony counts on the surface of Gruyère cheese were 2 log cycles lower than in situ and colony hybridization counts. This prompts the following question: do the bacterial counts reported in the present study and other studies (5, 6, 25) reflect the actual numbers of bacteria present in the smear, or are these values lower than the true numbers present? This question is currently being addressed using differential gradient gel electrophoresis to determine the other bacteria which may be present on the cheese.
The source of C. casei, C. mooreparkense, and M. gubbeenense on the cheese surface is not clear, but brine, shelving, and the cheesemakers' hands are probable sources. The most likely source may be the shelving, on which the cheese is held during ripening, but the cheesemakers' hands could also be important, as these cheeses receive a significant amount of manual handling during ripening and coryneforms are an important component of the microflora on human skin. The shelves were made of wood, which is difficult to clean and which could contain traces of cheese curd to sustain the growth of microorganisms, which would then inoculate the cheese surface when the cheese is placed on the shelf to ripen.
Except for C. flavescens, the same organisms were isolated at each time point during ripening (Table 1). These results show there is no real progression of organisms on the surface of the cheese during ripening. This is not surprising considering that the cheese surface is washed frequently during ripening. This results in the disruption of microcolonies and the spreading of the resulting cells on the cheese surface. There was a tendency for greater numbers of M. gubbeenense bacteria to be isolated late in ripening. M. gubbeenense is unable to grow below pH 5.8, and the pH of the cheese surface does not reach this value until 30 days of ripening (Fig. 2). The M. gubbeenense isolates recovered early in ripening may be just nongrowing contaminants. The fluctuations in each species during ripening can now be easily followed since the 16S rRNA sequences (1, 2) can be used to design species-specific probes for these organisms.
Compositional analysis of the cheese during ripening showed that the pH and salt content gradually increased during ripening and the moisture content decreased. The salt is dissolved in the moisture and the salt-in-moisture also increased during ripening, especially from day 23 of ripening. It is generally considered that yeast grow first on smear-ripened cheese, catabolizing the lactate produced by the starter bacteria to CO2 and H2O, which cause the pH to increase to a value at which the bacteria grow (6, 12, 19, 25). C. casei and C. mooreparkense are capable of growth at pH values below 4.9, in the presence of 8.0% NaCl. These values are much greater than those found in the cheese (Fig. 2). These organisms also metabolize lactate, implying that they can grow from the beginning of ripening and therefore may not be as dependent on significant yeast growth during the initial stages of ripening, as previously thought. At the end of ripening, yeast counts were 3 log cycles lower than the corresponding coryneform counts, indicating that, at the end of ripening, the microflora of the inoculated and noninoculated cheeses are dominated by bacteria. Keller and Puhan (12) also found that the yeast counts on Tilsit cheese during ripening were never greater than the bacterial counts. This shift from yeast dominance early in ripening to bacterial dominance is critical to the ripening process. Most of the isolates of C. casei, C. mooreparkense, and M. gubbeenense were able to produce methanthiol from methionine, which is one of the major flavor compounds in smear-ripened cheeses.
Previous studies on the surface microflora of smear-ripened cheese focused on the phenotypic identification of strains, usually at unspecified times during ripening. In the present study, both phenotypic and genotypic approaches were used to identify the bacteria on the surface of a farmhouse smear-ripened cheese throughout ripening. The results showed that the bacterial flora was dominated by single clones of three adventitious species of coryneforms, implying that the growth of these adventitious bacteria is more important than that of those, e.g., B. linens, which are added intentionally. This study is a detailed analysis of one batch of cheese, and it would be necessary to study other batches of this cheese and other smear cheeses to determine how similar or different they are. Such studies are currently under way.
Acknowledgments
Grateful thanks are extended to Giana and Tom Ferguson for facilitating this study of their cheese, Finbarr Drinan and Eddie Mulholland for their excellent technical help, Teagasc for the award of a Walsh Fellowship to N.B., and the National University of Ireland for the award of a Travel Bursary in Food Science and Technology to N.B. Mike Goodfellow and Tim Cogan acknowledge the European Community's project “Biodiversity and anti-listerial activity of surface microbial consortia from Limburger, Reblochon, Livarot, Tilsit and Gubbeen cheese,” QLK1-2001-O2228.
REFERENCES
- 1.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov., and Corynebacterium casei sp. nov. isolated from the surface of a smear-ripened cheese. Int. J. Syst. E vol. Microbiol. 51:843-852. [DOI] [PubMed] [Google Scholar]
- 2.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, M. Vancanneyt, T. M. Cogan, and P. F. Fox. 2001. Microbacterium gubbeenense sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 51:1969-1976. [DOI] [PubMed] [Google Scholar]
- 3.Busse, M. 1989. Die Oberflachenflora von Geschmierten Käse. Milchwirt. Berichte. 99:137-141. [Google Scholar]
- 4.Cure, G. L., and R. M. Keddie. 1973. Methods for the morphological examination of aerobic coryneform bacteria, p. 123-135. In R. G. Board and D. W. Lovelock (ed.), Sampling—microbiological monitoring of environments—1973. Academic Press, London, United Kingdom.
- 5.El-Erian, A. F. M. 1969. Bacteriological studies on limburger cheese. Ph.D. thesis. Agricultural University, Wageningen, The Netherlands.
- 6.Eliskases-Lechner, F., and W. Ginzinger. 1995. The bacterial flora of surface ripened cheese with special regard to coryneforms. Lait 75:571-584. [Google Scholar]
- 7.Eliskases-Lechner, F., and W. Ginzinger. 1995. The yeast flora of surface ripened cheese. Milchwissenschaft 50:458-462. [Google Scholar]
- 8.Engel, G. 1993. Hemmung von Hefe-und Schimmelpilzwachstum beim quantitativen Nachweis von Bakterien. Milchwissenschaft 48:325-327. [Google Scholar]
- 9.Fiedler, F., M. J. Schäffler, and E. Stackebrandt. 1981. Biochemical and nucleic acid hybridisation studies on Brevibacterium linens and related strains. Arch. Microbiol. 129:85-93. [Google Scholar]
- 10.Funke, G., G. Martinetti Lucchini, G. E. Pfyffer, M. Marchiani, and A. von Graevenitz. 1993. Characteristics of CDC group 1 and group 1-like coryneform bacteria isolated from clinical specimens. J. Clin. Microbiol. 31:2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irlinger, F., A. Morvan, N. El Solh, and J. L. Bergere. 1997. Taxonomic characterization of coagulase-negative staphylococci in ripening flora from traditional French cheeses. Syst. Appl. Microbiol. 20:319-328. [Google Scholar]
- 12.Keller, S., and Z. Puhan. 1985. Deacidification of smear cheese, e.g., Tilsit cheese. Schweiz. Milchwirtsch. Forsch. 14:3-11. [Google Scholar]
- 13.Kollöffel, B., L. Meile, and M. Teuber. 1999. Analysis of brevibacteria on the surface of Gruyère cheese detected by in situ hybridisation and by colony hybridisation. Lett. Appl. Microbiol. 29:317-322. [Google Scholar]
- 14.Lenoir, J. 1984. The surface flora and its role in the ripening of cheese. Int. Dairy Fed. Annu. Bull. 171:3-20. [Google Scholar]
- 15.Lynch, C. M., P. L. H. McSweeney, P. F. Fox, T. M. Cogan, and F. D. Drinan. 1997. Contribution of starter lactococci and non-starter lactobacilli to proteolysis in Cheddar cheese with a controlled microflora. Lait 77:441-459. [Google Scholar]
- 16.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fingerprints, p. 493-521. In M. Goodfellow and A. G. O'Donnell, (ed.), Chemical methods in prokaryotic systematics—1994. J. Wiley & Sons, Chichester, United Kingdom.
- 17.Pukro, M., W. O. Nelson, and W. A. Wood. 1951. The associative action between certain yeast and Bacterium linens. J. Dairy Sci. 24:699-705. [Google Scholar]
- 18.Rapps, M. 1974. Indikatorzusatze zur Keimdifferenzirerung auf Wurze-und Malzextrakt-agar. Milchwissenschaft 29:341-344. [Google Scholar]
- 19.Reps, A. 1993. Bacterial smear-ripened cheese, p. 137-172. In P. F. Fox (ed.), Cheese: chemistry, physics and microbiology, vol. 2. Major cheese groups, 2nd ed. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 20.Sauter, H. 1986. Die Oberflachenflora von Weinkase. Reinfungsfehler und ihre Ursachen. Ph.D. thesis. Technical University Munich, Munich, Germany.
- 21.Seiler, H. 1983. Identification key for coryneform bacteria derived by numerical taxonomic studies. J. Gen. Microbiol. 129:1433-1471. [DOI] [PubMed] [Google Scholar]
- 22.Seiler, H. 1986. Identification of cheese smear coryneform bacteria. J. Dairy Res. 53:39-441. [Google Scholar]
- 23.Seiler, H., R. Braatz, and G. Ohmayer. 1980. Numerical cluster analysis of the coryneform bacteria from activated sludge. Zentbl. Bakteriol. Mikrobiol. Hyg. I Abt. Orig. C 1:357-375. [Google Scholar]
- 24.Seiler, H., and R. Braatz. 1988. Computerprogramme MCOMPRE und CLUSUM zur Coryneformenidentifizierung. Bakteriologisches Institut, sudd versuchs-und Forschungsanstalt fur Milchwirtschaft, Weihenstephan, Germany.
- 25.Valdes-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeast and coryneform bacteria from the surface microflora of Brick cheese. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]
- 26.Wimpenny, J. W. T., and P. Waters. 1984. Growth of microorganisms in gel-stabilized two-dimensional diffusion gradient systems. J. Gen. Microbiol. 130:2921-2926. [DOI] [PubMed] [Google Scholar]