Abstract
It is possible that the low levels of production of exopolysaccharides (EPSs) by lactic acid bacteria could be improved by altering the levels of enzymes in the central metabolism that influence the production of precursor nucleotide sugars. To test this hypothesis, we identified and cloned the galU gene, which codes for UDP glucose pyrophosphorylase (GalU) in Streptococcus thermophilus LY03. Homologous overexpression of the gene led to a 10-fold increase in GalU activity but did not have any effect on the EPS yield when lactose was the carbon source. However, when galU was overexpressed in combination with pgmA, which encodes phosphoglucomutase (PGM), the EPS yield increased from 0.17 to 0.31 g/mol of carbon from lactose. A galactose-fermenting LY03 mutant (Gal+) with increased activities of the Leloir enzymes was also found to have a higher EPS yield (0.24 g/mol of carbon) than the parent strain. The EPS yield was further improved to 0.27 g/mol of carbon by overexpressing galU in this strain. However, the highest EPS yield, 0.36 g/mol of carbon, was obtained when pgmA was knocked out in the Gal+ strain. Measurements of the levels of intracellular metabolites in the cultures revealed that the Gal+ strains had considerably higher glucose 1-phosphate levels than the other strains, and the strain lacking PGM activity had threefold-higher levels of glucose 1-phosphate than the other Gal+ strains. These results show that it is possible to increase EPS production by altering the levels of enzymes in the central carbohydrate metabolism.
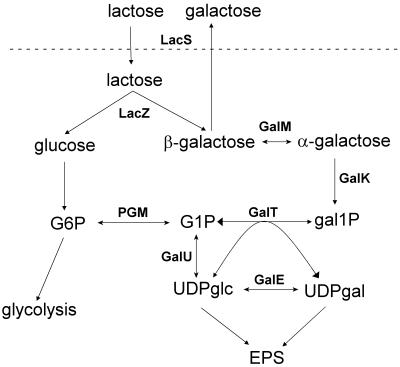
Exopolysaccharides (EPSs) produced by lactic acid bacteria are important for the texture of many fermented milk products. There is great diversity in the composition and structure of these EPSs, which results in different properties (for a review, see reference 12). A variety of EPSs with different repeating units and chain lengths have been found in Streptococcus thermophilus, and diversity is also found in the increasing number of eps operons that are being sequenced (1, 7, 32). The operons consist of a more conserved region with genes that are thought to be involved in regulation, chain length determination, and export, which is followed by a variable region with genes coding for glycosyltransferases and enzymes involved in polymerization and export of the EPSs. The information being accumulated could form the basis for the construction of new tailor-made polysaccharides by genetic engineering (35). However, before the variety of EPSs from lactic acid bacteria can be fully exploited, ways of enhancing the low levels of production must be found. For example, the amount of EPS produced by S. thermophilus normally is less than 1% of the amount of the carbohydrate source (12). EPSs are synthesized from activated nucleotide sugars, and a potential bottleneck in EPS production could be the low levels of these precursors. In S. thermophilus, which possesses the Leloir pathway, UDP glucose (UDPglc) and UDP galactose (UDPgal) can be formed from either the glucose or the galactose moiety of lactose (Fig. 1). The enzyme that links the Leloir pathway with glycolysis is phosphoglucomutase (PGM), which catalyzes interconversion of glucose 6-phosphate (G6P) and glucose 1-phosphate (G1P). We have recently found that S. thermophilus mutants lacking PGM activity produce the same amount of EPS as the parent strain when they are grown on lactose (20), which indicates that the Leloir pathway is important for supplying EPS precursors. LacS, the lactose transporter of S. thermophilus, can transport lactose either by lactose-galactose antiport or by proton symport (29), and in most dairy strains almost all galactose formed by β-galactosidase is secreted (33). The Leloir enzymes, galactokinase (GalK), galactose 1-phosphate uridyltransferase (GalT), and UDP glucose 4-epimerase (GalE), are present at low levels in these strains, but they can be upregulated by mutations in the promoter region since they are transcribed as a single unit (37). However, to produce a surplus of UDPglc and UDPgal from G1P or galactose 1-phosphate, UDP glucose pyrophosphorylase (GalU; EC 2.7.7.9) is also required.
FIG. 1.
Metabolic pathways. Abbreviations: LacS, lactose transporter; LacZ, β-galactosidase; GalM, mutarotase; GalK, galactokinase; PGM, α-phosphoglucomutase; GalT, galactose 1-phosphate udridyltransferase; GalE, UDP glucose 4-epimerase; GalU, UDP glucose pyrophosphorylase; gal1P, galactose 1-phosphate.
In this paper we describe cloning and characterization of galU, which encodes GalU, and its role in EPS production in S. thermophilus LY03. We show how altered expression of combinations of GalU, PGM, and the Leloir pathway affect metabolite levels and EPS production. Our results revealed two different successful metabolic engineering strategies for EPS overproduction in S. thermophilus.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The strains and plasmids used are listed in Table 1. Escherichia coli strains were cultivated in Luria-Bertani medium or Terrific broth. S. thermophilus strains were grown at 42°C in M17 (Oxoid) containing 5 g of lactose or galactose per liter. Spectinomycin was used at a concentration of 100 μg ml−1, and erythromycin was used at a concentration of 0.2 μg ml−1. Controlled batch experiments were performed in MST medium (21) containing (per liter) 50 g of lactose, 2.5 g of K2HPO4, 2.5 g of KH2PO4, 0.25 g of MgSO4 · 6H2O, 5 g of Casamino Acids (technical; Difco), 0.06 g of l-alanine, 0.2 g of l-serine, 0.03 g of l-tryptophan, 0.2 g of l-asparagine, 0.17 g of l-cysteine, 0.01 g of reduced glutathione, and 1 ml of Tween 80, as well as the nucleotides, trace elements, and vitamins that are in SD3 medium (36). Batch cultures were grown at least in duplicate in Biostat fermentors (B. Braun) at 42°C with an initial volume of 1.0 liter. The pH was maintained at 6.2 by automatic addition of 10 M NaOH, and anaerobic conditions were established by nitrogen flushing of the headspace. The first precultures were inoculated into M17 from stocks that were stored at −80°C, and this was followed by two more precultures inoculated (1%, vol/vol) into M17. The final precultures were grown for 9 h in M17 before inoculation (2%, vol/vol) of the fermentors.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli DH5α | Cloning host | Life Technologies Inc. |
| S. thermophilus strains | ||
| LY03 | Commercial yoghurt strain | IMDST01b |
| TMB 6003 | LY03 with pFL52, Specr | This study |
| TMB 6004 | LY03 with pFL46 | This study |
| TMB 6010 | Gal+ LY03 | This study |
| TMB 6011 | TMB 6010 with pFL52 | This study |
| TMB 6012 | TMB 6010 with integrated pFL41, Emr | This study |
| Plasmids | ||
| pUC19 | Cloning vector, AmprlacZ" | 26 |
| pFL41 | pG+host9 with internal pgmA fragment | 20 |
| pFL42 | pLZ12spec with pgmA with native promoter | 20 |
| pFL46 | pFL42 with galU after pgmA | This study |
| pFL52 | pFL42 with galU replacing pgmA | This study |
Emr, erythromycin resistant; Specr, spectinomycin resistant; Ampr, ampicillin resistant.
IMDST01, culture collection at Vrije University, Brussels, Belgium.
DNA techniques and cloning procedure.
DNA was manipulated by using standard procedures (3), and DNA enzymes were purchased from Roche (Roche Diagnostics Scandinavia AB). E. coli was transformed by using the protocol of Inoue et al. (19), and S. thermophilus was electroporated as described previously (27). Plasmid isolation was performed with Quantum kits (Bio-Rad Laboratories AB), with some modification of the protocol for S. thermophilus (20). A PCR was performed with Pwo DNA polymerase, and an inverse PCR was performed with Expand High Fidelity by using standard conditions, as described by the manufacturer. Sequencing was performed with an ABI Prism BigDye terminator cycle sequencing Ready Reaction kit (Perkin-Elmer, Warrington, Great Britain). Degenerate primers 5"-AACTGCAGCNYTNGCNAARGARATGYT-3" and 5"-CCCTCGAGMTCNGTNAARYTGNAYYTCRTT-3" (restriction sites are underlined) were used to amplify an internal fragment of galU. The fragment was sequenced and used for construction of primers 5"-GTAATCGCTCCTCAGGGTAAAGG-3" and 5"-GCTTCTTCAACGATAAATTGGATG-3", which were used for the inverse PCR performed with HindIII-digested and religated chromosomal DNA as the template. The whole gene was finally amplified from chromosomal DNA by PCR and was sequenced in both directions on two independent PCR amplicons.
Overexpression of galU.
The galU gene was amplified by PCR using primers 5"-TCTAAGGAGTTCTCATGAAAAATCA-3" and 5"-GCTCTAGATTCGATGGATTTTATATTAGCC-3" (restriction site is underlined) and was ligated into SmaI-cut pUC19. The gene was cut out with EcoRI and ScaI from a construct with galU in the reverse orientation and was ligated into pFL42 cut with EcoRI and partially with HincII, yielding pFL52. In this way, galU was expressed from the promoter of pgmA. For overexpression of both pgmA and galU, the PCR product was ligated into ScaI-cleaved pFL42, and the construct with both genes in the same orientation was designated pFL46. The galU gene in this construct was located immediately downstream of pgmA, which was expressed from its own promoter.
Selection and characterization of a galactose-positive (Gal+) strain.
S. thermophilus LY03 was grown continuously under lactose limitation conditions, as described previously (33), with some modifications. The cells were grown in 0.2 liter of M17 containing lactose (7 g liter−1). The pH was adjusted to 6.2 by automatic base addition, and the dilution rate was 0.25 h−1. The cells were harvested after 30 reactor volumes, plated on M17 plates containing galactose (4 g liter−1), and incubated overnight anaerobically at 42°C. One clone was chosen, and this strain was designated TMB 6010. Parts (350 bp) of the gal promoter regions from LY03 and TMB 6010 were amplified by PCR and sequenced in both directions by using primers 5"-CCGATTTCATCAGCGATAGTT-3" and 5"-TCCGTATGCTCACCAATCAA-3". A pgmA knockout derivative of TMB 6010 was constructed by transforming the strain with pFL41 at 30°C. The plasmid was integrated at 42°C, and the correct integration was verified by PCR (20).
Measurement of growth, substrate consumption, and product formation.
Cell growth was monitored by determining the optical density at 620 nm after appropriate dilution of samples. Samples were filtered through 0.2-μm-pore-size filters and used to determine lactose, galactose, and lactate concentrations by high-performance liquid chromatography, as described previously (20). EPS concentrations were determined twice after consumption of 27 ml of 10 M NaOH in batch cultures by using the method of Degeest and De Vuyst (10). The EPSs were quantified by weighing. Yields were calculated by using the number of moles of carbon of lactose consumed.
Enzyme assays.
Cells in the mid-exponential phase were harvested and washed twice in 50 mM potassium phosphate buffer (pH 7.0). The cells were ruptured in an X-press (AB Biox), and debris was removed by centrifugation at 15,000 × g for 15 min at 4°C. Protein concentrations were determined by the Micro BCA method (Pierce, Rockford, Ill.). All enzyme assays were carried out at 37°C with a Cobas Mira autoanalyzer (Roche) by monitoring the absorbance at 340 nm. The GalK assay mixture contained 100 mM triethanolamine (TEA) buffer (pH 7.8), 5 mM MgCl2, 2 mM phosphoenolpyruvate, 2 mM ATP, 0.25 mM NADH, 4 U of lactate dehydrogenase ml−1, and 3 U of pyruvate kinase ml−1, and the assay was started by adding 10 mM galactose (33). To compensate for NADH oxidase activity in the cell extracts, the activities were calculated from the difference between the slope when the assay was performed with galactose and slope when the assay was performed without galactose. GalT activity was measured in 100 mM TEA buffer (pH 7.8), and the reaction mixture contained 10 mM MgCl2, 1 mM NADP+, 0.25 mM glucose 1,6-diphosphate (GDP), 5 U of glucose 6-phosphate dehydrogenase (G6PDH) ml−1, and 3 U of PGM ml−1. The reaction was started by adding 1 mM UDPglc and 1 mM galactose 1-phosphate (33). GalU and PGM activities were measured in 50 mM TEA buffer (pH 7.2) with 5 mM MgCl2. The GalU assay mixture contained 0.4 mM NADP+, 1 mM UDPglc, 50 μM GDP, 2 U of G6PDH ml−1, and 2 U of PGM ml−1, and the assay was started with 5 mM pyrophosphate (4). The PGM assay mixture contained 0.4 mM NADP+, 65 μM GDP, and 2 U G6PDH ml−1, and the assay was started by adding 1 mM α-G1P (30).
Measurement of intracellular metabolites.
Five-milliliter culture samples were taken after consumption of 21 and 27 ml of 10 M NaOH, frozen in liquid nitrogen, and stored at −80°C. The methods used to extract metabolites and inactivate enzymes were based on the methods described by Gonzalez et al. (16). The frozen samples were placed in a mixture containing 21 ml of boiling 99% ethanol and 2 ml of 1 M HEPES (pH 7.5) and boiled for 5 min. The samples were then cooled and centrifuged for 10 min at 5,000 × g and 4°C, and the supernatants were each reduced to a volume of 4 ml with a Speedvac AES2000 (Savant) vacuum centrifuge. G1P, G6P, UDPglc, and UDPgal contents were measured photometrically by using NADPH-linked assays. Emission was measured at 460 nm after excitation at 350 nm with a TD-700 fluorometer (Turner Designs). In each case an internal standard was added to the sample after completion of the reaction, and the increases in fluorescence observed for the sample and for the standard were used to calculate the concentrations of the metabolites in micromoles per gram (dry weight) of cells. G6P and G1P contents were measured by using the assay of Garrigues et al. (14). Each reaction mixture contained 100 mM TEA (pH 7.8), 20 mM MgCl2, 0.2 mM NADP+, and 2 U of G6PDH ml−1 for the G6P measurement. When the reaction was completed, 1.8 U of PGM ml−1 was added to start G1P consumption. The UDPglc content was determined by mixing 40 mM TEA (pH 7.2), 4 mM MgCl2, 0.4 mM NADP+, 2 U of G6PDH ml−1, 0.05 mM GDP, 1.8 U of PGM ml−1, and 5 mM pyrophosphate. After consumption of G6P and G1P, 0.5 U of GalU ml−1 was added, and the UDPglc content was measured. Finally, 0.2 U of GalE ml−1 was added to measure the UDPgal content. High-performance anion-exchange chromatography (HPAEC) was also used to determine UDPglc and UDPgal contents. The method used was based on the protocol described by Palmieri et al. (28). Separation was performed at a flow rate of 0.6 ml min−1 on a PA10 column with a precolumn (Dionex, Sunnyvale, Calif.) by using a 27 to 35% 0.5 M KH2PO4 gradient from zero time to 20 min; 0.5 M NaOH (flow rate, 0.5 ml min−1) was added after the column before detection by pulsed amperometry with an ED40 detector (Dionex).
Bioinformatic methods.
Protein sequences were downloaded from, and Gapped Blast (2) was performed on, the homepage of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignment and construction of neighbor-joining trees were performed with ClustalX (34). Sequence data for S. thermophilus were obtained from the UCL Life Sciences Institute (ISV) website at http://www.biol.ucl.ac.be/gene/genome/.
Nucleotide sequence accession number.
The gene sequence described in this paper has been deposited in the EMBL database under accession number AJ300449.
RESULTS
Role of galU in EPS production.
A gene encoding GalU activity in S. thermophilus LY03 was identified and cloned by PCR after multiple alignment of GalU-like proteins from Streptococcus strains. The 304-amino-acid protein product of galU is 88% identical to GluA of Streptococcus mutans and is also closely related to other streptococcal proteins with the GalU function (Fig. 2). Furthermore, GalU aligns with the conserved nucleotidyl transferase domain Pfam 483 (E value, 10−24) (http://www.sanger.ac.uk/Software/Pfam/).
FIG. 2.
Phylogenetic tree based on GalU protein sequences. The tree is rooted with GalU of E. coli as the outgroup. Bootstrap values based on 1,000 trials are indicated at the nodes. The numbers to the right of the organism names are accession numbers. S., Streptococcus.
To assess the functionality of galU, the coding region and the putative ribosome binding site (5"-AAGGAG-3") (the latter is identical to the putative ribosome binding site of Lactococcus lactis [6]) were placed under control of the pgmA promoter (20) in pFL52. When this plasmid was transformed into S. thermophilus LY03, the resulting strain, TMB 6003, had GalU activity of 49 mU mg of protein−1, compared to 5 mU mg of protein−1 in LY03, which confirmed that galU encodes GalU activity.
To investigate if GalU activity in fact limits EPS production in S. thermophilus, TMB 6003 and LY03 were grown in pH-controlled batch cultures in MST medium. Both strains grew with a maximum specific growth rate of about 1.4 h−1, and no significant difference in EPS yields was observed (Table 2). The combined effect of PGM and GalU overexpression was also explored, since this could provide a way to shuttle G6P into UDPglc. Strain TMB 6004, in which galU was placed downstream of pgmA on plasmid pFL46, was constructed. This strain was also grown in batch cultures and compared to LY03 (Table 2). The growth rate of TMB 6004 was similar to that of LY03, but the EPS yield was higher in TMB 6004 (0.31 g mol of carbon−1) than in LY03 (0.17 g mol of carbon−1). Interestingly, the PGM and GalU activities were only 1.5 U mg of protein−1 and 9 mU mg of protein−1, respectively. When only PGM or only GalU was expressed from the same promoter on the same vector, the activities were 7 U mg of protein−1 (20) and 49 mU mg of protein−1, respectively. Plasmid preparations suggested that the lower levels of expression on pFL46 were due to a lower copy number of this plasmid (data not shown).
TABLE 2.
Effect of GalU and PGM on growth and EPS yields in MST medium
| Strain | PGM activity (U mg of protein−1) | GalU activity (mU mg of protein−1) | μmax (h−1)a | Ylactose, EPS (g mol of carbon−1)b |
|---|---|---|---|---|
| LY03 | 0.87 ± 0.03c | 5.0 ± 0.8 | 1.4 ± 0.2 | 0.17 ± 0.04 |
| TMB 6003 | 0.72 ± 0.01 | 49.1 ± 15.0 | 1.5 ± 0.4 | 0.15 ± 0.08 |
| TMB 6004 | 1.45 ± 0.02 | 9.3 ± 0.9 | 1.4 ± 0.2 | 0.31 ± 0.03 |
μmax, maximum specific growth rate.
Ylactose, EPS, yield of EPS when organisms are grown on lactose.
Average ± standard deviation based on the results obtained with at least two independent cultures.
Galactose metabolism and EPS production.
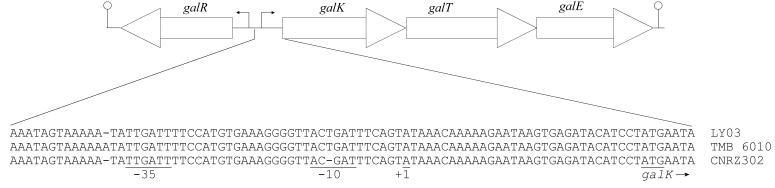
A galactose-fermenting (Gal+) strain of S. thermophilus was selected by growing LY03 under lactose-limiting conditions. The resulting strain, TMB 6010, was able to grow on M17 with galactose as the carbon source, which the parent strain was unable to do. The galactose-fermenting ability of the strain could be attributed to mutations in the galK promoter region, which enhanced transcription of the galKTE genes (37), resulting in higher activities of the corresponding enzymes (33). The GalK and GalT activities were therefore measured and were found to be about 10 times higher in TMB 6010 than in LY03 (Table 3). However, the accuracy of the GalK measurements suffered somewhat from significant NADH oxidase activities in the cell extracts (18). To further characterize the galactose-grown strain, the galK promoter regions of LY03 and TMB 6010 were sequenced, and a 1-bp insertion was found upstream of the −35 region in TMB 6010 (Fig. 3). This corresponded to a type I mutation as proposed by Vaughan et al. (37). Both strains also had an extra 1-bp insertion in the presumed −10 region compared to S. thermophilus CNRZ 302.
TABLE 3.
Effect of the Gal enzymes on growth and product yields in MST medium
| Strain | PGM activity (U mg of protein−1) | GalK activity (U mg of protein−1) | GalT activity (U mg of protein−1) | μmax (h−1)a | Ylactose, galactose (mol of carbon mol of carbon−1)b | Ylactose, EPS (g mol of carbon−1)c |
|---|---|---|---|---|---|---|
| LY03 | 0.87 ± 0.03d | 0.05 ± 0.02 | 1.3 ± 0.1 | 1.4 ± 0.2 | 0.39 ± 0.03 | 0.17 ± 0.04 |
| TMB 6010 | 0.98 ± 0.03 | 0.67 ± 0.04 | 17.3 ± 2.1 | 1.2 ± 0.0 | 0.18 ± 0.05 | 0.24 ± 0.02 |
| TMB 6011 | 0.79 ± 0.20 | 1.14 ± 0.07 | 16.4 ± 1.3 | 1.1 ± 0.1 | 0.12 ± 0.03 | 0.27 ± 0.02 |
| TMB 6012 | 0.01 ± 0.00 | 0.46 ± 0.11 | 12.9 ± 3.4 | 1.1 ± 0.1 | 0.27 ± 0.06 | 0.36 ± 0.02 |
μmax, maximum specific growth rate.
Ylactose, galactose, yield of galactose when organisms are grown on lactose.
Ylactose, EPS, yield of EPS when organisms are grown on lactose.
Average ± standard deviation based on the results obtained with at least two independent cultures.
FIG. 3.
Map of the mutation in the galK promoter. The promoter regions are defined as described by Vaughan et al. (37).
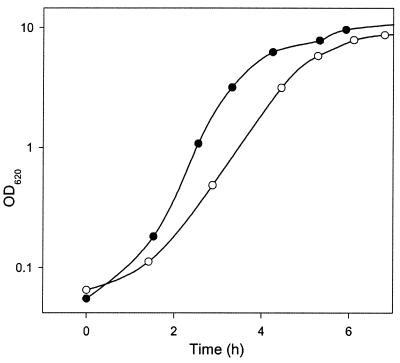
To investigate if the galactose-negative (Gal−) phenotype of S. thermophilus could be the result of a growth advantage, LY03 and TMB 6010 were grown in pH-controlled batch cultures with 50 g of lactose liter−1. Under these conditions, LY03 grew more rapidly initially, when lactose was abundant; the maximum specific growth rate was 1.4 h−1, compared to 1.2 h−1 for TMB 6010 (Fig. 4).
FIG. 4.
Growth patterns of LY03 and TMB 6010 in MST medium. Symbols: ○, LY03; •, TMB 6010. OD620, optical density at 620 nm.
To ascertain whether a higher flux through the Leloir pathway could increase EPS production, Gal+ strain TMB 6010 was also grown in batch cultures in MST medium, and product formation was measured (Table 3). The yield of galactose in the supernatant was lower in TMB 6010 cultures than in LY03 cultures, which confirmed that TMB 6010 catabolized the galactose moiety of lactose more effectively. Furthermore, the EPS yields were 0.17 g mol of carbon−1 in LY03 cultures and 0.24 g mol of carbon−1 in TMB 6010 cultures, which indicated that increased activity of the Leloir pathway could affect EPS production. Increased flux through the Leloir pathway should also increase the availability of intermediates (for example, G1P) in this pathway, and thus overexpression of GalU in TMB 6010 could be favorable for EPS production. We therefore transformed TMB 6010 with pFL52 and investigated product formation in the resulting strain, TMB 6011 (Table 3). A slight increase in EPS production was detected compared to the EPS production in TMB 6010, but it is likely that most of the higher galactose uptake seen in the Gal+ strains resulted only in an increase in galactose flux to glycolysis via PGM. To block this possibility, we constructed a knockout PGM mutant of TMB 6010 by single-crossover recombination. The PGM mutant, TMB 6012, was grown under the same conditions as the other strains, and the enzyme activities and product formation were measured (Table 3). This strain did not take up as much galactose as TMB 6010 and TMB 6011. Nevertheless, the EPS yield with this strain was significantly higher than the EPS yield with any of the other strains.
Intracellular metabolites.
There are many possible bottlenecks in the complex network of reactions leading to EPS biosynthesis, and to gain a better understanding of the metabolism, we measured the intracellular levels of G1P, G6P, UDPglc, and UDPgal in all the cultures mentioned above, both in the exponential phase (Table 4) and in the late exponential phase (Table 5). There were some variations in the metabolite levels between the two phases, but some common trends were observed. LY03 and the strain in which both pgmA and galU were overexpressed, TMB 6004, had similar levels of both G6P and G1P; the concentrations were about 2 to 5 μmol g of cells−1. TMB 6003, in which only galU was overexpressed, exhibited higher levels of both of these metabolites than the other Gal− strains, and there was more G6P than G1P. All the Gal+ strains had higher levels of G1P than the other strains. Furthermore, the pgmA knockout strain, TMB 6012, had clearly higher G1P levels than TMB 6010 and TMB 6011. UDPglc and UDPgal contents were measured both enzymatically and by HPAEC (Tables 4 and 5). The concentrations of these metabolites were low, and UDPgal contents could not be measured accurately by enzymatic methods. However, HPAEC revealed that the ratio of UDPglc to UDPgal was constant, about 3:1 (mol/mol), in all samples, and the UDPglc values obtained with the enzymatic and HPAEC methods were similar. It was found that the levels of UDPglc were highest in TMB 6003 among the Gal− strains and in TMB 6011 among the Gal+ strains. Thus, GalU overexpression from pFL52 seemed to have a positive effect on the levels of UDPglc and UDPgal.
TABLE 4.
Concentrations of intracellular metabolites after consumption of 21 ml of 10 M NaOH
| Strain | Concn (μmol g−1) ofa:
|
||
|---|---|---|---|
| G6P | G1P | UDPglc | |
| LY03 | 2.2 ± 0.4 | 2.2 ± 1.6 | 1.4 ± 0.3 |
| TMB 6003 | 7.5 ± 4.0 | 4.8 ± 1.2 | 2.1 ± 0.3 |
| TMB 6004 | 2.9 ± 0.7 | 2.4 ± 0.5 | 1.1 ± 0.3 |
| TMB 6010 | 12.1 ± 8.5 | 18.5 ± 2.4 | 0.6 ± 0.2 |
| TMB 6011 | 3.5 ± 0.5 | 6.8 ± 1.0 | 0.9 ± 0.0 |
| TMB 6012 | 1.2 ± 0.2 | 24.5 ± 5.2 | 0.7 ± 0.1 |
The concentrations are expressed in micromoles per gram (dry weight) and are averages ± standard deviations based on the results obtained with at least two independent cultures.
TABLE 5.
Concentrations of intracellular metabolites after consumption of 27 ml of 10 M NaOH
| Strain | Concn (μmol g−1) ofa:
|
||
|---|---|---|---|
| G6P | G1P | UDPglc | |
| LY03 | 2.5 ± 0.3 | 5.8 ± 1.2 | 0.9 ± 0.0 |
| TMB 6003 | 17.2 ± 9.9 | 7.0 ± 2.0 | 2.8 ± 0.0 |
| TMB 6004 | 2.9 ± 1.1 | 3.2 ± 0.7 | 1.0 ± 0.3 |
| TMB 6010 | 24.8 ± 16.4 | 19.4 ± 4.7 | 0.8 ± 0.3 |
| TMB 6011 | 7.9 ± 1.3 | 21.6 ± 3.6 | 1.3 ± 0.3 |
| TMB 6012 | 1.5 ± 0.3 | 57.6 ± 12.6 | 1.0 ± 0.5 |
The concentrations are expressed in micromoles per gram (dry weight) and are averages ± standard deviations based on the results obtained with at least two independent cultures.
DISCUSSION
We investigated how levels of enzyme activities in central carbohydrate metabolism affect EPS production in S. thermophilus LY03. As a first step, a gene coding for GalU activity in S. thermophilus was identified, cloned, and shown to be functional by homologous overexpression. galU is preceded by a gene encoding a putative NAD(P)+-dependent glycerol 3-phosphate dehydrogenase, and the protein product of the sequenced part exhibited 94% identity, over 102 amino acids, to GpsA (EC 1.1.1.8) of Streptococcus pyogenes M1 (accession no. AAK33309), a protein which has also been identified as GpdA (EC 1.1.1.94; accession no. P58143). The same gene organization is found in S. mutans (38), Streptococcus pneumoniae (23), and L. lactis (6). Furthermore, after the galU gene was cloned, preliminary data from sequencing of S. thermophilus LMG18311 became available. A BlastP search revealed a putative protein with 100% identity to GalU of S. thermophilus LY03 (contig 269___55, bases 7626 to 8546), which was also preceded by a GpsA/GpdA homologue. GpsA is a key enzyme for phospholipid biosynthesis in Bacillus subtilis, and it is regulated on the transcriptional level (25). Phospholipids are essential for multiple functions in the synthesis of fatty acids and in the cell membrane. Interestingly, a lipid carrier is needed for peptidoglycan biosynthesis and probably also for EPS biosynthesis, and this provides some rationale for the genetic organization of the gpsA and galU genes.
Once the galU gene had been identified, we investigated the effect of overexpressing the gene alone or in combination with other genes that have been shown to be crucial for polysaccharide production in various bacteria (6, 17). Tenfold overexpression of the genes coding for PGM (20) or GalU separately did not influence EPS production. However, EPS production was almost doubled when the specific activities of both enzymes were doubled at the same time. This indicates that there could be some kind of channeling of metabolites or other regulatory effects, since the measured levels of G6P, G1P, and UDPglc were not significantly different in TMB 6004 and LY03. It has also been observed previously that the levels of PGM, GalU, and GalE were high under conditions under which EPS production was high (9), but the flux-controlling function of these enzymes was not established. Our results also underline the fact that the result of overexpression of single genes in complex metabolic networks is often difficult to predict due to competition from different reactions for substrates and cofactors. Much of the recent work in metabolic engineering has also shown that single interventions in metabolic pathways are not sufficient to improve the yields of desired products, while multiple interventions are more likely to be successful (31). Boels et al. (6) found that homologous overexpression of galU in L. lactis NIZO B40 led to an eightfold increase in the levels of UDPglc and UDPgal. However, there was no effect on EPS production. The EPS produced by this strain contains rhamnose, and it was proposed that the availability of dTDP-rhamnose could limit EPS biosynthesis or that the level of expression of the eps gene cluster is the limiting factor. On the other hand, Gilbert et al. (15) found that heterologous expression of the S. pneumoniae type 3 capsule in L. lactis could be increased by coexpression of cps3U, which encodes a GalU enzyme. Our results suggest that there was an increase in the levels of UDPglc and UDPgal when galU was overexpressed and that the Glc1P/UDPglc ratios were lower (Tables 4 and 5). The EPS from LY03 has recently been shown to contain glucose, galactose, and N-acetylgalactosamine (22), and UDP-N-acetylgalactosamine could thus be a limiting precursor in addition to UDPglc and UDPgal. However, another recent study indicated that production of this precursor is not a limiting factor for EPS production in LY03 (11). It is therefore more probable that the lack of effect on EPS production in TMB 6003 is due to downregulation of EPS biosynthesis enzymes. Little is known about regulation of the eps operon in S. thermophilus, even though the product of the first gene, EpsA, shows similarity to the cps operon transcription activator CpsA in Streptococcus agalactiae (8). However, the physiological conditions that affect regulation of these enzymes are not known.
To obtain overexpression of the galKTE genes, we selected a strain under lactose-limited conditions. Interestingly, this strain did not grow as fast as the parent strain when lactose was abundant. This was probably due to the fact that lactose transport by LacS is energetically more favorable when LacS works as a galactose-lactose antiporter than when lactose uptake is driven by proton symport (13). The growth advantage could explain why Gal+ strains that are repeatedly transferred in milk, in which lactose is abundant, become Gal−.
As expected from increased flux through the Leloir pathway, all of the Gal+ strains contained higher levels of G1P than the other strains. However, the concentrations of the UDP sugars were apparently not affected, although the EPS levels were higher. It should be emphasized that the levels of UDP sugars were low with significant standard deviations. Thus, it was difficult to determine whether the observed positive effect on EPS levels was due to elevated UDP sugar levels or upregulated EPS biosynthesis. However, the fact that the Gal+ strain lacking PGM activity, TMB 6012, accumulated very high levels of G1P and also produced more EPS than all of the other strains indicates that G1P is forced towards EPS biosynthesis in this strain.
The results of this study clearly show that it is possible to enhance production of EPSs in S. thermophilus by altering the expression of enzymes that are involved in supplying EPS precursors. Higher levels of PGM and GalU together led to a proportional increase in the EPS yield, although no effect could be seen when either enzyme was overexpressed alone. It will be interesting to see if further co-overexpression of pgmA and galU leads to even higher levels of EPSs. However, if strains are to be used for in situ production in food, it may be difficult to achieve simultaneous overexpression of these two genes by food-grade genetic methods. More promising is the strategy of selecting a galactose-fermenting strain and then knocking out pgmA by classical mutagenesis or by internal deletion in the pgmA gene with the help of a food-grade system for gene deletion (5, 24). Nevertheless, to further improve the EPS yields for production of polysaccharides as additives, further studies of the regulation of EPS biosynthesis on the transcription and enzyme levels are required.
Acknowledgments
This work was supported by European Community FAIR Programme contract CT-98-4267.
REFERENCES
- 1.Almiron-Roig, E., F. Mulholland, M. J. Gasson, and A. M. Griffin. 2000. The complete cps gene cluster from Streptococcus thermophilus NCFB 2393 involved in the biosynthesis of a new exopolysaccharide. Microbiology 146:2793-2802. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology. John Wiley & Sons, Boston, Mass.
- 4.Bernstein, R. L., and P. W. Robbins. 1965. Control aspects of uridine 5"-diphoshate glucose and thymidine 5"-diphosphate glucose synthesis by microbial enzymes. J. Biol. Chem. 240:391-397. [PubMed] [Google Scholar]
- 5.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgoin, F., A. Pluvinet, B. Gintz, B. Decaris, and G. Guedon. 1999. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene 233:151-161. [DOI] [PubMed] [Google Scholar]
- 8.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 9.Degeest, B., and L. De Vuyst. 2000. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degeest, B., and L. De Vuyst. 1999. Indication that the nitrogen source influences both amount and size of exopolysaccharides produced by Streptococcus thermophilus LY03 and modelling of the bacterial growth and exopolysaccharide production in a complex medium. Appl. Environ. Microbiol. 65:2863-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degeest, B., F. Vaningelgem, A. P. Laws, and L. De Vuyst. 2001. UDP-N-acetylglucosamine 4-epimerase activity indicates the presence of N-acetylgalactosamine in exopolysaccharides of Streptococcus thermophilus strains. Appl. Environ. Microbiol. 67:3976-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vuyst, L., and B. Degeest. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 23:153-177. [DOI] [PubMed] [Google Scholar]
- 13.Foucaud, C., and B. Poolman. 1992. Lactose transport system of Streptococcus thermophilus. Functional reconstitution of the protein and characterization of the kinetic mechanism of transport. J. Biol. Chem. 267:22087-22094. [PubMed] [Google Scholar]
- 14.Garrigues, C., P. Loubiere, N. D. Lindley, and M. Cocaign-Bousquet. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, C., K. Robinson, R. W. Le Page, and J. M. Wells. 2000. Heterologous expression of an immunogenic pneumococcal type 3 capsular polysaccharide in Lactococcus lactis. Infect. Immun. 68:3251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, B., J. Francois, and M. Renaud. 1997. A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast 13:1347-1355. [DOI] [PubMed] [Google Scholar]
- 17.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutkins, R., H. A. Morris, and L. L. McKay. 1985. Galactokinase activity in Streptococcus thermophilus. Appl. Environ. Microbiol. 50:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 20.Levander, F., and P. Rådström. 2001. Requirement for phosphoglucomutase in exopolysaccharide biosynthesis in glucose- and lactose-utilizing Streptococcus thermophilus. Appl. Environ. Microbiol. 67:2734-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levander, F., M. Svensson, and P. Rådström. 2001. Small-scale analysis of exopolysaccharides from Streptococcus thermophilus grown in a semi-defined medium. BMC Microbiol. 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall, V. M., A. P. Laws, Y. Gu, F. Levander, P. Rådström, L. De Vuyst, B. Degeest, F. Vaningelgem, H. Dunn, and M. Elvin. 2001. Exopolysaccharide-producing strains of thermophilic lactic acid bacteria cluster into groups according to their EPS structure. Lett. Appl. Microbiol. 32:433-437. [DOI] [PubMed] [Google Scholar]
- 23.Mollerach, M., R. Lopez, and E. Garcia. 1998. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J. Exp. Med. 188:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollet, B., J. Knol, B. Poolman, O. Marciset, and M. Delley. 1993. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J. Bacteriol. 175:4315-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morbidoni, H. R., D. de Mendoza, and J. E. Cronan, Jr. 1995. Synthesis of sn-glycerol 3-phosphate, a key precursor of membrane lipids, in Bacillus subtilis. J. Bacteriol. 177:5899-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 27.O'Sullivan, T. F., and G. F. Fitzgerald. 1999. Electrotransformation of industrial strains of Streptococcus thermophilus. J. Appl. Microbiol. 86:275-283. [DOI] [PubMed] [Google Scholar]
- 28.Palmieri, M. J., G. T. Berry, D. A. Player, S. Rogers, and S. Segal. 1991. The concentration of red blood cell UDP glucose and UDP galactose determined by high-performance liquid chromatography. Anal. Biochem. 194:388-393. [DOI] [PubMed] [Google Scholar]
- 29.Poolman, B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125-147. [DOI] [PubMed] [Google Scholar]
- 30.Qian, N., G. A. Stanley, B. Hahn-Hägerdal, and P. Rådström. 1994. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J. Bacteriol. 176:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephanopoulos, G., and J. Kelleher. 2001. Biochemistry: how to make a superior cell. Science 292:2024-2025. [DOI] [PubMed] [Google Scholar]
- 32.Stingele, F., J. R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, T. D., and V. L. Crow. 1984. Selection of galactose-fermenting Streptococcus thermophilus in lactose-limited chemostat cultures. Appl. Environ. Microbiol. 48:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL___X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kranenburg, R., I. C. Boels, M. Kleerebezem, and W. M. de Vos. 1999. Genetics and engineering of microbial exopolysaccharides for food: approaches for the production of existing and novel polysaccharides. Curr. Opin. Biotechnol. 10:498-504. [DOI] [PubMed] [Google Scholar]
- 36.van Niel, E. W. J., and B. Hahn-Hägerdal. 1999. Nutrient requirements of lactococci in defined growth media. Appl. Microbiol. Biotechnol. 52:617-627. [Google Scholar]
- 37.Vaughan, E. E., P. T. van den Bogaard, P. Catzeddu, O. P. Kuipers, and W. M. de Vos. 2001. Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita, Y., Y. Tsukioka, Y. Nakano, K. Tomihisa, T. Oho, and T. Koga. 1998. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology 144:1235-1245. [DOI] [PubMed] [Google Scholar]