Abstract
A norB gene encoding a putative nitric oxide reductase is present in the genome of the nondenitrifying cyanobacterium Synechocystis sp. strain PCC6803. The gene product belongs to the quinol-oxidizing single-subunit class of nitric oxide reductases, discovered recently in the denitrifier Ralstonia eutropha. Heterologous complementation of a nitric oxide reductase-negative mutant of R. eutropha with norB from Synechocystis restored nitric oxide reductase activity. With reduced menadione as the electron donor, an enzymatic activity of 101 nmol of NO per min per mg of protein was obtained with membrane fractions of Synechocystis wild-type cells. Virtually no nitric oxide reductase activity was present in a norB-negative mutant of Synechocystis. Growing cells of this mutant are more sensitive toward NO than wild-type cells, indicating that the presence of a nitric oxide reductase is beneficial for Synechocystis when the cells are exposed to NO. Transcriptional fusions with the chloramphenicol acetyltransferase reporter gene were constructed to monitor norB expression in Synechocystis. Transcription of norB was not enhanced by the addition of the NO-generating agent sodium nitroprusside.
Nitric oxide is an important molecule in cell signaling and host defense mechanisms of eukaryotes. In prokaryotes, NO is a true intermediate of denitrification and is produced from nitrite by the dissimilatory nitrite reductase. In the course of denitrification, NO is further reduced to nitrous oxide by nitric oxide reductase. Bacterial nitric oxide reductases are integral membrane proteins which are connected to an electron transport chain. Nitric oxide reductases purified from denitrifying bacteria are classified as cytochrome cb heterodimers, consisting of a catalytic subunit, NorB, and a small subunit, NorC (NorCB enzyme) (12, 36, 42).
The prototype of a novel class of nitric oxide reductases was discovered in the β-proteobacterium Ralstonia eutropha (5). The carboxy terminus of this monomeric NorB protein corresponds to the catalytic subunit of NorCB enzymes. The amino terminus of NorB is extended and contains a large, probably periplasmic domain flanked by two additional transmembrane segments (3). The purified protein accepts electrons from quinols but fails to oxidize cytochrome c (3). To distinguish between the two types of nitric oxide reductases, the NorCB enzymes are designated cNor, and the single-component enzymes are referred to as qNor (12). Database analyses of unfinished genome sequences suggest that several bacteria harbor qNor enzymes. The majority of these hosts are classified as nondenitrifying pathogens.
A norB gene homologue was discovered in the genome sequence of the unicellular cyanobacterium Synechocystis sp. strain PCC6803 (16). Recently, the question was raised as to whether the putative norB gene product is a member of the class of qNor proteins, since its sequence predicts an N-terminal extension similar to that of qNor from R. eutropha and a norC-like gene is absent in the genome of Synechocystis (5). In this study, we show that the product of norB from Synechocystis is indeed physiologically active with reduced menadione. We demonstrate that an active nitric oxide reductase confers on its host an elevated tolerance to toxic NO.
MATERIALS AND METHODS
Bacterial strains and culturing.
R. eutropha H16 (ATCC 17699) is a wild-type strain harboring endogenous megaplasmid pHG1. HF420 is a nitric oxide reductase-negative mutant of strain H16 (5). Escherichia coli S17-1 (29) served as the donor in conjugative transfers. E. coli XL1-blue (Stratagene) was used as a host in standard cloning procedures. Synechocystis sp. strains M320 and M321 are norB-negative mutants of wild-type Synechocystis sp. strain PCC6803.
E. coli strains were grown in Luria-Bertani broth at 37°C. R. eutropha strains were cultivated at 30°C in mineral salts medium (27) with 0.4% (wt/vol) fructose as the carbon source (FN medium). Under denitrifying conditions, the cells were cultivated in 150-ml glass flasks sealed with a rubber septum and containing 100 ml of FN medium supplemented with 0.1% (wt/vol) sodium nitrate. The gas phase consisted of helium. Synechocystis sp. strain PCC6803 (wild type) and mutant strains were grown under continuous illumination at 30°C in BG-11 medium (30) supplemented with either 18 mM nitrate or 5 mM ammonia. Media for photoheterotrophic cultivation of Synechocystis strains were amended with 5 mM glucose.
Solid media contained 1.5% (wt/vol) agar. Antibiotics were added as follows: for R. eutropha, kanamycin (360 μg/ml) and tetracycline (15 μg/ml); for E. coli, kanamycin (50 μg/ml), tetracycline (20 μg/ml), and ampicillin (50 μg/ml); and for Synechocystis, kanamycin (40 μg/ml).
Nucleic acid manipulations.
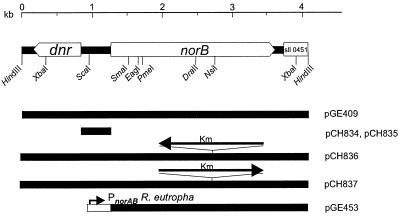
Isolation of plasmids, transformation, and cloning were carried out by standard methods (26). Southern hybridization was performed by using a digoxigenin-11-dUTP kit (Roche, Mannheim, Germany) for primer labeling and detection. Genomic DNA of Synechocystis was isolated as described by Franche and Damerval (10). A cosmid clone (cs0502) containing a 39-kb genomic DNA fragment of Synechocystis sp. strain PCC6803 was obtained from the Kazusa Research Institute (Kazusa, Japan). Plasmids pCH670, pCH681, and pGE409 (Fig. 1) were constructed by cloning a 4-kb HindIII fragment of cs0502 into pBluescript KS(+) (Stratagene), pBluescript SK(+) (Stratagene), and pVK101 (17), respectively. Transcriptional fusions were constructed by using vector pSB2A (22), which contains a promoterless chloramphenicol acetyltransferase (cat) reporter gene. The intergenic region between dnr and norB of Synechocystis was amplified from plasmid pCH681 by PCR with primers 445 (5"-TGCGCATGGGATTTAGGAATTAAG-3") and 446 (5"-TGCGCATACACAAAGAATGAAAAA-3"), which generate FspI sites at both ends of the PCR product. The resulting 426-bp fragment was digested with FspI and inserted into the HpaI site of vector pSB2A in both orientations, yielding plasmids pCH834 (Pdnr-cat) and pCH835 (PnorB-cat) (Fig. 1).
FIG. 1.
Synechocystis norB gene region. A physical map and relevant restriction sites are shown. DNA fragments of subclones are indicated by black bars. The orientation of a kanamycin resistance cartridge (Km) is indicated by arrows. The norAB promoter of R. eutropha is depicted by a white box with an arrow.
An NdeI site was introduced 5" of the start codon of norB by PCR with primers 451 (5"-TCTTTGCATATGGCAAATTCAACCTTTGATCTCCC-3") and 452 (5"-CCATCGTAGAGCACACCGGCCGTAGCCAGC-3") and plasmid pCH670 as a template. The 327-bp PCR product was cloned as an NdeI-EagI fragment into pET22b(+) (Novagen), yielding plasmid pCH838. The complete norB gene was restored in plasmid pCH839 by inserting a 2.46-kb EagI-XhoI fragment from pCH670 into pCH838. NdeI and BglII sites were introduced in the norAB promoter region of R. eutropha by PCR with primers 453 (5"-CGCAAGATCTCGTCGACCGCCACCGGCCGCAGGTG-3") and 454 (5"-GCAGGGTCATATGGTTTCTCCGTCTGAAGTTACGC-3") and plasmid pCH510 (5) as a template. The resulting 321-bp NdeI-BglII fragment containing PnorAB was cloned into pCH839. The resulting plasmid, pCH840, contains a fusion of PnorAB of R. eutropha with the complete norB gene of Synechocystis. For complementation of R. eutropha strains, a 3.1-kb BglII-HindIII fragment of pCH840 was cloned into HindIII-BamHI-linearized broad-host-range vector pVDZ"2 (8), yielding pGE453 (Fig. 1). For inactivation of the norB gene of Synechocystis, a 1.5-kb PstI kanamycin resistance cartridge from pRME1 (W. Messer, Berlin, Germany) was cloned in both orientations into the NsiI site of plasmid pCH670, yielding pCH836 and pCH837, respectively (Fig. 1).
Analytical procedures.
Protein was determined as described by Lowry et al. (20). N2O and N2 were analyzed by gas chromatography from the headspace of the culture bottles as described previously (4). Cat protein formed by cells containing the cat reporter gene was measured by an enzyme-linked immunosorbent assay according to the manufacturer's recommendations (Roche) by using 4-nitrophenyl phosphate as the substrate for alkaline phosphatase. Assays were calibrated by using serial dilutions of Cat enzyme.
Nitric oxide reductase activity was assayed at 30°C as described previously (5) by using a Clark electrode. The reaction mixture (2 ml) contained 50 mM sodium phosphate buffer (pH 7.0), 20 mM d-glucose, 10 U of glucose oxidase, and 250 U of catalase. NO reduction was measured by using 10 μmol of ascorbate plus 0.25 μmol of phenazine methosulfate. NADH-dependent NO reduction was measured by using 0.25 μmol of NADH and 0.35 μmol of 2-methyl-1,4-naphthoquinone (menadione). Quinols were formed from the corresponding quinones in the presence of 40 U of diaphorase from Clostridium kluyveri. After an incubation period of 5 min, NO-saturated buffer (100 nmol of NO) was added. The reaction was started by the addition of membrane extracts.
Inactivation of Synechocystis norB.
Synechocystis cells were transformed with plasmids pCH836 and pCH837 by following the protocol of Ermakova and coworkers (9). Cells were grown for 6 h on nonselective agar plates and subsequently underlaid with a kanamycin solution (final concentration, 10 μg/ml). Homozygous strains were obtained after four serial streak purifications of single colonies on plates containing increasing concentrations of kanamycin to a final concentration of 40 μg/ml (37) and were confirmed by Southern analysis.
RESULTS
Complementation of a norB-negative mutant of R. eutropha.
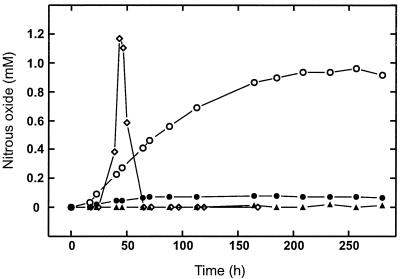
To test whether the norB gene product of Synechocystis is physiologically active, complementation studies were conducted with norB-negative mutant HF420 of R. eutropha as a recipient. Due to the high toxicity of NO, the absence of the nitric oxide reductase is lethal to denitrifying mutant cells of R. eutropha (5). Complementation with a physiologically active norB gene should therefore rescue the cells, and transconjugants derived from such experiments should produce nitrous oxide under anaerobic conditions with nitrate as the electron acceptor. A gene region including norB and dnr of Synechocystis was cloned into broad-host-range vector pVK101, yielding plasmid pGE409. In a parallel approach, the norB gene of Synechocystis was cloned under the control of the norAB promoter of R. eutropha, yielding plasmid pGE453. Plasmid pGE409 restored nitrous oxide production, although to a moderate degree, presumably due to the low expression of norB. An increase in nitrous oxide production was obtained with HF420(pGE453), reaching almost the R. eutropha wild-type level (Fig. 2). In contrast to the wild-type cells, however, the transconjugant cells did not consume nitrous oxide. A similar phenotype is occasionally observed for several R. eutropha mutants which have an altered nitric oxide reductase expression pattern (unpublished observations).
FIG. 2.
Nitrous oxide production by R. eutropha strains. Symbols: ◊, wild-type H16; ▴, nitric oxide reductase-negative mutant HF420; •, HF420 complemented with the norB gene region from Synechocystis(pGE409); ○, HF420 complemented with Synechocystis norB under the control of the norAB promoter of R. eutropha(pGE453).
Nitric oxide reductase activity in Synechocystis membranes.
Complementation analysis indicated that the norB gene from Synechocystis encodes a functional nitric oxide reductase. To investigate whether the norB gene is also active in its natural host, a knockout mutation in Synechocystis was constructed. A kanamycin resistance cartridge was inserted into norB in both orientations with the aid of plasmids pCH836 and pCH837 (Fig. 1). The corresponding mutants, M320 and M321, were assayed for nitric oxide reductase activity by monitoring NO consumption with membrane fractions and a Clark electrode. With ascorbate and phenazine methosulfate as the electron-donating agents, 50 nmol of NO per min per mg of protein was consumed by wild-type membranes of Synechocystis, corresponding to 10% of the level obtained with membrane fractions of R. eutropha. The addition of NADH-reduced 2-methyl-1,4-naphthoquinone (menadione) to membrane extracts of Synechocystis enhanced NO-reducing activity twofold (101 nmol of NO per min per mg of protein). Virtually no NO-reducing activity was detected with mutants M320 and M321. This result shows that NO reduction in Synechocystis is mediated by the norB gene product. Furthermore, the enzymatic activity seen with menadione supports the notion that the enzyme belongs to the group of qNor proteins.
Promoter analysis of PnorB and Pdnr.
norB of Synechocystis is preceded by a dnr gene, which is transcribed in the opposite direction. The putative dnr gene product belongs to a subfamily of Fnr-like transcriptional activators which presumably respond to effectors other than oxygen. To assess a possible effect of NO on the expression of norB in Synechocystis, the intergenic region between dnr and norB was cloned in both orientations into vector pSB2A. This construct should allow the identification of promoter activities exerted from Pdnr as well as PnorB if fused to an appropriate reporter gene. In this instance, transcriptional fusions were constructed with the chloramphenicol acetyltransferase-encoding cat gene on plasmid pSB2A. Plasmids pCH834 (Pdnr-cat) and pCH835 (PnorB-cat) and control vector pSB2A were transferred by conjugation into wild-type cells of Synechocystis. Transconjugant cells were grown photoautotrophically by using either ammonia or nitrate as the nitrogen source. The rationale for using nitrate, which is converted to nitrite by the assimilatory nitrate reductase, was to provide to the cells nitrogen oxide compounds, which may have an inducing effect on norB transcription.
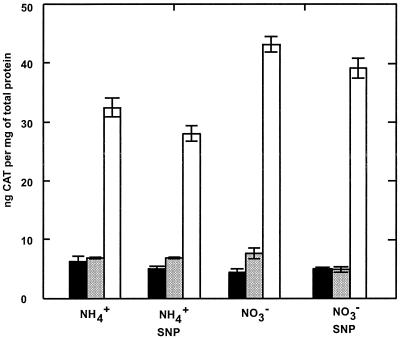
The Pdnr-cat fusion showed weak promoter activity that was not affected by the nitrogen source. An approximately three-fold-higher level of promoter activity was obtained with the PnorB-cat fusion (Fig. 3). Cells grown with nitrate showed a slightly higher level of promoter activity than did cell cultures amended with ammonia. Therefore, we cannot exclude the possibility that nitrate and/or nitrite modulates the expression of norB in Synechocystis.
FIG. 3.
cat expression conferred by the intergenic region between dnr and norB of Synechocystis. Media were supplemented with either 18 mM nitrate or 5 mM ammonia. When the cells reached an optical density at 750 nm of 1.0, 5 mM SNP was added. Activities were measured after an additional 4 h of growth. Error bars represent standard errors from three replicates. Symbols: black bars, pSB2A; gray bars, dnr"-cat; white bars, norB"-cat.
The Dnr/Nnr regulators from Paracoccus denitrificans (34) and Rhodobacter sphaeroides (19) and the sigma N-dependent regulator NorR from R. eutropha (24) have been shown to respond to the NO-generating (1) and nitrosylating (25) agent sodium nitroprusside (SNP). To investigate possible induction of Synechocystis norB by exogenous NO, photoautotrophically grown cultures of Synechocystis were amended with 5 mM SNP in the presence of ammonia as the nitrogen source. As shown in Fig. 3, the transcription of neither dnr nor norB was enhanced by the addition of SNP. Similar results were obtained with lower SNP concentrations, in the range of 0.5 to 4 mM (data not shown). Cells cultivated under oxygen limitation or grown photoheterotrophically were also not susceptible to induction by SNP-released NO (data not shown).
Sensitivity of Synechocystis to NO.
So far the data indicate that Synechocystis forms a catalytically active nitric oxide reductase even in the absence of its substrate, NO. This behavior enables the cells to respond immediately to the toxic compound if encountered in the environment. To examine the hypothesis of a protective role of NorB, the effects of exogenously added SNP and NO were investigated with growing cells of Synechocystis.
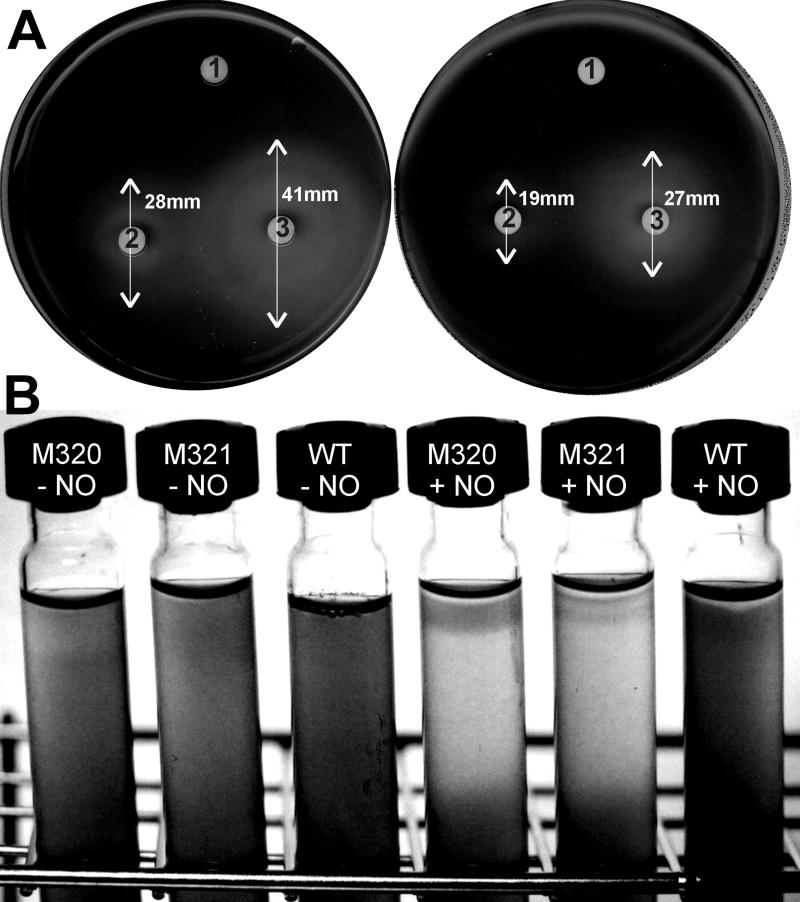
In a first attempt, paper disks soaked with SNP were placed on agar plates spread with a layer of Synechocystis cells. After 36 h of incubation, a clearance zone was visible around the paper disks containing SNP, indicating inhibition of growth by the NO-donating agent (Fig. 4A). The diameter of the area of inhibition was clearly enlarged when cells of NorB-deficient mutant M321 were used, indicating an increased sensitivity to the toxic compound.
FIG. 4.
Effect of NO on growing cells of Synechocystis. (A) Paper disks soaked with water (1) or 5 μl (2) or 15 μl (3) of a 0.5 mM SNP solution were placed on agar plates spread with a layer of Synechocystis cells. Left plate, mutant M321; right plate, wild type. Inhibition zones occurred after incubation for 36 h. (B) Suspensions of Synechocystis cells in BG-11 medium containing 0.6% agar were grown in glass tubes sealed with a rubber septum. WT, wild type; M320 and M321, norB-negative mutant strains. NO gas (10 mM) was injected before incubation (+NO).
To test if gaseous NO affects the growth of Synechocystis, wild-type and norB mutant cells were mixed with BG-11 soft agar in gastight tubes and subsequently incubated with or without NO in the headspace. A similar method was used by Cross and coworkers (6) to show increased NO sensitivity of Rhodobacter capsulatus cytochrome c" mutants. After 36 h of incubation at 30°C in light, the mutant cells showed an inhibition of growth in the upper layer of the agar directly exposed to NO (Fig. 4B). No growth inhibition occurred in tubes with wild-type cells or in tubes without NO in the headspace. On the basis of these experiments, we concluded that the norB gene product is beneficial for Synechocystis, since it protects the cells to a certain degree from the detrimental effect of NO.
DISCUSSION
Several bacteria have been described to contain incomplete denitrification pathways. The lack of nitrous oxide reductase, which mediates the production of dinitrogen, is most common (41). Wollinella succinogenes appears to be devoid of an NO-producing nitrite reductase but, on the other hand, is able to convert NO and nitrous oxide to the final product, dinitrogen (40). Some strains of Campylobacter fetus have the capacity to reduce nitrous oxide despite the lack of a nitrous oxide-producing activity (23). The nondenitrifying strain R. sphaeroides 2.4.1 contains a nor operon and displays nitric oxide reductase activity upon the introduction of a heterologous nitrite reductase (18). Finally, nondenitrifying Rhizobium hedysari represents so far a unique example of an organism that produces NO endogenously via a nitrite reductase without possessing a nitric oxide reductase (31). In this study, we show that Synechocystis joins the lists of organisms that contain a single nitrogen oxide-metabolizing activity.
Interestingly, the NO-converting enzyme from Synechocystis belongs to the qNor class of nitric oxide reductases. The analysis of unfinished genomes available to the public suggests a relatively wide distribution of qNor enzymes, predominantly in pathogens, e.g., Neisseria meningitidis, Neisseria gonorrhoeae, Corynebacterium diphtheriae, Mycobacterium avium, Bacillus stearothermophilus, Staphylococcus aureus, Legionella pneumophila, and Salmonella enterica serovar Paratyphi. Many intracellular pathogens may encounter NO during host defense. Thus, qNor enzymes may be a tool to escape host defense by detoxification of NO. Recently, it was shown that the qNor of N. gonorrhoeae is active in its host, suggesting a function of qNor in immune system evasion (13). One must consider, however, that N. gonorrhoeae harbors a nitrite reductase, implying that the organism also needs self-defense against endogenously produced NO.
The presence of a nitric oxide reductase and the absence of an NO-producing nitrite reductase in Synechocystis resemble the enzymatic pattern seen in R. sphaeroides 2.4.1. The regulation of these two systems, however, seems to be different. The expression of the nitric oxide reductase in R. sphaeroides 2.4.1 is induced by exogenously or endogenously provided NO (18), whereas the qNor of Synechocystis is expressed independently of NO. A largely nonregulated phenotype was unexpected, since norB is neighbored by a dnr gene whose product likely is a regulator of the Dnr/Nnr subgroup of Fnr-like proteins that lack the N-terminal cysteines critical for oxygen sensing. Dnr/Nnr proteins have been shown to control the NO-sensitive transcription of nor genes in R. sphaeroides (18, 19, 32), Pseudomonas stutzeri (35), and Paracoccus denitrificans (33, 34). Interestingly, a change of the conserved residue Tyr93 to Phe in Nnr of P. denitrificans resulted in severe downregulation, explained by less tight contact of the mutant protein with RNA polymerase (14). In Dnr of Synechocystis, the corresponding residue in the native sequence is Phe. Therefore, the lack of induction by SNP in Synechocystis indicates that Dnr, if involved in regulation at all, responds to a different signal or is nonfunctional.
Most, if not all, organisms that contain a cNor protein also contain sets of accessory genes for the NorD, NorE, NorF, and NorQ/NirQ proteins, which affect the activity, assembly, or stability of the NorCB heterodimer in P. denitrificans (7). None of these genes is present in the genome of Synechocystis, suggesting that the function of qNor enzymes does not rely on these accessory genes. This observation is surprising, since qNor and the NorB subunit of cNor share similar overall structures and equipment with prosthetic groups. The broad phylogenetic distribution of qNor-encoding genes among proteobacteria, cyanobacteria, firmicutes (12), and even archaea (28) may reflect a selective advantage of qNor whose function relies only on a single gene transfer.
We have demonstrated the function of the NorB product of Synechocystis by complementation of a nitric oxide reductase-negative mutant of R. eutropha. Wild-type cells of Synechocystis tolerate high concentrations of approximately 10 mM NO in the headspace. Steady-state levels of NO in the environment of denitrifying organisms are supposed to be in the nanomolar range (11, 15, 41). Therefore, Kwiatkowski and coworkers (18) have proposed that the nitric oxide reductase of R. sphaeroides 2.4.1 uses NO from the environment to gain energy rather than to protect cells from the toxic compound. The same function may account for the nitric oxide reductase of Synechocystis. Moreover, the recent discovery of NO formation by nitrate-grown cells of the unicellular cyanobacterium Synechococcus leopoliensis (21) raises the question of whether Synechocystis produces NO endogenously under certain conditions. In addition to denitrification, NO is also released into the environment by nitrifying bacteria (2), cyanobacteria, green algae (21), and even higher plants (38, 39). Thus, detoxification of exogenous NO may become relevant for Synechocystis when it grows in a natural community of NO-producing organisms.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
We are grateful to Satoshi Tabata for providing clone cs0502 and to Franck Chauvat for providing plasmid pSB2A.
REFERENCES
- 1.Bates, J. N., M. T. Baker, R. Guerra, Jr., and D. G. Harrison. 1991. Nitric oxide generation from nitroprusside by vascular tissue. Evidence that reduction of the nitroprusside anion and cyanide loss are required. Biochem. Pharmacol. 42:157-165. [DOI] [PubMed] [Google Scholar]
- 2.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cramm, R., A. Pohlmann, and B. Friedrich. 1999. Purification and characterization of the single-component nitric oxide reductase from Ralstonia eutropha H16. FEBS Lett. 460:6-10. [DOI] [PubMed] [Google Scholar]
- 4.Cramm, R., R. A. Siddiqui, and B. Friedrich. 1994. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J. Biol. Chem. 269:7349-7354. [PubMed] [Google Scholar]
- 5.Cramm, R., R. A. Siddiqui, and B. Friedrich. 1997. Two isofunctional nitric oxide reductases in Alcaligenes eutrophus H16. J. Bacteriol. 179:6769-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross, R., D. Lloyd, R. K. Poole, and J. W. Moir. 2001. Enzymatic removal of nitric oxide catalyzed by cytochrome c" in Rhodobacter capsulatus. J. Bacteriol. 183:3050-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, A. P., J. van der Oost, W. N. Reijnders, H. V. Westerhoff, A. H. Stouthamer, and R. J. van Spanning. 1996. Mutational analysis of the nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur. J. Biochem. 242:592-600. [DOI] [PubMed] [Google Scholar]
- 8.Deretic, V., S. Chandrasekharappa, J. F. Gill, D. K. Chatterjee, and A. M. Chakrabarty. 1987. A set of cassettes and improved vectors for genetic and biochemical characterization of Pseudomonas genes. Gene 57:61-72. [DOI] [PubMed] [Google Scholar]
- 9.Ermakova, S. Y., I. V. Elanskaya, K.-U. Kallies, A. Weihe, T. Börner, and S. V. Shestakov. 1993. Cloning and sequencing of mutant psbB genes of the cyanobacterium Synechocystis PCC6803. Photosynth. Res. 37:139-146. [DOI] [PubMed] [Google Scholar]
- 10.Franche, C., and T. Damerval. 1988. Tests on nif probes and DNA hybridizations. Methods Enzymol. 122:69-78. [Google Scholar]
- 11.Goretski, J., O. C. Zafiriou, and T. C. Hollocher. 1990. Steady-state nitric oxide concentrations during denitrification. J. Biol. Chem. 265:11535-11538. [PubMed] [Google Scholar]
- 12.Hendriks, J., A. Oubrie, J. Castresana, A. Urbani, S. Gemeinhardt, and M. Saraste. 2000. Nitric oxide reductases in bacteria. Biochim. Biophys. Acta 1459:266-273. [DOI] [PubMed] [Google Scholar]
- 13.Householder, T. C., E. M. Fozo, J. A. Cardinale, and V. L. Clark. 2000. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect. Immun. 68:5241-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchings, M. I., N. Shearer, S. Wastell, R. J. van Spanning, and S. Spiro. 2000. Heterologous NNR-mediated nitric oxide signaling in Escherichia coli. J. Bacteriol. 182:6434-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalkowski, I., and R. Conrad. 1991. Metabolism of nitric oxide in denitrifying Pseudomonas aeruginosa and nitrate-respiring Bacillus cereus. FEMS Microbiol. Lett. 66:107-111. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, T., A. Tanaka, S. Sato, H. Kotani, T. Sazuka, N. Miyajima, M. Sugiura, and S. Tabata. 1995. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 2:153-166, 191-198. [DOI] [PubMed]
- 17.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 18.Kwiatkowski, A. V., W. P. Laratta, A. Toffanin, and J. P. Shapleigh. 1997. Analysis of the role of the nnrR gene product in the response of Rhodobacter sphaeroides 2.4.1 to exogenous nitric oxide. J. Bacteriol. 179:5618-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwiatkowski, A. V., and J. P. Shapleigh. 1996. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Biol. Chem. 271:24382-24388. [DOI] [PubMed] [Google Scholar]
- 20.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 21.Mallick, N., L. C. Rai, F. H. Mohn, and C. J. Soeder. 1999. Studies on nitric oxide (NO) formation by the green alga Scenedesmus obliquus and the diazotrophic cyanobacterium Anabaena doliolum. Chemosphere 39:1601-1610. [DOI] [PubMed] [Google Scholar]
- 22.Marraccini, P., S. Bulteau, C. Cassier-Chauvat, P. Mermet-Bouvier, and F. Chauvat. 1993. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 23:905-909. [DOI] [PubMed] [Google Scholar]
- 23.Payne, W. J., M. A. Grant, J. Shapleigh, and P. Hoffman. 1982. Nitrogen oxide reduction in Wolinella succinogenes and Campylobacter species. J. Bacteriol. 152:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pohlmann, A., R. Cramm, K. Schmelz, and B. Friedrich. 2000. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol. Microbiol. 38:626-638. [DOI] [PubMed] [Google Scholar]
- 25.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775-783. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: wachstumsphysiologische Untersuchungen. Arch. Microbiol. 38:209-222. [PubMed] [Google Scholar]
- 28.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-De Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 30.Stanier, R. Y., R. Kunisawa, M. Mandel, and G. Cohen-Batire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35:171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toffanin, A., Q. Wu, M. Maskus, S. Caselia, H. D. Abruna, and J. P. Shapleigh. 1996. Characterization of the gene encoding nitrite reductase and the physiological consequences of its expression in the nondenitrifying Rhizobium “hedysari” strain HCNT1. Appl. Environ. Microbiol. 62:4019-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosques, I. E., A. V. Kwiatkowski, J. Shi, and J. P. Shapleigh. 1997. Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179:1090-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Spanning, R. J., A. P. De Boer, W. N. Reijnders, S. Spiro, H. V. Westerhoff, A. H. Stouthamer, and J. Van der Oost. 1995. Nitrite and nitric oxide reduction in Paracoccus denitrificans is under the control of NNR, a regulatory protein that belongs to the FNR family of transcriptional activators. FEBS Lett. 360:151-154. [DOI] [PubMed] [Google Scholar]
- 34.Van Spanning, R. J., E. Houben, W. N. Reijnders, S. Spiro, H. V. Westerhoff, and N. Saunders. 1999. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J. Bacteriol. 181:4129-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vollack, K. U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watmough, N. J., G. Butland, M. R. Cheesman, J. W. Moir, D. J. Richardson, and S. Spiro. 1999. Nitric oxide in bacteria: synthesis and consumption. Biochim. Biophys. Acta 1411:456-474. [DOI] [PubMed] [Google Scholar]
- 37.Wilde, A., H. Härtel, T. Hübschmann, P. Hoffmann, S. V. Shestakov, and T. Börner. 1995. Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wildt, J., D. Kley, A. Rockel, P. Rockel, and H. J. Segschneider. 1997. Emission of NO from several higher plant species. J. Geophys. Res. 102:5919-5927. [Google Scholar]
- 39.Yamasaki, H., and Y. Sakihama. 2000. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 468:89-92. [DOI] [PubMed] [Google Scholar]
- 40.Zumft, W. G. 1992. The denitrifying prokaryotes. Springer Verlag, Berlin, Germany.
- 41.Zumft, W. G. 1993. The biological role of nitric oxide in bacteria. Arch. Microbiol. 160:253-264. [DOI] [PubMed] [Google Scholar]
- 42.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]