Abstract
Ralstonia eutropha JMP134(pJP4) and several other species of motile bacteria can degrade the herbicide 2,4-dichlorophenoxyacetate (2,4-D), but it was not known if bacteria could sense and swim towards 2,4-D by the process of chemotaxis. Wild-type R. eutropha cells were chemotactically attracted to 2,4-D in swarm plate assays and qualitative capillary assays. The chemotactic response was induced by growth with 2,4-D and depended on the presence of the catabolic plasmid pJP4, which harbors the tfd genes for 2,4-D degradation. The tfd cluster also encodes a permease for 2,4-D named TfdK. A tfdK mutant was not chemotactic to 2,4-D, even though it grew at wild-type rates on 2,4-D.
Most motile bacteria can sense and respond to low concentrations of organic compounds in their environment by the process of chemotaxis. There is evidence that chemotaxis can enhance biodegradation (11), presumably by rapidly bringing cells into close contact with degradable substrates. 2,4-Dichlorophenoxyacetate (2,4-D) is a widely used herbicide (Industry Task Force on 2,4-D research data [http://www.24d.org]) that can be degraded by a number of species of motile bacteria (12).
The bacterium Ralstonia eutropha JMP134(pJP4) grows on 2,4-D by using genes present on a self-transmissible plasmid called pJP4 (3). Plasmid pJP4 also encodes a permease for 2,4-D named TfdK (10). The predicted TfdK protein has 12 membrane-spanning regions and is a member of the aromatic acid:H+ symporter family of the major facilitator superfamily of transport proteins (15). It is 33% identical in amino acid sequence to PcaK, a 4-hydroxybenzoate permease from Pseudomonas putida that is also required for chemotaxis to 4-hydroxybenzoate (2, 7, 13). The similarity of TfdK to PcaK led us to investigate if R. eutropha is chemotactically attracted to 2,4-D and, if so, whether tfdK is required for the chemotactic response.
Bacterial strains and experimental procedures.
Wild-type R. eutropha [strain JMP134(pJP4)], a plasmid pJP4-cured derivative (strain JMP289), and a tfdK mutant [strain JMP134(pJP4::cba79)] (10) were obtained from J. R. van der Meer. Strains were grown on mineral salts medium (7) at 30°C with shaking at 250 rpm. Analysis by gas chromatography-mass spectrometry as previously described (17) indicated that the 2,4-D (Sigma-Aldrich, St. Louis, Mo.) used in these studies contained no detectable contaminants. Bacterial transformations were carried out according to the method of Hanahan (6).
Plasmid DNA was prepared from Escherichia coli with a QIAprep spin miniprep kit (Qiagen Inc., Chatsworth, Calif.). Plasmid DNA was prepared from R. eutropha as described previously (18). DNA fragments were purified from agarose gels with the QIAquick gel extraction kit (Qiagen Inc.).
Plasmid pHAH108, a broad-host-range plasmid designed for expression of TfdK, was constructed in several steps. First, a 1,388-bp fragment of DNA encompassing the tfdK gene was amplified from pJP4 by PCR and cloned into pAF7 (A. Ferrandez and C. S. Harwood, unpublished data) to form pHAH107. Plasmid pAF7 is a derivative of pT7-6 (20) that carries the P. putida pcaK gene fused at its predicted N terminus to a hemagglutinin (HA) epitope sequence derived from the X47 virus hemagglutinin (14) behind a transcriptional enhancer and a ribosome-binding site (RBS).
The PCR-amplified tfdK gene was cloned into the NaeI and BamHI sites of pAF7, replacing the pcaK gene. An EcoRI/HindIII fragment from pHAH107 containing HA-tfdK, the transcriptional enhancer, and the consensus RBS was then subcloned into the broad-host-range vector pBBR1MCS-5 (9) to create pHAH108. Plasmids were introduced into R. eutropha by conjugation (1) from Escherichia coli DH5α in triparental matings using E. coli CC118(pRK600) (8) to provide the transfer functions. Gentamicin (20 μg per ml), chloramphenicol (100 μg per ml), and ampicillin (100 μg per ml) were added to growth media to select for plasmids in E. coli and R. eutropha. Western blot analysis (20) with anti-HA antiserum (Roche Molecular Biochemicals, Indianapolis, Ind.) was used to show that HA-TfdK was expressed in R. eutropha JMP134(pJP4::cba79, pHAH108).
R. eutropha JMP134(pJP4) cells are attracted to 2,4-D, and this is a plasmid-encoded, inducible trait.
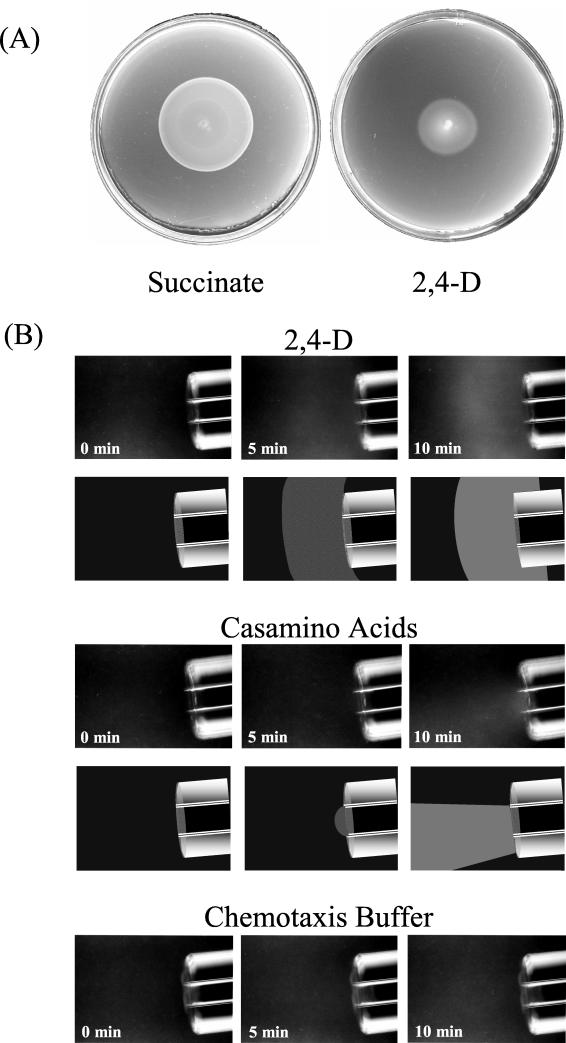
Chemotaxis was tested with a soft agar swarm plate assay and a modified capillary assay. For the soft agar swarm plate assay, an R. eutropha colony was stabbed into the center of a mineral salts medium plate that contained 0.25 mM 2,4-D or 1.0 mM succinate and was solidified with 0.3% Noble agar (Difco Laboratories, Detroit, Mich.). Wild-type cells had a positive chemotactic response to succinate and to 2,4-D, as evidenced by a sharp ring of growth that formed when cells responded to the gradient of attractant that was created as they metabolized the carbon source in the plates (Fig. 1A).
FIG. 1.
Chemotactic response of R. eutropha JMP134(pJP4). (A) Chemotactic rings formed by wild-type cells on soft agar swarm plates containing 0.5 mM succinate (left) or 0.25 mM 2,4-D (right). Plates were inoculated by stabbing motile cells in the center of the plate at a point corresponding to the center of the swarm ring and incubated for 24 to 48 h at 30°C. (B) Attraction of wild-type cells in a modified capillary assay. Cells were grown on 3 mM 2,4-D. Capillaries contained 10 mM 2,4-D, 1% Casamino Acids, or no attractant (chemotaxis buffer). Chemotactic responses are shown in photographic form (top) and illustrative form (bottom). Illustrations for chemotaxis buffer were omitted because no response was observed.
Modified capillary assays allowed qualitative assessment of chemotaxis with a phase contrast microscope (5). With this method, chemotaxis can be visualized in the absence of metabolism of the attractant. Capillaries (1 μl) contained attractant in 1% low-melting-temperature agarose dissolved in chemotaxis buffer (50 mM potassium phosphate [pH 7.0], 20 μM EDTA, 0.05% glycerol). Cells that had been grown in mineral medium (7) containing 3 mM 2,4-D, 10 mM succinate, or 10 mM succinate plus 0.5 mM 2,4-D were harvested in the mid-logarithmic phase of growth and suspended in chemotaxis buffer to a final A660 of approximately 0.1. Samples of the cell suspension were placed in a chamber formed by a microscope slide, a glass U-tube, and a cover slip, and the capillary containing the attractant was inserted into the suspension. Cell behavior around the tip of the capillary was observed at a final magnification of ×40 and photographed at 0 min, 5 min, and 10 min. Modified capillary assays were repeated at least five times for each condition tested with reproducible results.
R. eutropha wild-type cells formed visible clouds of turbidity as they accumulated around the open tips of capillaries that contained either 10 mM 2,4-D or 1% Casamino Acids (Fig. 1B). Wild-type cells also accumulated around the open tips of capillaries that contained 1 mM 2,4-D, but this response was less dramatic and difficult to photograph (data not shown). Chemotaxis buffer, used as a control, did not elicit a chemotactic response from R. eutropha in the modified capillary assay. R. eutropha JMP134(pJP4) cells that had been grown in 10 mM succinate did not respond to 2,4-D in modified capillary assays. However, these cells were attracted to 2,4-D when grown on 10 mM succinate plus 0.5 mM 2,4-D (data not shown). This indicates that chemotaxis to 2,4-D was induced by growth with 2,4-D. R. eutropha cells that had been cured of the pJP4 plasmid (strain JMP289) and grown on 10 mM succinate plus 0.5 mM 2,4-D showed little or no attraction to 2,4-D in a modified capillary assay (data not shown). This indicates that 2,4-D chemotaxis is a plasmid-encoded trait.
tfdK gene is required for chemoattraction to 2,4-D.
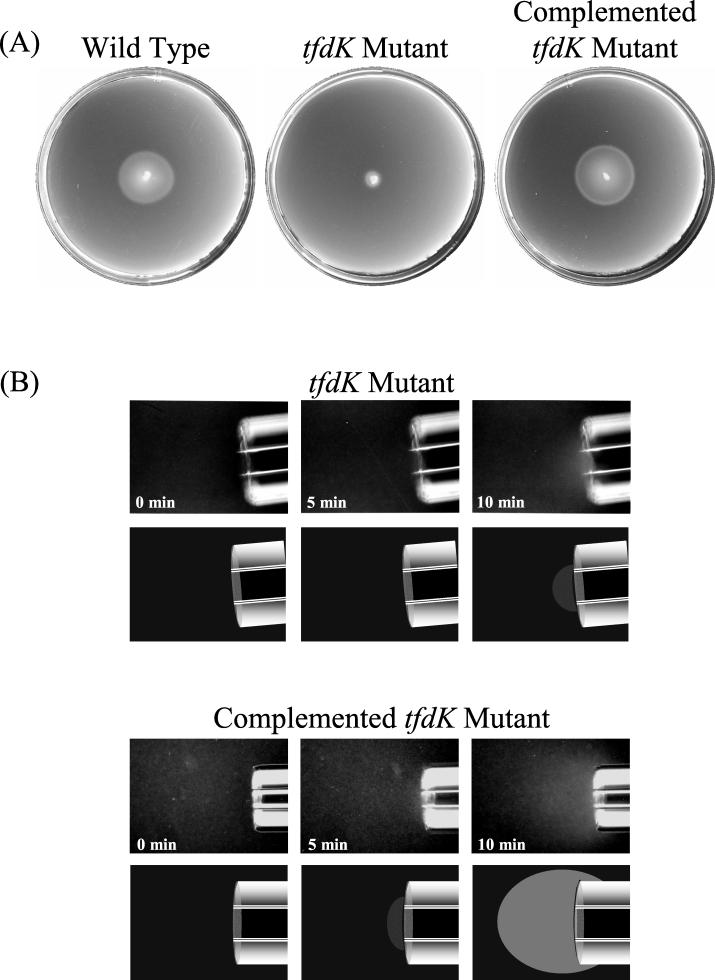
TfdK is a membrane-spanning protein that catalyzes the active transport of 2,4-D into cells. The TfdK permease allows cells to take up very low concentrations of 2,4-D, but it is not essential for entry of 2,4-D into cells, as 2,4-D can cross the cell membrane of R. eutropha by simple diffusion (10). When 2,4-D was supplied at a concentration of 3 mM, the tfdK mutant had a specific growth rate with 2,4-D (0.324 ± 0.064 h−1) that was nearly identical to that of the wild type (0.333 ± 0.044 h−1). A tfdK mutant did not form chemotactic rings in soft agar swarm plates containing 2,4-D, even after incubation for 2 days (Fig. 2A). Instead, the cells formed a fuzzy, slow-moving ring that reflected random movement of motile cells.
FIG. 2.
Response of a tfdK mutant to 2,4-D. (A) Chemotactic swarms formed by wild-type cells, the tfdK mutant, and the tfdK mutant expressing the tfdK gene in trans on swarm plates containing 0.25 mM 2,4-D. (B) Modified capillary assays showing the response of tfdK mutant cells and complemented tfdK mutant cells to 2,4-D. Both strains were grown on 3 mM 2,4-D. Results are shown in photographic form (top) and illustrative form (bottom).
tfdK mutant cells that had been grown on 3 mM 2,4-D were attracted to 1% Casamino Acids but not to 10 mM 2,4-D in modified capillary assays (Fig. 2B). The tfdK mutant sometimes had a slight positive response to 2,4-D, but this response was no greater than that exhibited by cells of the pJP4-cured strain, JMP289, that had been grown under the same conditions (10 mM succinate plus 0.5 mM 2,4-D).
When the TfdK protein was expressed in trans from plasmid pHAH108 in the tfdK mutant, cells formed chemotactic rings on 2,4-D swarm plates (Fig. 2A) and were attracted to 2,4-D in modified capillary assays (Fig. 2B). The vector pBBR1MCS-5 did not complement the tfdK mutant in chemotaxis assays.
Conclusions.
The coordinate regulation of 2,4-D chemotaxis with 2,4-D degradation suggests that chemotaxis may be an integral feature of the biodegradation of 2,4-D by R. eutropha. Various strains of Pseudomonas can sense and swim towards the pollutants benzene, toluene, trichloroethylene, and naphthalene as well as to the aromatic acid 4-hydroxybenzoate (4, 5, 7, 16). Chemotaxis to these compounds requires prior growth with toluene (for the benzene, toluene, and trichloroethylene responses), naphthalene, or 4-hydroxybenzoate. The coregulation of chemotaxis and degradation often reflects the coordinate induction of a chemoreceptor gene with degradation genes.
Recently, a methyl-accepting chemotaxis protein (MCP) gene required for chemotaxis to naphthalene was found to be cotranscribed with meta-cleavage pathway genes present on the NAH7 catabolic plasmid for naphthalene degradation (5). MCPs have been well studied in E. coli and Salmonella spp., in which they serve as cell surface receptors for chemoattractants. On binding an attractant, an MCP undergoes a conformational change that initiates sensory signal transduction by altering the activity of CheA, an associated sensor kinase. CheA-P then acts as a phosphodonor for the response regulator, CheY, which interacts with rotational switch proteins in the flagellar motors. This causes a change in swimming behavior so that cells migrate towards chemoattractants (19).
The data presented here suggest that plasmid pJP4 for 2,4-D degradation also encodes a chemoreceptor protein that is coordinately regulated with 2,4-D degradation genes. However, this protein, TfdK, does not resemble an MCP. Instead, it is homologous to PcaK, a protein that in P. putida functions in the transport of 4-hydroxybenzoate as well as in chemotaxis to 4-hydroxybenzoate (2, 13). TfdK is thus a second example of a major facilitator superfamily transport protein that has dual roles in transport and in chemotaxis.
We do not understand how either PcaK or TfdK functions in chemotaxis. These permeases could play a direct role in signaling and communicate a conformational change in protein structure that occurs on ligand binding or transport to a physically associated MCP or other chemosensory protein. Alternatively, they may have a more indirect role in chemotaxis. Since 2,4-D can diffuse into R. eutropha at rates sufficient to support wild-types rates of growth under the conditions in which 2,4-D chemotaxis was measured, simple accumulation of this aromatic acid within cells does not appear to be sufficient for chemotaxis to occur; TfdK must be present. A more likely scenario for an indirect role is that TfdK functions in chemotaxis by delivering a high local concentration of chemoattractant to chemosensory proteins present on the cytoplasmic side of the cell membrane.
Acknowledgments
We thank Jan Roelof van der Meer for supplying strains used in this study, Rebecca Parales for helpful discussions, and Juan Parales for assistance with GC-MS analysis.
This work was supported by Public Health Service grant GM56665 from the National Institute of General Medical Sciences. A.C.H. has been supported by a National Science Foundation Research Training Grant (DBI9602247) and by a fellowship from the University of Iowa Center for Biocatalysis and Bioprocessing.
REFERENCES
- 1.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn 5- and Tn 10-derived mini-transposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 2.Ditty, J. L., and C. S. Harwood. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J. Bacteriol. 181:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Don, R. H., and J. M. Pemberton. 1985. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J. Bacteriol. 161:466-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimm, A. C., and C. S. Harwood. 1997. Chemotaxis of Pseudomonas spp. to the polyaromatic hydrocarbon naphthalene. Appl. Environ. Microbiol. 63:4111-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm, A. C., and C. S. Harwood. 1999. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 181:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 7.Harwood, C. S., N. N. Nichols, M. K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 9.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, 2nd, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 10.Leveau, J. H., A. J. Zehnder, and J. R. van der Meer. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180:2237-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marx, R. B., and M. D. Aitken. 2000. Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system. Environ. Sci. Technol. 34:3379-3383. [Google Scholar]
- 12.McGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols, N. N., and C. S. Harwood. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niman, H. L., R. A. Houghten, L. E. Walker, R. A. Reisfeld, I. A. Wilson, J. M. Hogle, and R. A. Lerner. 1983. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 80:4949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parales, R. E., J. L. Ditty, and C. S. Harwood. 2000. Toluene-degrading bacteria are chemotactic towards the environmental pollutants benzene, toluene, and trichloroethylene. Appl. Environ. Microbiol. 66:4098-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resnick, S. M., and D. T. Gibson. 1996. Regio- and stereospecific oxidation of 9,10-dihydroanthracene and 9,10-dihydrophenanthrene by naphthalene dioxygenase: structure and absolute stereochemistry of metabolites. Appl. Environ. Microbiol. 62:3355-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, and M. Schaechter (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 20.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]