Abstract
Purification of xylulose 5-phosphate phosphoketolase (XpkA), the central enzyme of the phosphoketolase pathway (PKP) in lactic acid bacteria, and cloning and sequence analysis of the encoding gene, xpkA, from Lactobacillus pentosus MD363 are described. xpkA encodes a 788-amino-acid protein with a calculated mass of 88,705 Da. Expression of xpkA in Escherichia coli led to an increase in XpkA activity, while an xpkA knockout mutant of L. pentosus lost XpkA activity and was not able to grow on energy sources that are fermented via the PKP, indicating that xpkA encodes an enzyme with phosphoketolase activity. A database search revealed that there are high levels of similarity between XpkA and a phosphoketolase from Bifidobacterium lactis and between XpkA and a (putative) protein present in a number of evolutionarily distantly related organisms (up to 54% identical residues). Expression of xpkA in L. pentosus was induced by sugars that are fermented via the PKP and was repressed by glucose mediated by carbon catabolite protein A (CcpA) and by the mannose phosphoenolpyruvate phosphotransferase system. Most of the residues involved in correct binding of the cofactor thiamine pyrophosphate (TPP) that are conserved in transketolase, pyruvate decarboxylase, and pyruvate oxidase were also conserved at a similar position in XpkA, implying that there is a similar TPP-binding fold in XpkA.
Lactic acid bacteria (LAB) are capable of generating energy by homo- or heterofermentative degradation of sugars. During anaerobic growth of obligately homofermentative LAB in the presence of excess substrate, energy sources like glucose are converted into pyruvate via the Embden-Meyerhoff-Parnas pathway, and the pyruvate is further metabolized to lactate. During heterofermentative degradation sugars are metabolized via the phosphoketolase pathway (PKP), which results in equimolar amounts of CO2, lactate, and acetate-ethanol. Heterolactic LAB can be divided into obligately heterofermentative species, in which both hexoses and pentoses are fermented via the PKP, and facultatively heterofermentative organisms, which degrade hexoses via the Embden-Meyerhoff-Parnas pathway and pentoses via the PKP.
Xylulose 5-phosphate phosphoketolase is the central enzyme of the PKP. In the presence of inorganic phospate this enzyme converts xylulose 5"-phosphate (X5P) into glyceraldehyde 3-phosphate and acetylphosphate. Phosphoketolase activity was first detected in heterofermentative lactobacilli (7, 9, 12, 25), but it has also been found in other organisms, such as Acetobacter xylinum (20, 23), yeasts (6, 21), Thiobacillus novellus (8), and Butyrivibrio fibrisolvens (16), as well as Fibrobacter succinogenes and Fibrobacter intestinalis (17). In bifidobacteria two types of phosphoketolase have been described, a fructose 6-phosphate (F6P)-specific enzyme and an enzyme with specificity for F6P and X5P. The gene encoding the enzyme with dual specificity was recently cloned from Bifidobacterium lactis and characterized (18).
Fermentation of pentoses (e.g., xylose) can be divided into the following two parts: a specific part, in which pentose is taken up and converted into X5P; and a general part, in which further metabolism of X5P follows the PKP. In Lactobacillus pentosus xylose is transported by the mannose phosphoenolpyruvate phosphotransferase system (PTS) and is metabolized by xylose isomerase and xylulose kinase, which are encoded by xylA and xylB (2, 15). Expression of these genes is induced in the presence of xylose and is repressed by carbon catabolite repression (CCR) (13, 14).
CCR in gram-positive bacteria, including lactobacilli, is mediated by global catabolite control protein A (CcpA), which can bind to specific operator sequences, the catabolite responsive elements (cre), that are found within or near promoter sequences of some catabolic genes and either repress or activate transcription (11, 24). CcpA-mediated CCR is linked to PTS activity by HPr in Bacillus and Lactobacillus. In these organisms HPr can be phosphorylated at two sites, His15 (by enzyme I) and Ser46 (by an ATP-dependent HPr kinase). The ATP-dependent HPr kinase is activated by metabolic intermediates, the most important of which is fructose 1,6-biphosphate, and is inhibited by inorganic phosphate. HPr(SerP) (HPr phosphorylated at Ser46) interacts with CcpA, which increases the affinity for cre sequences (5).
To study xylose metabolism in more detail and especially to learn more about the central enzyme of the PKP, we purified phosphoketolase from L. pentosus and isolated and characterized the encoding gene, xpkA. The functionality of xpkA was verified through expression in Escherichia coli. To study the regulation of synthesis of XpkA, activity assays were performed with wild-type and mutant strains of L. pentosus. Furthermore, to investigate the effect of inactivating XpkA activity on the capacity of a strain to utilize sugars that are fermented via the PKP, we constructed an xpkA knockout mutant of L. pentosus which did not have XpkA activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli was cultivated on Luria-Bertani (LB) agar or in LB broth. Ampicillin was added at a final concentration of 100 μg/ml when necessary. Lactobacilli were cultivated without agitation at 37°C in MRS medium (3) (Difco Laboratories, Detroit, Mich.) or M medium (14) containing a sugar at a concentration of 1% (wt/vol). Erythromycin was added at a final concentration of 5 μg/ml when necessary. The xpkA knockout mutant LPE179 was cultivated at 40°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Lactobacillus strains | ||
| L. pentosus MD353 | Wild-type strain | 15 |
| L. pentosus MD363 | Wild-type strain | 13 |
| L. pentosus LPE4 | MD363, ccpA::pE12 Emr | 13 |
| L. pentosus LPE5 | Deoxyglucose-resistant mutant of MD353 | 2 |
| L. pentosus LPE179 | MD363, xpkA::pLPA30 Emr | This study |
| Escherichia coli JA221 | F−hsdR lacY leuB6 trpE5 recA1 lambda− | LCa |
| Plasmids | ||
| pIN15E | 5.8-kb integration plasmid for Lactobacillus, Emr Apr, temperature-sensitive replicon | 13 |
| pBCP367 | E. coli expression vector, Apr | 26 |
| pLPA22 | pUC18 with a 2-kb internal PCR fragment of xpkA inserted between BamHI and EcoRI sites | This study |
| pPLA28 | pUC18 with a 2.5-kb inverse PCR fragment of the C terminus of xpkA inserted in StuI site | This study |
| pLPA30 | pIN15E, 850-bp internal PCR fragment of xpkA inserted between BamHI and PstI sites | This study |
| pLPA31 | pUC18 with 0.4-kb PCR fragment of upstream region of xpkA inserted between EcoRI and BamHI sites | This study |
| pLPA32 | pUC18 with PCR-amplified xpkA (complete gene) inserted between NdeI and BamHI sites | This study |
| pLPA33 | pUC18 with PCR-amplified xpkA (complete gene) inserted between NdeI and BamHI sites | This study |
| pLPA36 | pBCP367 xpkA PCR amplified (expression in E. coli) and inserted between NdeI and BamHI sites | This study |
| pLPA41 | pUC18 with 0.4-kb EcoRI-BamHI PCR fragment of upstream region of xpkA | This study |
LC, laboratory culture collection.
Materials.
X5P was obtained from Fluka (Buchs, Switzerland). The enzymes used for the XpkA activity assay and for DNA manipulation were obtained from Boehringer Mannheim or New England Biolabs and were used according to the specifications of the manufacturer. [α-32P]dATP (3,000 Ci/mmol) was obtained from Amersham.
DNA manipulation.
Recombinant DNA procedures, Southern blot analysis, and transformation of E. coli were performed by using standard methods (22). Chromosomal DNA of Lactobacillus strains were isolated as described previously (15). DNA fragments were isolated from agarose gels with a QIAEX II gel extraction kit (QIAGEN GmbH). PCRs were performed by using the Expand high-fidelity PCR system (Boehringer Mannheim), unless indicated otherwise. Automated sequencing was performed by BaseClear, Leiden, The Netherlands. Chromosomal DNA sequencing was performed by Gendika, Veendam, The Netherlands.
Cloning and sequencing of xpkA.
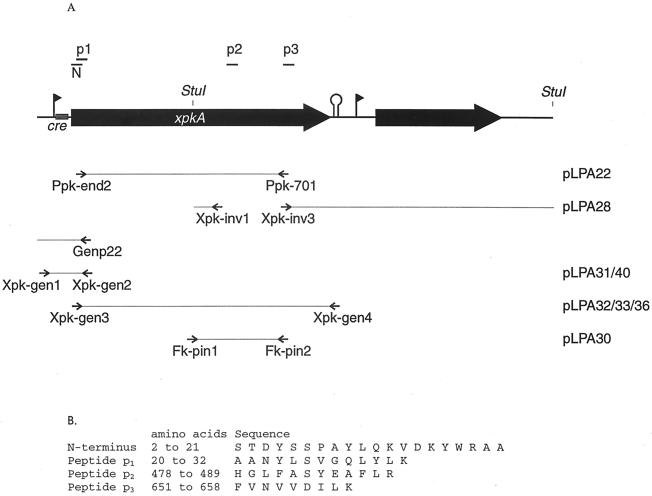
The positions of the peptides and primers used to clone and sequence xpkA are shown in Fig. 1A. Degenerate primers were designed based on the N-terminal amino acid sequence of XpkA (Ppk-end2; CGCGGATCCCARAARGTTGAYAARTAYTGG; amino acids 12 to 18; BamHI site underlined) and the sequence of one internal peptide of XpkA (Ppk-701; CCGGAATTCATRTCNACNACRTTNACRAA; amino acids 657 to 651; EcoRI site underlined) and were used in a touchdown PCR experiment (4); in this PCR experiment we started with an annealing temperature of 55°C for 5 cycles, decreased the temperature to 40°C in 15 cycles, and ended with 5 cycles at 40°C. The resulting 2-kb internal PCR fragment of xpkA was cloned in pUC18 to obtain pLPA22, which was used for sequencing. To obtain an inverse PCR fragment of the C terminus, chromosomal DNA was digested by StuI and used in a PCR with primers Xpk-inv1 (CGCGGATCCCCAAGACCCGGCCAGCAG; amino acids 456 to 451; BamHI site underlined) and Xpk-inv3 (CCGGAA TTCCGCTTGAAACGTTGGCTGC; amino acids 630 to 636; EcoRI site underlined). The amplified fragment (about 2.5 kb) was cloned in pUC18 to obtain pLPA28 and sequenced. The upstream region of xpkA was sequenced directly from chromosomal DNA by using primer Genp22 (GACCGATTGGGTGAACCTTA; amino acids 53 to 47). The complete xpkA gene was amplified twice in two independent PCR with primers Xpk-gen3 (GGAATTCCATATGTCTACAGATTACTCATCACC; amino acids 1 to 8; NdeI site underlined) and Xpk-gen4 (CGCGGATCCGCGGTTCAGTTATCTTAAATGAC; downstream of xpkA; BamHI site underlined) and cloned in pUC18 to obtain pLPA32 and pLPA33. The upstream region of xpkA was amplified twice in two independent reactions by using primers Xpk-gen1 (CCGGAATTCAGACCGTATAAGTGATCAAGTTC; upstream of xpkA; EcoRI site underlined) and Xpk-gen2 (CGCGGATCCTTGGCCGGCAATCGTGCCC; amino acids 60 to 55; BamHI site underlined), cloned in pUC18 (to obtain pLPA31 and pLPA41), and sequenced. The sequences of pLPA31, pLPA32, pLPA33, and pLPA41 were used to verify the assembled sequence. For the definite sequence, we chose nucleotides that occurred in two of three sequences.
FIG. 1.
(A) Organization of the xpkA locus of L. pentosus MD363. The positions of the peptides that were sequenced (N, p1, p2, and p3), of the primers and the plasmids that were used for cloning and sequencing of xpkA, and of the StuI restriction sites are indicated. Solid triangles indicate putative transcription start sites, and the stem-loop structure indicates a putative transcription stop site. cre, catabolite responsive element. (B) Amino acid sequences of the N terminus and the three internal peptides of XpkA from L. pentosus.
Construction of xpkA knockout mutant.
Plasmid pLPA30 (xpkA integration vector) was constructed as follows: an 850-bp internal fragment of xpkA was amplified by PCR by using primers Fk-pin1 (AAAACTGCAGAACTGGCGATGCACGGAT; amino acids 352 to 357; PstI site underlined) and Fk-pin2 (CGCGGATCCGCAGCCAACGTTTCAAGCG; amino acids 636 to 630; BamHI site underlined) and cloned in pIN15E, which contains a temperature-sensitive origin of replication. pLPA30 was used to transform L. pentosus MD363 by electroporation (14), and Emr transformants were isolated. Integration of the plasmid into the chromosome was forced at 40°C as described previously (13). Correct integration was checked by PCR by using combinations of primers Ery-atg (TAATGAACGAGAAAAATATAAAACACAG), Ery-taa2 (CTTAACTTACTTATTAAATAATTTATAGC), Xpk-gen3, and Xpk-gen4 (see above).
Expression of xpkA in E. coli.
xpkA was amplified by using Xpk-gen3 and Xpk-gen4 (see above) and was cloned between the NdeI and BamHI sites under control of the trc promoter of the E. coli expression vector pBCP367 (26), which yielded pLPA36. E. coli JA221 cells harboring pLPA36 were cultivated aerobically in 200 ml of LB broth containing ampicillin at 25°C in order to reduce the formation of inclusion bodies. Cells were induced at an A600 of 0.4 with 7.5 μM isopropyl β-d-thiogalactopyranoside (IPTG). Every 30 min, 50 ml of the culture was harvested until 2 h after induction. The harvested cells were used for preparation of cell extracts. JA221 containing pBCP367 was used as a negative control.
Purification of XpkA.
L. pentosus MD363 was cultivated in M medium containing 1% (wt/vol) xylose until the mid-log phase. Cells were harvested by centrifugation and washed once with 20 mM HEPES-HCl (pH 7). The pellet was frozen at −20°C until it was used. The entire purification procedure was performed at 4°C by using buffer B, which contained 25 mM MES (morpholineethanesulfonic acid)-NaOH (pH 6), 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, and 0.1 mM NaN3. About 30 g (wet weight) of cells was resuspended in 120 ml of buffer B. The cells were disrupted with a Dynomill (Bachofen) by using glass beads (diameter, 0.5 mm). Cell debris was removed by centrifugation for 1 h at 20,000 × g. The supernatant was loaded on a Q-Sepharose column (6 by 9 cm; Pharmacia) equilibrated with buffer B. Proteins were eluted with a 4-liter linear 0 to 0.8 M NaCl gradient in buffer B, and fractions (about 30 ml) were collected. XpkA-containing fractions (which eluted at 0.35 to 0.52 M NaCl) were identified by measuring the XpkA activity. Ammonium sulfate was added to the pooled fractions at a final concentration of 30% (wt/vol). After centrifugation (13,000 × g, 1 h), the supernatant was applied to a Butyl-TSK column (5 by 22 cm; Tosohaas) that had been preequilibrated with 30% (wt/vol) ammonium sulfate. XpkA activity was eluted with a 2-liter linear 30 to 0% (wt/vol) ammonium sulfate gradient, and fractions (about 24 ml) were collected. XpkA eluted at 15.6 to 12.0% (wt/vol) ammonium sulfate. The pooled fractions were concentrated about sixfold by pressure dialysis (Amicon YM-2 membrane) to obtain about 40 ml and were applied to a Sephadex G-200 gel filtration column (5 by 90 cm; Pharmacia), which was eluted with 2 liters of buffer B, and fractions (about 24 ml) were collected. The fractions containing XpkA activity were pooled and applied to a DEAE-TSK column (2.2 by 16 cm; Merck) equilibrated with buffer B. XpkA was eluted with a 700-ml linear 0 to 0.5 M NaCl gradient, and fractions (about 6 ml) were collected. Pooled fractions (which eluted at 0.22 to 0.23 M NaCl) were applied to a MonoQ column (HR5/5 fast protein liquid chromatography; Pharmacia Biotech) equilibrated with buffer B as the final purification step. A linear 0 to 0.5 M NaCl gradient was applied, and fractions (about 0.5 ml) were collected. XpkA activity eluted at 0.36 to 0.45 M NaCl. Aliquots of the purified protein were electrophoretically separated from contaminating impurities by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto nitrocellulose for sequencing of the N-terminal amino acids, and used for preparation of internal peptides by trypsin digestion; the internal peptides were also sequenced.
Protein analysis procedures.
The amino acid sequences of purified XpkA and of trypsin fragments were determined by automatic Edman degradation with an Applied Biosystems pulsed liquid phase sequencer (model 477A). Antibodies against purified XpkA were raised in rabbits by conventional procedures. Western blot analysis was performed by using standard procedures.
XpkA activity assay.
XpkA activity was determined by a coupled assay which monitored NADP-dependent oxidation of glyceraldehyde 3-phosphate formed from X5P in a two-step procedure, essentially as described by Goldberg et al. (method A) (7). XpkA activities are expressed below in nanomoles per minute per milligram of protein. The decrease in absorption at 340 nm was linearly dependent on the protein concentration.
Nucleotide sequence accession number.
The nucleotide sequence of xpkA has been deposited in the EMBL/GenBank database under accession number AJ309011.
RESULTS
Purification and amino acid sequencing of phosphoketolase.
To determine the structure of xylulose 5-phosphate phosphoketolase from L. pentosus and to study the regulation of expression of the gene coding for the enzyme, we attempted to clone the gene using PCR probes derived from conserved regions of various transketolases. Since all these attempts failed, we purified phosphoketolase to apparent homogeneity in order to obtain a partial amino acid sequence of the protein which could be used to clone the encoding gene. To do this, L. pentosus MD363 was cultivated in the presence of xylose, and phosphoketolase was purified from cell extracts by column chromatography in several steps. The eluted fractions were assayed for enzymatic activity (Table 2) and were analyzed by SDS-PAGE (data not shown). Since the phosphoketolase activity in the fractions from the MonoQ column corresponded with the intensity of a dominant 90-kDa band on SDS-PAGE gels (data not shown), the 90-kDa protein was assumed to be phosphoketolase. At the end of the purification procedure, phosphoketolase activity was purified ∼40-fold. Since phosphoketolase in partially and highly purified fractions was stable, these results suggest that this enzyme accounts for between 2 and 3% of the total cell protein. The N-terminal sequences of purified phosphoketolase and of three trypsin peptides were determined (Fig. 1B). The N-terminal sequence and the sequence of peptide p3 were used to design two degenerate primers, Ppk-end2 and Ppk-701, respectively.
TABLE 2.
Purification of XpkA from L. pentosus MD363a
| Prepn | Total activity (μmol/min) | Sp act (nmol/min/mg) | Yield (%) | Enrichment (fold) |
|---|---|---|---|---|
| Cell extract | 133 | 106 | 100 | 1 |
| Q-Sepharose | 60 | 35 | 45 | 0.3 |
| Butyl-TSK | 9b | 174b | 7b | 1.6b |
| Sephadex G-200 | 17 | 798 | 13 | 7.6 |
| DEAE-TSK | 2 | 1,277 | 1.5 | 12.1 |
| MonoQ | 0.06 | 4,455 | 0.5 | 42.2 |
The total activity, specific activity, yield, and enrichment factor were determined for different fractions during the purification procedure. The results obtained for the pooled XpkA-containing fractions by using Q-Sepharose (anion-exchange), butyl-TSK (hydrophobic interaction), Sephadex G-200 (gel filtration), and MonoQ (anion-exchange) procedures are shown. Only the fraction with the highest specific activity that eluted from the DEAE (anion-exchange) column was loaded on the MonoQ column in this purification procedure.
The activity was underestimated because of the presence of (NH4)2SO4.
Cloning and sequencing of xpkA, the gene encoding phosphoketolase.
Using appropriate primers in PCR and inverse PCR, we obtained two DNA fragments from L. pentosus MD363. When fused, these fragments contained two open reading frames (ORF) with the same polarity. Sequence analysis revealed that the deduced N-terminal amino acid sequence encoded by one of these ORF, ORF1, is the same as the deduced N-terminal amino acid sequence of the purified protein, except for the initial Met, which is probably processed. Also, the three internal peptides whose amino acid sequences were determined were found in the deduced amino acid sequence, and the sequence of one of these peptides appeared to overlap the N-terminal amino acid sequence (Fig. 1B). These results indicated that ORF1 encodes the purified protein, and this ORF was designated xpkA. The region upstream of xpkA was sequenced directly from chromosomal DNA. A physical map of the xpkA locus and the cloning strategy used are shown in Fig. 1A.
xpkA consists of 2,364 bp and encodes a 788-amino-acid protein with a calculated Mr of 88,705, which is very similar to the Mr of the purified protein estimated by SDS-PAGE. A putative promoter (−35 [TTGATT] and −10 [TAGAAT]) was identified 70 nucleotides before the ATG start codon of xpkA, and a Shine-Dalgarno element was identified 10 nucleotides before the ATG start codon of xpkA. A fully conserved cre element (10) was found 6 nucleotides downstream of the −10 element of xpkA. An inverted repeat 21 nucleotides downstream of the stop codon of xpkA could represent a transcription terminator. About 420 nucleotides downstream of xpkA, ORF2 starts; this ORF encodes a 384-amino-acid protein. A database search with the deduced amino acid sequence encoded by this ORF revealed that about 25% of the residues were identical to residues of sugar transferases.
A database search showed that XpkA is very similar to an F6P/X5P-specific phosphoketolase from B. lactis (18). Significant levels of similarity (up to 54% identical residues) were also observed with several ORF present in evolutionarily distantly related organisms (Fig. 2). The highest level of similarity was with an ORF from Sinorhizobium meliloti (70% conserved amino acids). The high level of sequence conservation suggests that the ORF encode an Xpk, although phosphoketolase activity has not been described for these organisms. The similarity between XpkA and the transketolases of Saccharomyces cerevisiae and Bacillus subtilis is rather low (about 15% identical residues and 45% conserved residues) despite the fact that XpkA and transketolases have the same cofactor (thiamine pyrophosphate [TPP]) and bind X5P and glyceraldehyde 3-phosphate. Besides, transketolases contain about 680 amino acids and thus are considerably smaller than XpkA from L. pentosus.
FIG. 2.
Multiple-sequence alignment of XpkA of L. pentosus MD363 (Lpe-XpkA), Xfk from B. lactis (Bla-Xfk), and an ORF from S. meliloti (Sme-ORF). The S. meliloti ORF represents a number of ORF identified in finished and unfinished genome sequencing projects. Conserved residues involved in binding of TPP are indicated by asterisks. Identical residues are shaded.
Expression of xpkA in E. coli.
To obtain additional evidence that xpkA actually encodes the protein with XpkA activity, the gene was expressed in E. coli. xpkA was PCR amplified and cloned in expression vector pBCP367 containing an IPTG-inducible trc promoter (25), which yielded pLPA36. Induction of XpkA in E. coli JA221 containing pLPA36 or pBCP367 (empty vector) was monitored by SDS-PAGE and by measuring XpkA activity in cell extracts. A Coomassie blue-stained SDS-PAGE gel showed that a 90-kDa protein appeared in JA221 harboring pLPA36 as a function of the time after induction with IPTG. Western blotting of the same samples with antibodies raised against purified XpkA clearly showed that there was an increase in the amount of the 90-kDa protein, which was not observed with the negative control (data not shown). The XpkA activities in cell extracts from the cultures increased from 78 nmol min−1 mg−1 at 30 min to 173 nmol min−1 mg−1 at 120 min after induction with IPTG. A background activity of 20 nmol min−1 mg of protein−1 was present in JA221 containing pBCP367. These results indicate that xpkA does encode XpkA.
Regulation of XpkA synthesis.
L. pentosus MD363 was cultivated on different energy sources, and XpkA activity was measured in cell extracts (Table 3). Low levels of activity were found during growth on glucose and fructose (17 and 25 nmol min−1 mg of protein−1, respectively). In contrast, growth on sugars that are fermented via the PKP resulted in higher XpkA activities, which ranged from 157 nmol min−1 mg of protein−1 for xylose to 607 nmol min−1 mg of protein−1 for gluconate.
TABLE 3.
XpkA activities in cell extracts of L. pentosus strains grown in M + medium supplemented with sugars at a concentration of 1%
| Sugar(s) | XpkA sp act (nmol min−1 mg of protein−1) in:
|
||
|---|---|---|---|
| MD363 (wild type) | LPE4 (CcpA) | LPE5 (2DGr)b | |
| d-Glucose | 17 ± 4a | 16 ± 4 | 86 ± 23 |
| d-Fructose | 25 ± 9 | 27 ± 10 | 25 ± 7 |
| d-Gluconate | 607 ± 62 | 611 ± 226 | 328 ± 48 |
| d-Glucose + d-gluconate | 30 ± 10 | 214 ± 7 | 345 ± 12 |
| d-Xylose | 157 ± 19 | 166 ± 24 | NG |
| d-Glucose + d-xylose | 27 ± 8 | 27 ± 2 | 79 ± 11 |
| d-Ribose | 456 ± 81 | 452 ± 53 | 560 ± 14 |
| d-Glucose + d-ribose | 10 ± 1 | 110 ± 9 | 114 ± 11 |
| l-Arabinose | 254 ± 50 | 259 ± 50 | 318 ± 35 |
Mean ± standard deviation based on the results of two independent experiments.
2DGr, deoxyglucose resistant; NG, no growth.
The low levels of XpkA activity during growth on glucose or fructose could have been the result of the absence of an inducing sugar and/or the result of CCR. XpkA activities were also measured in cell extracts of two mutants of L. pentosus, ccpA knockout mutant LPE4 and mutant LPE5, which lacks mannose PTS activity (2). The XpkA activities of LPE4 were comparable to those of the wild-type strain, irrespective of the sugar used (Table 3). In mutant LPE5, the XpkA activities were comparable to those of the wild-type strain when the organisms were cultivated on fructose, gluconate, or one of the pentoses. However, the XpkA activity of LPE5 on glucose was fivefold greater than the XpkA activity of the wild-type strain (Table 3).
After growth of the wild-type strain on mixtures of sugars, the XpkA activities were comparable to those after growth on glucose as the sole energy source. For the two mutants, however, growth on a combination of glucose and gluconate or a combination of glucose and ribose resulted in elevated XpkA activities. Growth of both mutants on a mixture of glucose and xylose resulted in levels of XpkA activity that were the same as the levels observed after growth of the mutants on glucose alone.
The results of the activity measurements were confirmed by a Western blot experiment in which XpkA-specific antibodies were used. No antibody reaction was seen after growth of wild-type or mutant bacteria on glucose or fructose, while a strong signal was observed after growth on sugars fermented via the PKP. After growth on a combination of gluconate and glucose a strong signal was observed for either LPE4 or LPE5 but not for wild-type bacteria (data not shown).
xpkA knockout mutant.
Additional experiments were performed to determine whether sugars can be metabolized in L. pentosus by a route different from the PKP. To study the effect of deletion of XpkA activity on growth on various energy sources, an xpkA knockout mutant of L. pentosus was constructed, as described in Materials and Methods. The knockout mutant was designated LPE179. Correct integration of the vector, pLPA30, in xpkA was verified by PCR (data not shown).
Growth of LPE179 was tested on plates supplemented with several energy sources. Integration of pLPA30 into xpkA resulted in an inability to grow on d-gluconate, d-xylose, d-ribose, and l-arabinose, whereas growth on glucose or fructose was not affected. XpkA activity was measured in LPE179 cell extracts after growth on glucose and on a mixture of glucose and gluconate. No XpkA activity was detected in either extract (less than 0.5 nmol min−1 mg of protein−1). Our data indicate that XpkA is essential for fermentation of pentoses and gluconate via this pathway. Apparently, there is no other route for metabolism of these compounds in Lactobacillus.
DISCUSSION
Xylulose 5-phosphate phosphoketolase is the key enzyme of the PKP. This enzyme connects the upper (oxidative) part of the PKP with the lower part of the glycolytic pathway through the formation of glyceraldehyde 3-phosphate from X5P, enabling the metabolism of a variety of pentoses and pentitols and, under aerobic growth conditions, the metabolism of glucose in heterolactic acid bacteria. XpkA differs from transketolase, which is used by many other bacterial species (e.g., E. coli) for conversion of X5P. XpkA utilizes inorganic phosphate instead of a phosphorylated aldose as an acceptor during cleavage of the ketol linkage. In this paper we describe for the first time cloning and nucleotide sequence analysis of a phosphoketolase gene from a heterolactic acid bacterium. In addition, our data show that expression of the phosphoketolase gene is subject to different regulatory mechanisms.
Xylulose 5-phosphate phosphoketolase (XpkA) was purified to apparent homogeneity, and xpkA, the gene encoding this protein in L. pentosus MD363, was cloned and characterized. Based on a sequence analysis, a molecular mass of 89 kDa was calculated, which was in close agreement with the molecular mass of the purified protein determined by SDS-PAGE. Since the molecular mass of phosphoketolase from the closely related species Lactobacillus plantarum was shown to be 550 kDa (10), XpkA is probably a hexamer. The same size and subunit structure were observed for the F6P/X5P-specific phosphoketolase from B. lactis (18).
The XpkA activity in wild-type strain L. pentosus MD363 was high when cells were cultivated in the presence of gluconate, ribose, xylose, or arabinose, but cells cultivated on glucose or fructose showed very low activities. In the presence of glucose and gluconate, xylose, or ribose, there was no XpkA activity or the level of activity was very low in wild-type bacteria, while elevated levels of activity were found for the CcpA mutant strain (except in the presence of xylose [see below]). Apparently, induction of XpkA activity is dependent on substrates that are fermented via the PKP. It is unlikely that these compounds have separate routes for induction of XpkA activity, and we therefore believe that the common intermediate, X5P, might be the inducer. Since no regulatory protein for expression of xpkA has been identified, the exact mechanism by which induction of xpkA takes place remains to be elucidated.
The presence of a cre element in the promoter region of xpkA suggested that expression of xpkA is subject to catabolite repression. Indeed, repression of xpkA expression was partially relieved in the ccpA knockout mutant LPE4 compared to wild-type bacteria when the organisms were grown in a mixture of glucose and gluconate or ribose. No relief of CCR was found when the bacteria were cultivated in a mixture of xylose and glucose. Our findings indicated that xylulose kinase activity is absent under these conditions (unpublished data), suggesting that conversion of xylose into X5P does not take place in LPE4. This could explain why no induction of XpkA activity was observed in this mutant during growth on a mixture of glucose and xylose.
The observation that XpkA activity in LPE4 was three- to fourfold lower during growth on a combination of sugars than during growth on the inducing sugar as the sole energy source suggests that another mechanism of CCR might be active in addition to the mechanism involving CcpA. Previous observations indicated that the mannose PTS of L. pentosus also plays a role in CCR of catabolic enzymes, such as β-galactosidase and β-glucuronidase (1). Indeed, CCR of xpkA expression was relieved in LPE5 during growth on a mixture of ribose or gluconate and glucose, indicating that the mannose PTS is also involved in CCR of xpkA.
In the presence of glucose, XpkA activity was fourfold higher in LPE5 than in wild-type bacteria. Glucose is predominantly transported by the mannose PTS in L. pentosus (1). Since mannose PTS activity is not present in LPE5, glucose must be transported by an alternative pathway, probably by a glucose permease, after which glucose is phosphorylated by glucokinase. Growth of LPE5 on glucose might result in a lower internal glucose 6-phosphate concentration than that in wild-type bacteria, possibly leading to a shift from the glycolytic pathway to the PKP. This would result in formation of the putative inducer X5P and thus in increased XpkA activity.
After cultivation on a mixture of glucose and xylose, the XpkA activity of LPE5 was comparable to the activity found during growth on glucose as the sole energy source. In L. pentosus, xylose is transported by the mannose PTS without concomitant phosphorylation. Thus, mutant LPE5 is not able to transport xylose and to grow on xylose as a sole energy source (2). As a consequence, an insufficient quantity of inducer might be formed, explaining why cultivation of LPE5 on glucose and xylose does not lead to higher levels of XpkA activity compared to the levels when glucose is the sole energy source.
Besides XpkA (10), several enzymes require TPP as a cofactor; some of these enzymes are transketolase, pyruvate oxidase, and pyruvate decarboxylase. A general TPP-binding fold was identified based on a comparison of the crystal structures of transketolase from S. cerevisiae, pyruvate oxidase from L. plantarum, and pyruvate decarboxylase from S. cerevisiae (19). The overall levels of similarity among transketolase, pyruvate oxidase, and pyruvate decarboxylase are low, but several amino acids that were identified as amino acids involved in binding of TPP appeared to be conserved (19). Alignment of transketolase from S. cerevisiae with XpkA and the XpkA-like proteins showed that nine of the residues (Gly68, His69, Gly116, Gly156, Asp157, Gly158, Asn187, Val217, and Glu418; the numbers refer to positions in transketolase) are fully conserved in these proteins (Fig. 2). Two other residues involved in TPP binding are partially conserved; these are a Glu residue in XpkA and XpkA-like proteins that is found at position 202 and an Ile residue at position 244 of transketolase. Only the Leu177 residue is not conserved in XpkA and the XpkA-like proteins. Four of the conserved residues in XpkA and XpkA-like proteins, Gly156, Asp157, Gly158, and Asn187, are part of the common TTP-binding motif that was detected by aligning TPP-requiring enzymes (9). This result strongly suggests that TPP is bound in a similar manner in XpkA.
Acknowledgments
We thank J. W. Back and A. O. Muijsers for performing the amino acid sequence analysis of XpkA and M. Hendrix for preparing antibodies.
REFERENCES
- 1.Chaillou, S., P. W. Postma, and P. H. Pouwels. 2001. Contribution of the phosphoenolpyruvate:mannose phosphotransferase system to carbon catabolite repression in Lactobacillus pentosus. Microbiology 147:671-679. [DOI] [PubMed] [Google Scholar]
- 2.Chaillou, S., P. H. Pouwels, and P. W. Postma. 1999. Transport of d-xylose in Lactobacillus pentosus, Lactobacillus casei, and Lactobacillus plantarum: evidence for a mechanism of facilitated diffusion via the phosphoenolpyruvate:mannose phosphotransferase system. J. Bacteriol. 181:4768-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:217-237. [Google Scholar]
- 4.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dossonnet, V., V. Monedero, M. Zagorec, A. Galinier, G. Perez-Martinez, and J. Deutscher. 2000. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J. Bacteriol. 182:2582-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans, C. T., and C. Ratledge. 1984. Induction of xylulose-5-phosphate phosphoketolase in a variety of yeasts grown on d-xylose: the key to efficient xylose metabolism. Arch. Microbiol. 139:48-52. [Google Scholar]
- 7.Goldberg, M., J. M. Fessenden, and E. Racker. 1966. Phosphoketolase. Methods Enzymol. 9:515-520. [Google Scholar]
- 8.Greenley, D. E., and D. W. Smith. 1979. A novel pathway of glucose catabolism in Thiobacillus novellus. Arch. Microbiol. 122:257-261. [Google Scholar]
- 9.Hawkins, C. F., A. Borges, and R. N. Perham. 1989. A common structural motif in thiamine pyrophosphate-binding enzymes. FEBS Lett. 255:77-82. [DOI] [PubMed] [Google Scholar]
- 10.Heath, E. C., J. Hurwitz, B. L. Horecker, and A. Ginsburg. 1958. Pentose fermentation by Lactobacillus plantarum. I. The cleavage of xylulose-5-phosphate by phosphoketolase. J. Biol. Chem. 231:1009-1029. [PubMed] [Google Scholar]
- 11.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15:395-401. [DOI] [PubMed] [Google Scholar]
- 12.Hurwitz, J. 1958. Pentose phosphate cleavage by Leuconostoc mesenteroides. Biochim. Biophys. Acta 28:599-602. [DOI] [PubMed] [Google Scholar]
- 13.Lokman, B. C., M. Heerikhuisen, R. J. Leer, A. van den Broek, Y. Borsboom, S. Chaillou, P. W. Postma, and P. H. Pouwels. 1997. Regulation of expression of the Lactobacillus pentosus xylAB operon. J. Bacteriol. 179:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lokman, B. C., R. J. Leer, R. van Sorge, and P. H. Pouwels. 1994. Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol. Gen. Genet. 245:117-125. [DOI] [PubMed] [Google Scholar]
- 15.Lokman, B. C., P. van Santen, J. C. Verdoes, J. Krüse, R. J. Leer, M. Posno, and P. H. Pouwels. 1991. Organization and characterization of three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol. Gen. Genet. 230:161-169. [DOI] [PubMed] [Google Scholar]
- 16.Marounek, M., and O. Petr. 1995. Fermentation of glucose and xylose in ruminal strains of Butyrivibrio fibrisolvens. Lett. Appl. Microbiol. 21:272-276. [DOI] [PubMed] [Google Scholar]
- 17.Matheron, C., A. M. Delort, G. Gaudet, and E. Forano. 1997. Re-investigation of glucose metabolism in Fibrobacter succinogenes, using NMR spectroscopy and enzymatic assays. Evidence for pentose phosphate phosphoketolase and pyruvate formate lyase activities. Biochim. Biophys. Acta 1355:50-60. [DOI] [PubMed] [Google Scholar]
- 18.Meile, L., L. M. Rohr, T. A. Geissmann, M. Herensperger, and M. Teuber. 2001. Characterization of the d-xylulose 5-phosphate/d-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J. Bacteriol. 183:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller, Y. A., Y. Lindqvist, W. Furey, G. E. Schulz, F. Jordan, and G. Schneider. 1993. A thiamine diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure 1:95-103. [DOI] [PubMed] [Google Scholar]
- 20.Racker, E. 1962. Fructose-6-phosphate phosphoketolase from Acetobacter xylinum. Methods Enzymol. 5:276-280. [PubMed] [Google Scholar]
- 21.Ratledge, C., and J. E. Holdsworth. 1985. Properties of a pentulose-5-phosphate phosphoketolase from yeasts grown on xylose. Appl. Microbiol. Biotechnol. 22:217-221. [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Schramm, M., V. Klybas, and E. Racker. 1958. Phosphorolytic cleavage of fructose-6-phosphate by fructose-6-phosphate phosphoketolase from Acetobacter xylinum. J. Biol. Chem. 233:1283-1288. [PubMed] [Google Scholar]
- 24.Stülke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Veiga-da-Cunha, M., H. Santos, and E. Van Schaftingen. 1993. Pathway and regulation of erythritol formation in Leuconostoc oenos. J. Bacteriol. 175:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velterop, J. S., M. A. Dijkhuizen, R. van't Hof, and P. W. Postma. 1995. A versatile vector for controlled expression of genes in Escherichia coli and Salmonella typhimurium. Gene 153:63-65. [DOI] [PubMed] [Google Scholar]