Abstract
In W303-derived strains, disruption of FIG2 increased agglutinability of α cells, but not a cells, and did not alter expression of α-agglutinin, binding of 125I-labeled α-agglutinin, or mating efficiency. Fig2p overexpression led to α-cell-specific suppression of agglutinability. These results imply that Fig2p is an indirect masker of the active sites in α-agglutinin.
The FIG2 gene encodes a pheromone-induced Ser/Thr-rich cell surface protein similar to the agglutinin subunit Aga1p. Disruption of FIG2 affects mating frequency and zygote morphology and increases agglutination in an S288C-derived strain of Saccharomyces cerevisiae (3, 9, 16). The disruption also reduces mating frequency at 16°C and increases it three- to sevenfold at 30°C, with greater effect if the gene is disrupted in both mating partners.
In a sigma strain, FIG2 or AGA1 is required in cells of each mating type for efficient mating, but deletion of either alone has little effect on mating efficiency under standard conditions (4). The ability of mating mixtures to adhere to and invade agar is FIG2 dependent (4, 12).
Thus, Fig2p is a cell surface protein that can modulate cell-cell interactions in S. cerevisiae. We report here that the modulation of sexual agglutination is indirect.
Strains and methods.
Yeast strains included W303-1A (MATa ade2-1 can1-100 ura3-1 leu2-3,112 trp1-1 his3-11,15), W303-1B (isogenic MATα), and the diploid strain W303 (MATa/MATα, isogenic and homozygous for other markers; Rodney Rothstein, Columbia University). All chemicals were from commercial suppliers and were reagent grade. Sequences of all primers and details of PCR are available from the corresponding author (7). RNA protection assays (RPAs) were carried out according to published procedures (2, 13, 15). Probes for FIG2, SAG1/AGα1, and ACT1 were constructed by PCR, radiolabeled (Ambion Maxiscript kit), and purified by polyacrylamide gel electrophoresis. Agglutination and quantitative mating assays were carried out according to published procedures (8, 14).
FIG2 plasmids.
A 5-kb yeast genomic DNA fragment containing upstream sequences and the first 1 kb of the 4.8-kb open reading frame (ORF) of FIG2 was identified by hybridization of a yeast clone bank with a FIG2-specific probe, followed by sequencing (5). This fragment was subcloned into pUC18 to create p102. p102 was digested with XcmI and Eco47III to delete 34 bp within the ORF of FIG2 (nucleotides 524 to 559). YEp24 was digested with HindIII to isolate a 1.1-kb URA3 fragment. Both the digested p102 and URA3 were blunt ended with Taq polymerase and then ligated to create plasmid p103. This construct should express the first 174 (including the signal sequence, but no cell wall localization sequences) of the total 1,609 residues in Fig2p and then 15 residues encoded by a normally noncoding region of Yep24 before the first stop codon. For gene disruptions, p103 was digested with SalI to release yeast DNA from the vector, transformed into W303-1A and W303-1B, and then selected on synthetic medium lacking uracil. Strains carrying the expected insertions of URA3 were identified by genomic Southern (data not shown) and Northern analyses (Fig. 1A).
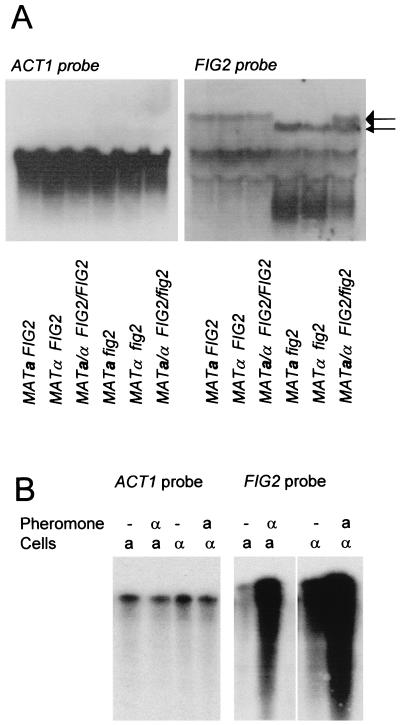
FIG. 1.
Expression of FIG2. (A) Northern blot analysis of expression of FIG2 in wild-type and gene-disrupted cells. The blot was exposed for 10 days with the FIG2-specific probe and then stripped, rehybridized with the ACT1 probe, and exposed for 40 min. The large arrow on the right shows the calculated position of FIG2 mRNA; the smaller arrow shows the position of the mRNA from the fig2::URA3 disruption allele. (B) RPA analysis of effects of pheromone treatment on FIG2 expression.
PRS423-FIG2 is a multicopy plasmid containing the YCR088(w)-FIG2 intergenic region, the entire FIG2 ORF, and 200 bp of 3′ sequence. The plasmid was constructed in a three-way ligation of the SalI-AgeI insert fragment from p102, the AgeI/NotI fragment from the PCR-generated complete ORF of FIG2, and pRS423 after digestion with SalI and NotI.
FIG2 expression.
Northern analysis and RPAs identified FIG2 transcripts in noninduced W303-1A, W303-1B, and W303 cells (Fig. 1A). Expression was approximately 3,600-fold weaker than actin expression. Pheromone treatments increased expression about 30-fold in α cells after treatment with a-factor or in a cells after treatment with α-factor (Fig. 1B).
FIG2 disruption phenotypes.
In the W303 background, disruption of FIG2 in either mating type did not affect growth kinetics or culture yield. There was little effect on mating efficiency at 30 or 16°C (1.05- to 1.3-fold enhancement) in liquid or solid medium mating protocols (8). Careful comparison of micrographs showed no detectable differences in morphologies of fig2 shmoos nor zygotes compared to those of FIG2 cells. Quantitative cell lysis tests showed no effect of disruption of FIG2 on wall structure in haploid or diploid cells, whether in the stationary or exponential growth phase (data not shown) (10, 11).
Agglutination effects of FIG2.
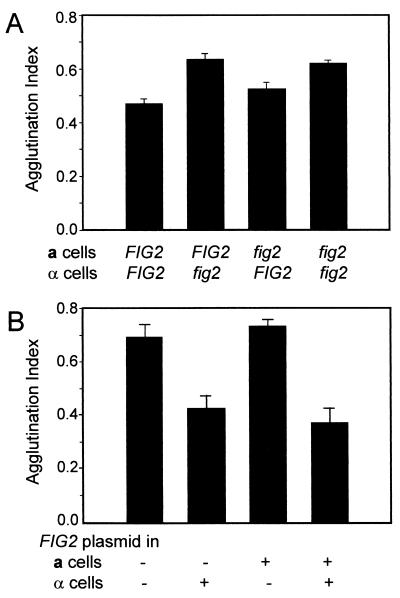
In agglutinating pairs with fig2 disruption in the α cells, the agglutination index increased by about 35%. Conversely, fig2 disruption in the a cells alone increased agglutinability by only 8%. Therefore, agglutination was potentiated by inactivating FIG2 in α cells (Fig. 2A).
FIG. 2.
Agglutination effects of FIG2. (A) Effects of disruption of FIG2 on agglutination. (B) Effects of overexpression of FIG2. fig2::URA3 cells of each mating type were transformed with the FIG2 overexpression plasmid or the vector pRS423.
When FIG2 was expressed from its own promoter in the multicopy plasmid pRS423, the agglutinability of α cells was decreased relative to the wild type, while overexpression of FIG2 in a cells produced little change in agglutination index (Fig. 2B). These results show a FIG2-mediated, α-specific attenuation of agglutinability.
Possible roles for FIG2 in agglutination.
We tested several hypotheses to explain the effect of disruption or overexpression of FIG2 on agglutination. fig2 disruptions did not affect the rate of cellular response to pheromone nor the concentrations of pheromone required to induce increased expression of agglutinins. An RPA experiment showed that the presence of the FIG2 overexpression plasmid did not alter amounts of α-agglutinin (SAG1/AGα1) transcripts in W303-1B cells treated without or with the sex pheromone a-factor (data not shown).
Fig2p might inhibit α-agglutinin activity. To test this idea, purified 125I-labeled α-agglutinin was bound to a-factor-treated α cells that had intact FIG2 or the fig2 disruption allele. The cells and the labeled material were allowed to bind for 1 h at room temperature or at 4°C. The cells were collected and washed three times with binding buffer. There was no significant difference in binding to the two cell types.
There appear to be strain-specific differences in the cellular roles of FIG2 (3, 4). In all strains tested, sex pheromones induce FIG2 by 30-fold or more (3, 4; Saccharomyces GenomeDatabase, http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl). Nevertheless, in a sigma strain, the efficiencies of mating were similar with and without Fig2p (in reference 4, see Fig. 2A, patch b and bar b), and there was no significant effect under a variety of mating conditions in W303 strains. These results contrast with those for strain Y800, in which fig2 disruptions increased mating efficiency at 30°C and decreased it at 16°C (3, 16).
Roles in agglutination.
FIG2 attenuated agglutination when the gene was expressed on cells of the α mating type. A fig2 disruption potentiated agglutination (also reported in reference 16), and FIG2 overexpression decreased agglutination. This phenotype might result if FIG2 down-regulated expression of the α-agglutinin structural gene SAG1, but no such effect was seen. Also, there was no difference in binding of 125I-α-agglutinin to α cells with intact or disrupted FIG2, a result that implies that α-agglutinin does not bind to Fig2p. The simplest model for the α-specific effect of Fig2p is based on the predicted structures of that protein and α-agglutinin. Much of Fig2p consists of a Ser/Thr-rich sequence of about 900 amino acid residues that are predicted to be highly glycosylated and therefore in an extended conformation. Such glycosylated “stalks” extend from the cell surface with an average elevation of about 2 Å per residue (1, 6). Thus, Fig2p should extend up to 1,800 Å (180 nm) from the surface, whereas the 300-residue stalk of α-agglutinin is only about 60 nm long. Therefore, a high cell surface concentration of Fig2p on α cells may sterically “overshadow” α-agglutinin. In contrast, Fig2p would not hinder access on MATa cells, because the stalk of Aga1p is of comparable length to that in Fig2p. Thus, after initial adhesion of an α cell with an a cell, newly expressed Fig2p would decrease the ability of α-agglutinin on the surface of the α cell to bind to other a cells in the vicinity, helping to ensure mating between a single pair of cells.
Acknowledgments
We thank Rafael Ovalle and Anne Dranginis for helpful discussions. We thank Jeanne Hirsch for the yeast clone bank and Fred Naider for the very kind gift of synthetic a-factor.
This work was supported by SCORE Program grant SO6-GM60654 and grant R01-GM47176 to Janet Kurjan, University of Vermont, from the National Institute of General Medical Science. Support from the Research Centers in Minority Institutions Program of NIH (RR-03037) is also appreciated.
REFERENCES
- 1.Cappellaro, C., C. Baldermann, R. Rachel, and W. Tanner. 1994. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 13:4737-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crotchfelt, K. A., B. Pare, C. Gaydos, and T. C. Quinn. 1998. Detection of Chlamydia trachomatis by the Gen-Probe AMPLIFIED Chlamydia Trachomatis Assay (AMP CT) in urine specimens from men and women and endocervical specimens from women. J. Clin. Microbiol. 36:391-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erdman, S., L. Lin, M. Malczynski, and M. Snyder. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140:461-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch, J. P., and F. R. Cross. 1993. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated G alpha protein. Genetics 135:943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jentoft, N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15:291-294. [DOI] [PubMed] [Google Scholar]
- 7.Jue, C. K. 2001. Ph.D. thesis. City University of New York, New York, N.Y.
- 8.Lipke, P. N., D. Wojciechowicz, and J. Kurjan. 1989. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 9:3155-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver, S. G., Q. J. van der Aart, M. L. Agostoni-Carbone, M. Aigle, L. Alberghina, D. Alexandraki, G. Antoine, R. Anwar, J. P. Ballesta, P. Benit et al. 1992. The complete DNA sequence of yeast chromosome III. Nature 357:38-46. [DOI] [PubMed] [Google Scholar]
- 10.Ovalle, R., S. T. Lim, B. Holder, C. K. Jue, C. W. Moore, and P. N. Lipke. 1998. A spheroplast rate assay for determination of cell wall integrity in yeast. Yeast 14:1159-1166. [DOI] [PubMed] [Google Scholar]
- 11.Ovalle, R., M. Spencer, M. Thiwanont, and P. N. Lipke. 1999. The spheroplast lysis assay for yeast in microtiter plate format. Appl. Environ. Microbiol. 65:3325-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 13.Samani, N. J., K. Morgan, W. J. Brammar, and J. D. Swales. 1987. Detection of renin messenger RNA in rat tissues: increased sensitivity using an RNAse protection technique. J. Hypertens. Suppl. 5:S19-S21. [DOI] [PubMed] [Google Scholar]
- 14.Terrance, K., and P. N. Lipke. 1981. Sexual agglutination in Saccharomyces cerevisiae. J. Bacteriol. 148:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodcock, D. M., M. R. Williamson, and J. P. Doherty. 1996. A sensitive RNase protection assay to detect transcripts from potentially functional human endogenous L1 retrotransposons. Biochem. Biophys. Res. Commun. 222:460-465. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, M., D. Bennett, and S. E. Erdman. 2002. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell 1:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]