Abstract
The unicellular green alga Chlamydomonas reinhardtii is a valuable model for studying metal metabolism in a photosynthetic background. A search of the Chlamydomonas expressed sequence tag database led to the identification of several components that form a copper-dependent iron assimilation pathway related to the high-affinity iron uptake pathway defined originally for Saccharomyces cerevisiae. They include a multicopper ferroxidase (encoded by Fox1), an iron permease (encoded by Ftr1), a copper chaperone (encoded by Atx1), and a copper-transporting ATPase. A cDNA, Fer1, encoding ferritin for iron storage also was identified. Expression analysis demonstrated that Fox1 and Ftr1 were coordinately induced by iron deficiency, as were Atx1 and Fer1, although to lesser extents. In addition, Fox1 abundance was regulated at the posttranscriptional level by copper availability. Each component exhibited sequence relationship with its yeast, mammalian, or plant counterparts to various degrees; Atx1 of C. reinhardtii is also functionally related with respect to copper chaperone and antioxidant activities. Fox1 is most highly related to the mammalian homologues hephaestin and ceruloplasmin; its occurrence and pattern of expression in Chlamydomonas indicate, for the first time, a role for copper in iron assimilation in a photosynthetic species. Nevertheless, growth of C. reinhardtii under copper- and iron-limiting conditions showed that, unlike the situation in yeast and mammals, where copper deficiency results in a secondary iron deficiency, copper-deficient Chlamydomonas cells do not exhibit symptoms of iron deficiency. We propose the existence of a copper-independent iron assimilation pathway in this organism.
While iron is abundant in the environment, it is present in the insoluble ferric [Fe(III)] state, so that its bioavailability is low (16). Yet iron is an essential micronutrient for all organisms because it functions as a cofactor in enzymes that catalyze redox reactions in fundamental metabolic processes. Iron exhibits stable, redox-interchangeable ionic states with the potential to generate less stable electron-deficient intermediates during multielectron redox reactions involving oxygen chemistry (16). Therefore, organisms are challenged with the acquisition of sufficient iron to meet cellular metabolic requirements while avoiding uncontrolled intracellular chemistry. This is accomplished via the operation of iron homeostatic mechanisms. The essential features of iron metabolism include assimilation and distribution, storage and sequestration, and utilization and allocation. The assimilatory pathway can be further subdivided into reduction of insoluble ferric species to more soluble ferrous species and uptake into the cell, followed by intracellular transport and intraorganellar distribution. The storage and sequestration of iron involve loading of cellular proteins as well as compartmentalization into organelles like vacuoles and plastids, which in turn requires proteins for transport into and out of these compartments and organellar iron binding and metabolizing proteins. Photosynthetic organisms have requirements for iron beyond those of heterotrophs because of the abundance of iron in the photosynthetic apparatus and its occurrence in many metabolic pathways within the plastid. Therefore, in a eukaryotic phototroph, there must be an additional layer of complexity in iron metabolism.
Mobilization by redox chemistry.
Saccharomyces cerevisiae uses different assimilatory pathways depending on the chemical source of iron and its concentration. Reduction of Fe3+ to Fe2+ (68, 91) is a key step in uptake, either to solubilize the ion or to release it from ferric specific chelates. Under iron-limiting conditions, high-affinity uptake is mediated by the inducible multicopper oxidase (MCO) (Fet3p)/iron permease (Ftr1p) complex at the plasma membrane (6, 91, 100), where Fet3p oxidizes Fe2+ to Fe3+ which is then delivered to Ftr1p for transport of Fe3+ into the cell (91, 100). Highly related pathways operate in other fungi, including Schizosaccharomyces pombe (7) and the fungal pathogen Candida albicans (21, 35, 56, 92). Under iron-replete conditions, low-affinity systems operate, such as Fet4p (19, 37) or Smf1/Smf2 (15). Iron can also enter the cell complexed with siderophores, mediated by the ARN family of siderophore transporters through an endocytic pathway (40, 41, 62, 63, 86, 118, 119).
In mammals, dietary Fe3+ is reduced to Fe2+ by Dcytb (69) and perhaps other as-yet-unidentified ferrireductases and transported across the apical surface of enterocytes by the divalent metal ion transporter DMT1 (DCT1, Nramp2) (2, 3, 97, 101). The basolateral transporter ferroportin1/IREG1/MTP1 (1, 20, 70) mediates the transport of iron out of enterocytes into the blood for distribution to other organs. Iron is moved through the circulatory system bound to the plasma protein transferrin. The loading of iron onto transferrin requires oxidation from Fe2+ to Fe3+, which may be mediated by either or both of the MCOs hephaestin and ceruloplasmin. Hephaestin may act together with ferroportin1 at the basolateral surface of enterocytes to oxidize Fe2+ to Fe3+ prior to export into the plasma (53). Alternatively or in addition, plasma ceruloplasmin oxidizes Fe2+ to Fe3+ subsequent to export into blood plasma for loading onto transferrin. An additional important role of ceruloplasmin is the mobilization of iron from organs like the liver where ceruloplasmin is synthesized (24). Therefore, unlike yeast, where the MCO/iron permease complex mediates iron uptake, the analogous complex(es) in mammals is involved in iron release from cells. Nevertheless, the principle of transport in conjuction with redox chemistry holds.
Iron acquisition by all plants begins with the reduction of Fe3+ from insoluble Fe3+ complexes in the soil (8, 33, 93, 115). In addition to iron reduction, iron solubility in the rhizosphere is increased through iron-deficiency-induced activation of a specific H+-ATPase (29). In dicotyledons and nongraminaceous monocotyledons, IRT1 (23, 32, 57) and members of the NRAMP family (17, 103) are responsible for iron uptake. Grasses secrete phytosiderophores, which chelate Fe3+, and the resulting complex is taken up by iron-deficiency-induced siderophore transporters represented by the prototypical member YS1 (9, 18).
Ferritin.
Ferritin has long been known as an iron storage protein in vertebrates. Accordingly, its expression is increased in cells supplied with high iron through translational regulation (38). Vertebrate ferritin is composed of multiple subunits of two types of chains, called heavy and light. Together these chains function to oxidize iron and bind up to 4,500 ferric atoms within the core of a multimeric structure. In plants, the 24-subunit ferritin multimer consists of only one type of subunit, whose sequence is most similar to that of the heavy subunit of vertebrate ferritins that contains the ferroxidase active site, although the carboxylates of the light subunit that promote and stabilize the mineralized core are also present (reviewed in reference 10). Ferritin is found in plastids and is a source of iron during plastid development (reviewed in reference 9). Preferritin, with a plastid targeting sequence, is encoded by a multigene family in plants, as is the case in animals (113). Increased ferritin production in high-iron-supplied cells is accomplished, at least in part, by transcriptional regulation of one or more ferritin genes (10, 61). This pattern of expression is consistent with a role for ferritin as an iron storage molecule under conditions of iron overload.
Role of copper in iron assimilation.
Since the MCOs Fet3p and ceruloplasmin and hephaestin in S. cerevisiae and mammals, respectively, require copper for activity, copper is an essential cofactor for high-affinity iron uptake. Also essential are copper-metabolizing components such as the copper chaperone ATX1 or ATOX1 in S. cerevisiae and mammals, respectively, that deliver copper to the copper-transporting ATPase Ccc2p or the Wilson (WND)/Menkes (MNK) proteins, respectively. The ATPases transport copper into the lumen of secretory pathway vesicles for incorporation into apo-Fet3p (66, 117) or apoceruloplasmin and apohephaestin, respectively. Therefore, in S. cerevisiae or mammals, copper deficiency leads to iron deficiency. In contrast, plants do not appear to manifest a similar dependence on copper for iron assimilation. In Arabidopsis species, both types of iron transporters, IRT1 and NRAMP, take up Fe2+, so that a role for a ferroxidase is not evident, nor has one been identified. However, ATX1 and WND/MNK homologues occur in Arabidopsis and Synechocystis species, and they carry out analogous functions (44, 45, 47, 105, 106).
Chlamydomonas model.
Significant insights into plant metal metabolism can be gained from studying other model photosynthetic organisms such as the unicellular green alga Chlamydomonas reinhardtii. With its simple growth requirements C. reinhardtii is a valuable experimental model for the study of metalloprotein biosynthesis (72, 75) and metal-responsive gene regulation in photosynthetic organisms (71). As in other organisms, iron uptake by Chlamydomonas involves reductases (22, 42, 67, 114) that are induced in iron deficiency and may be the same enzyme as that induced in copper-deficient (−Cu) cells (42). Iron uptake is also induced, although to a lesser extent than Fe3+ reduction (22). The molecular components of iron assimilation in C. reinhardtii have not as yet been identified.
In this work, we report the identification, expression, and functional characterization of Chlamydomonas homologues of a multicopper ferroxidase, an iron permease, a copper chaperone, and ferritin and propose that together these components form part of an iron assimilation pathway related to the high-affinity iron uptake pathway of S. cerevisiae. Yet, while copper-requiring components appear to be required for iron assimilation in C. reinhardtii, copper deficiency in Chlamydomonas does not lead to a secondary iron deficiency, indicating that backup systems are in place to accommodate simultaneous copper and iron deficiency.
MATERIALS AND METHODS
Strains and culture conditions.
Escherichia coli DH5α was used for cloning, maintenance, and propagation of plasmids and was cultured in Luria-Bertani medium (U.S. Biologicals) at 37°C. Ampicillin was added to a 100-μg/ml concentration where appropriate. E. coli clones containing plasmid-borne expressed sequence tags (ESTs) were obtained from the Kazusa DNA Research Institute, Chiba, Japan. S. cerevisiae strains YPH250 (wild type [wt]) (98) and SL215 (atxΔ) (66) (kind gifts from V. C. Culotta) and DBY746 (wt) and EG118 (sod1Δ) (kind gifts from J. S. Valentine and E. Gralla) were maintained on standard yeast extract-peptone-dextrose medium at 30°C (14). Tests for iron-dependent growth were carried out as described previously (100) with synthetic dextrose (SD) minimal medium plates containing 1.5 mM ferrozine [3-(2-pyridyl)-5,6-bis(4-phenylsulfonic acid)-1,2,4-triazine; Sigma] and supplemented with 350 μM ferrous ammonium sulfate where required. Agarose was used instead of agar to minimize contamination with iron. Plates were incubated at 30°C for 5 days. Tests for complementation of lysine and methionine auxotrophy of the sod1Δ mutant were carried out on SD plates lacking either lysine or methionine. Plates were incubated at 30°C for 5 days. C. reinhardtii strain CC125 was used for growth experiments and was cultured in Tris-acetate-phosphate (TAP) medium prepared with copper-free, iron-free trace elements (89) and supplemented with either Fe-EDTA prepared as described previously (79) or CuCl2 at the required concentrations where appropriate. Cell density was determined by counting cells, and the chlorophyll content was measured spectrophotometrically at 652 nm as described previously (79).
Chemicals and reagents.
Enzymes were from New England Biolabs, and vectors were from Stratagene. Chemicals were purchased from Fisher, Sigma, and Gibco BRL. DNA purification kits were from Qiagen. Oligonucleotide primers were synthesized by Genosys or Gibco BRL.
DNA manipulation and sequence analysis.
Standard procedures were used for plasmid DNA isolation from E. coli and for DNA cloning and manipulation (95).
Amplification.
Oligonucleotide primers used in amplification reactions were derived from ESTs (Table 1). For preparation of first-strand cDNA, C. reinhardtii total RNA was used as the template in a reaction mixture containing random hexamers or oligo-(dT)20 (Gibco BRL), 10 mM deoxynucleoside triphosphates, 0.1 M dithiothreitol, Moloney murine leukemia virus reverse transcriptase (Gibco BRL), 5× reverse transcription buffer, and RNasin RNase inhibitor (Promega). For amplification of specific sequences from the C. reinhardtii cDNA template, the reaction mixture contained 1.25 μM concentrations of the appropriate primers, 0.2 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 10× PCR buffer (Promega), and Taq polymerase purified as described previously (25). The same conditions were used with primers 5′-gcccgcggccgcTGTAGTGCATTTTTGAGCTTC-3′ and 5′-gcccgcggccgcCTTTTTTCTTCGTAAGCATCC-3′ to amplify S. cerevisiae ATX1 from genomic DNA. Thermal cycling conditions for Atx1, Fox1, and the copper ATPase cDNAs were as follows: 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min (4 times); 94°C for 1 min, 55°C for 45 s, and 72°C for 1 min (26 times); and 72°C for 10 min (1 time). For Fer1 and Ftr1 the conditions were the same except that annealing temperatures of 45 and 55°C, respectively, were used. PCR products were purified with Qiaquick columns (Qiagen) and cloned into the BamHI site of pBluescript SK II(+) (Stratagene), pGEM-T Easy (Promega) in the case of the copper ATPase, or the NotI site of pFL61 (78) in the case of C. reinhhardtii Atx1 and S. cerevisiae ATX1. All plasmids are available from the Chlamydomonas culture collection under the names pFox1-594, pFox1-852, pFox1-TrxA, pFtr1-358, pAtx1-410, pFer1-399, and pCcc2-229.
TABLE 1.
Oligonucleotides used for PCR amplification of gene-specific fragments
| Gene | Upstream primer (5′-3′)c | Downstream primer (5′-3′)c | Inserta | Accession no., source, or reference |
|---|---|---|---|---|
| Fox1 | cgggatcc946GGGCAGACAGTCACCTACCT965 | cgggatcc1539GTCTCCGGTGACGGTGTAGA1520 | 594 bp, 946 to 1539 | AV395796, AV394462 |
| Fox1 (5-RACE) | (CUA)4GGCCACGCGTCGACTAGTAC | cggaattc853GGTTGTTCTTGAACACCACGCGAAT829 | 852 bp, 1 to 852 | Gibco BRL (UAPb), AF450137 |
| Fox1 (TrxA-Fox1 fusion) | cgggatccccactg1476CCACTGCCACGTTAACTTCCACAT1500 | cgggatcc2212CCACCACGGTAAAGAAGATGACAAT2237 | 761 bp, 1476 to 2237 | Fig. 6 |
| Ftr1 | cgggatcc131GTCGTTCTGGCGATAGAGTTACC153 | cgggatcc488AACAGTTTGCTTTGAGCTGTGTAG465 | 358 bp, 131 to 488 | AV395492 |
| Atx1 | gcccgcggccgc17CTTGCTTGCGAACAGTATCCA37 | gcccgcggccgc426GTGTCACCCACGAGGTTTACC406 | 410 bp, 17 to 426 | AV388156 |
| Fer1 | cgggatcc451TGCTCATGGAGTACCAGAACC471 | cgggatcc849ATCTCAACCAACCGAGCTGT830 | 399 bp, 451 to 849 | AV395232 |
| Cu ATPase | cgcggatcc221GTTGGCAACCGGCTGCTGCTGAT240 | tgctctaga449AGTTGTCGTCCGTGAGCATC430 | 229 bp, 221 to 449 | BE761354 |
Inserts were cloned into the BamHI site of pBluescript SK II(+) or pTrxFus, the NotI site of pFL61, or pGEM-T Easy. The numbers correspond to sequences shown in Fig. 2, 3, 6, and 9. For the Cu ATPase, the numbers correspond to EST accession BE761354.
UAP, universal amplification primer.
Additional sequences added to introduce restriction enzyme sites for cloning are lowercased; gene-specific sequence is uppercased.
5′-RACE.
To amplify the 5′ end of the Fox1 mRNA, the 5′-rapid amplification of cDNA ends (RACE) system from Gibco BRL was used. Gene-specific primer 1 (5′-GCCACAGGCGGCTGGTCACAGTGGCGTT-3′), designed to correspond to positions 1027 to 1000 of the cDNA sequence shown in Fig. 6, was used to prime first-strand cDNA synthesis from total RNA. Gene-specific primer 2, corresponding to positions 853 to 829 and containing an EcoRI site at the 5′ end to facilitate cloning (Table 1), was designed for amplification of the desired fragment from the cDNA with the universal amplification primer from the kit. The reactions were carried out according to the manufacturer's instructions. A diffuse band of approximately 8 × 102 bp was purified and cloned into the SalI (5′ end of PCR product) and EcoRI (3′ end of PCR product) sites of pBluescript KS II(+) (Stratagene), and DNA from individual clones was sequenced with the T7 primer to determine if the clones overlapped with the 5′ end of clone CL48f10 corresponding to EST accession no. AV395796. The longest overlapping clone, designated Fox1-RACE9 (∼8 × 102 bp), was sequenced completely on both strands by Qiagen genomics and assembled with the sequence of the cDNA insert in clone CL48f10 to generate the sequence shown in Fig. 6.
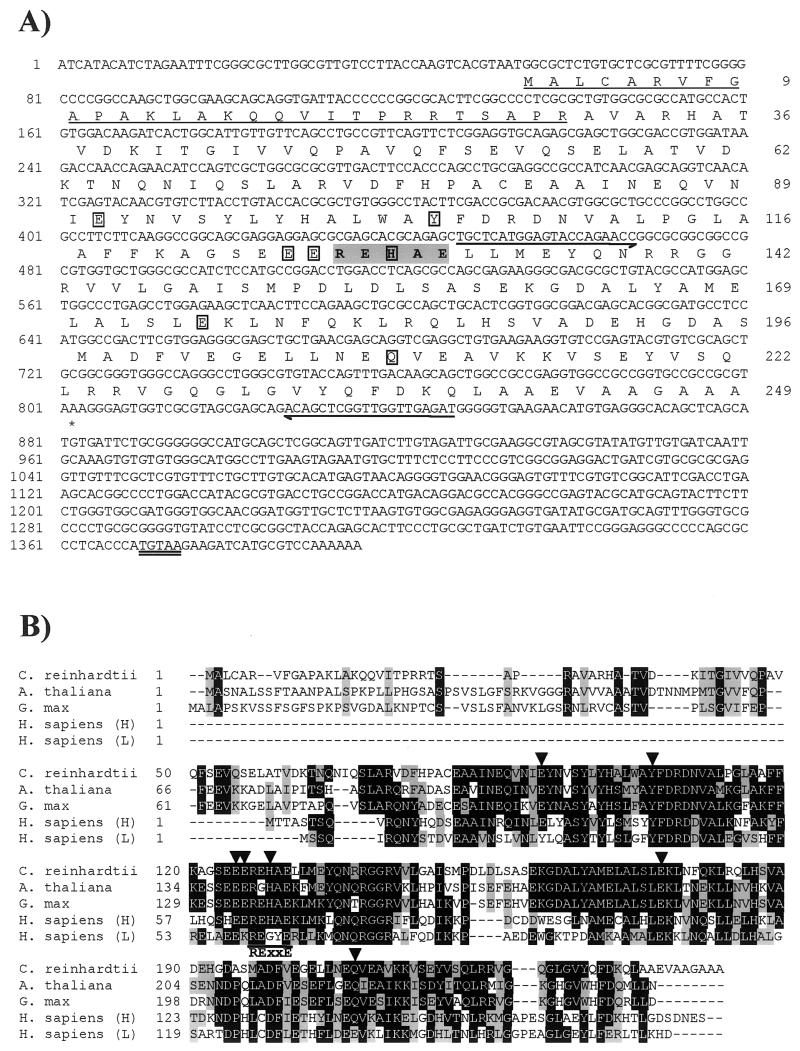
FIG. 6.
Sequence analysis of Fox1. The nucleotide sequence shown is derived from the 5′-RACE product (positions 1 to 852) and the insert in clone CL48f10 corresponding to EST AV395796 (Kazusa DNA Research Institute) (positions 787 to 4908). The numbers on the left refer to the nucleotide sequence, which is numbered +1 from the first nucleotide of the GenBank entry. The deduced amino acid sequence of the longest ORF is given below the nucleotide sequence. The numbers on the right indicate the positions of the amino acids in the reading frame. The first methionine is numbered +1. The first 40 amino acids are shown in lowercase since the relationship of Fox1 with other MCOs suggests that the second methionine residue is likely to represent the initiator methionine. The putative N-terminal signal peptide is underlined, and the C-terminal transmembrane region is boxed. The polyadenylation signal is double underlined. Gray shading denotes sequences corresponding to MCO signature 1, while MCO signature 2 is given in boldface. The His-Xaa-His motifs are given in boldface and underlined. Half-arrows denote the sequences of primers used for 5′-RACE and PCR.
Sequence analysis.
The cloned inserts were sequenced by using primers derived from the multiple cloning site of pBluescript (M13 reverse, 5′-CCCAGTCACGACGTTGTAAAACG-3′, and M13 forward, 5′-AGCGGATAACAATTTCACACAG-3′) and gene-specific internal primers that were designed from known sequences. Reactions for automated sequencing were carried out with the BigDye Terminator reaction mix (Perkin-Elmer Applied Biosystems), and unincorporated dye terminators were removed by gel filtration cartridges (Edge BioSystems). Reaction mixtures were analyzed on an automated sequencer (ABI Prism 377 DNA sequencer). Raw sequence data were analyzed with Sequence Analysis 2.0.1 for Macintosh, and contigs were assembled with Autoassembler 1.4.0 for Macintosh (Perkin-Elmer Applied Biosystems). The inserts in clone CM017g07 (Kazusa DNA Research Institute) corresponding to the Atx1 cDNA, clone CL42d10 (Kazusa DNA Research Institute) corresponding to the Ftr1 cDNA, and clones Fox1-RACE9-852 and CL48f10 (Kazusa DNA Research Institute) corresponding to positions 1 to 852 and 787 to 4908 of the Fox1 cDNA, respectively, were sequenced by Qiagen Genomics. Database searches were carried out with the BLAST software on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/). Protein alignments were carried out with ClustalW (http://www.ebi.ac.uk/clustalw/). Protein sequences were analyzed with BioEdit software (34), Prosite (13, 48), SignalP V1.1 (80), TMpred (49), and TopPred2 (110).
RNA blot analysis.
C. reinhardtii total RNA was isolated and analyzed by hybridization as described previously (43). Five micrograms of RNA was loaded per lane. Gene-specific probe fragments were isolated after BamHI or NotI digestion of the cloned amplification products described above (Table 1), purified with Qiaquick columns (Qiagen), and labeled with [α-32P]dCTP by random priming. The probes for Fox1, Atx1, and Ftr1 recognize a single band in Southern hybridization analysis of digested genomic DNA. The probe used to detect Cox17 transcripts was generated by NotI digestion of clone CM080h02 (Kazusa DNA Research Institute; GenBank accesssion no. AV392030). The probe used to monitor RbcS2 encoding the small subunit of ribulose-bisphosphate carboxylase-oxygenase or the Cβlp gene (for normalization between samples) was described previously (90, 96). Specific activities of probes ranged from 3 × 108 to 6 × 108 cpm/μg of DNA. Hybridization signals were detected by exposure of membranes to Biomax MS film (Eastman-Kodak Co.) at −80°C with two intensifying screens and developed typically after overnight exposure. Hybridization signals from the same blots were quantitated with a Molecular Dynamics PhosphorImager and Image QuaNT (version 4.2a) software (Sunnyvale, Calif.).
Yeast complementation experiments.
PCR products encoding C. reinhardtii Atx1 and S. cerevisiae Atx1p were cloned into the NotI site of the yeast expression vector pFL61 (78) under the control of the PGK1 promoter. Plasmid DNA was transformed into the appropriate strain of S. cerevisiae (30). Transformants were selected on SD plates lacking uracil. Several independent transformants were analyzed for each experiment.
Preparation of Fox1 antiserum.
A thioredoxin (TrxA)-Fox1 fusion protein was generated for the production of Fox1 antiserum. A PCR with Fox1-specific primers (Table 1) resulted in the amplification of a 761-bp product (nucleotides 1476 to 2237; amino acids His394 to Val646) that was digested with BamHI, cloned in frame into the 3′ terminus of the TrxA-encoding sequence of the expression vector pTrxFus, and introduced into E. coli for tryptophan-inducible expression (Invitrogen). The majority (approximately 90%) of the expressed TrxA-Fox1 fusion protein was present in inclusion bodies. E. coli cells expressing the fusion protein were cultured and harvested as described in the manufacturer's protocols. Briefly, flasks containing 100 ml of prewarmed induction medium were inoculated with 2 ml of an overnight culture of the TrxA-Fox1-expressing strain, grown to an A600 of 0.7 (30°C, 200 rpm), and then induced (with tryptophan) for 4 h at 37°C. Cultures were chilled in ice-water; collected by centrifugation (4,300 × g, 15 min); washed once with a solution of cold 10 mM Tris-Cl (pH 8.0), 1 mM EDTA (pH 8.0), and 100 mM NaCl (TEN); resuspended in 10 ml of cold 50 mM Tris HCl (pH 7.5) containing 5 mM EDTA; subjected to three quick-freeze (dry ice ethanol)-quick-thaw (37°C) cycles; and disrupted by sonication (Fisher Scientific model 550 Sonic Dismembrator; microtip probe, amplitude setting 4, 12 cycles of 30 s of sonication followed by 60 s of cooling). Samples were kept cold by immersion in an ice-water bath. The inclusion bodies were collected by centrifugation (5 min at 14,000 × g, 4°C), washed three times with cold TEN, solubilized with sample buffer (50 mM Tris-HCl [pH 6.8], 5% [vol/vol] 2-mercaptoethanol, 2% [wt/vol] sodium dodecyl sulfate [SDS], 0.1% [wt/vol] bromophenol blue, 10% [wt/vol] glycerol), and subjected to preparative SDS-polyacrylamide gel electrophoresis (12% acrylamide). The region of the gel containing the fusion protein (visualized by zinc-imidazole staining) was excised and used directly for antiserum production in rabbits (service provided by Covance Research Products, Denver, Pa.).
Immunoblot analysis.
Immunoblotting was performed as described previously (79). Chlamydomonas cells (107 cells ml−1 in 200 ml) were collected by centrifugation (3,800 × g, 5 min), washed once with 0.01 M sodium phosphate (pH 7.0), and resuspended in the same buffer such that samples were matched for number of cells per unit volume. For Fox1 immunoblotting, aliquots of 50 μl were subjected to three cycles of freezing to −80°C and thawing to 4°C and centrifuged (5 min at 14,000 × g, 4°C). The supernatant was discarded, and the pellets were washed once with 100 μl of ice-cold phosphate-buffered saline before resuspension in 50 μl of 0.01 M sodium phosphate. For analysis of Fox1 expression in cells grown under various iron concentrations, washed and resuspended cells were analyzed for CF1 by immunoblotting and densitometric quantitation of the signals for the α and β subunits. Sample loadings for the Fox1 immunoblot were then adjusted such that samples were matched for CF1 content. The abundance of CF1 in C. reinhardtii is not affected by iron nutritional status.
Nucleotide sequence accession number.
The newly published sequences and their GenBank accession numbers are as follows: Fox1, AF450137; Ftr1, AF478411; Atx1, AY120936 and AF280056; Fer1, AF503338; and Cox17, AF280543.
RESULTS
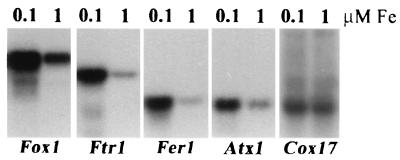
To identify components of iron assimilation pathways in C. reinhardtii, the EST database was searched by BLAST for candidate homologues of the proteins that are known to be involved in iron or copper metabolism in fungi, plants, or animals. Sequences corresponding to ferritin and copper chaperones (Atx1 and Cox17) were identified with excellent probability scores (AV395232, 1e−31 for soybean ferritin input; AV388156, 5e−15 for Arabidopsis CCH input; and AV392030, 4e−15 for human Cox17 input). A candidate Ftr1-like sequence with a weak probability score (0.004 for S. cerevisiae Ftr1p input) but containing a conserved RExxE motif was identified (accession no. AV395492), as were several sequences corresponding to the copper binding sites of MCOs (input sequence, laccase or ascorbate oxidase) or the ATP binding sites of P-type ATPases (input sequence, Ccc2p, MNK, or WND). The genes corresponding to these sequences were eventually named as follows: Fer1 for ferritin; Atx1, Cox17, Ftr1, and Ccc2 on the basis of a sequence relationship to the equivalent yeast proteins; and Fox1 for ferroxidase. The relevance of candidate sequences to iron metabolism was tested by RNA blot analysis (Fig. 1). RNA was prepared from cells grown in TAP medium containing either 0.1 μM supplemental iron, under which condition they were evidently iron deficient as assessed by chlorophyll accumulation (i.e., Fe-deficiency chlorosis), or 1 μM supplemental iron, under which condition chlorosis was not evident. Cox17 mRNA was not affected by medium iron concentrations, but Fer1, Atx1, Ftr1, and Fox1 RNAs were each induced severalfold (Fig. 1) relative to total RNA and were, therefore, of interest for further analysis.
FIG. 1.
Increased accumulation of RNAs encoding iron metabolism components in iron-deficient Chlamydomonas cells. C. reinhardtii cells from a late-log culture in copper-free TAP medium were harvested and resuspended in 90 ml of −Cu TAP with 0.1 μM iron chelate. One milliliter was used to inoculate 100-ml cultures of +Cu TAP with either 0.1 μM iron chelate (cells severely chlorotic) or 1 μM iron chelate (chlorophyll content relatively unaffected compared to that of iron-replete cells). Cultures were grown to late log phase and transferred, and this process was repeated twice more to adapt cells to 1 or 0.1 μM iron chelate. Total RNA was prepared from late-log-phase cultures after three rounds and analyzed by hybridization with gene-specific probes as indicated. For quantitation, the signals were normalized to total RNA loaded. Specifically, the relative intensities (10−5) after object average background correction were as follows: Fox1, 25.2 and 8.7; Ftr1, 5.2 and 1.2; Fer1, 3.7 and 0.9; Atx1, 1.2 and 0.5; Cox17, 0.05 and 0.05. For RbcS2, the signal was actually decreased in 0.1 μM iron samples. The relative intensities (10−5) were 20.6 and 44.8. Transcript sizes were as follows: Fox1, 5.2 kb; Ftr1, 2.8 kb; Fer1, 1.3 kb; and Atx1, 1.0 kb. They were estimated from a standard curve of the relative mobility of each marker (Gibco BRL; 0.24- to 9.5-kb RNA marker) versus log10 of its size in bases.
Ferritin (Fer1).
The cDNA clone (LC007f05) corresponding to EST AV619384 was obtained from the Kazusa DNA Research Institute. The 1.4-kb cDNA insert was sequenced completely (AF503338) (Fig. 2A). A 750-bp open reading frame (ORF) encoding a predicted polypeptide of 249 amino acids was identified and was flanked by 52 bp of 5′ untranslated region (UTR) and 593 bp of 3′ UTR with a putative polyadenylation signal (TGTAA) located 15 nucleotides upstream of the poly(A) tail. The corresponding gene was designated Fer1. The putative polypeptide encoded by the ORF identified within the assembled sequence showed a high level of similarity to mammalian and plant ferritins (Fig. 2B), which increased slightly if only the mature subunits were considered (28 and 47% amino acid identity, respectively). All of the plant ferritins showed greater similarity with the human heavy chain subunits than with the light chain subunit. Not surprisingly, the C. reinhardtii mature sequence showed less similarity with either of the Arabidopsis thaliana and Glycine max sequences (47%) than the similarity that exists between these latter two sequences (72%). In addition, there was less similarity between the C. reinhardtii sequence and the human ferritin subunits than between the other plant sequences and the human subunits. The N terminus had features that are consistent with the presence of a chloroplast transit peptide (12), in agreement with the ChloroP software, which predicted its targeting to the plastid. The C. reinhardtii sequence also appeared to possess an extension peptide, a plant-specific sequence thought to be involved in stabilizing the protein in vitro (59, 109). This peptide, from approximately amino acid position 30 to 69, is located between the predicted transit peptide cleavage site and the region that shows a high level of conservation with both plant and human sequences, and it is less conserved in the C. reinhardtii sequence than in the other plant sequences.
FIG.2.
Sequence analysis of Fer1. (A) Nucleotide and amino acid sequences of Fer1. The nucleotide sequence of 1.4 kb of the Fer1 cDNA is shown. The sequence was determined for clone LC007f05 (Kazusa DNA Research Institute) by Qiagen Genomics. The numbers on the left refer to the nucleotide sequence, which is numbered +1 from the first nucleotide of the assembled sequence. The deduced amino acid sequence of the longest ORF is given below the nucleotide sequence. The numbers on the right indicate the positions of the amino acids in the reading frame. The putative transit peptide is underlined. Boxes denote conserved amino acids required for ferroxidase activity. The iron-binding RExxE motif is given in boldface and shaded gray. Half-arrows denote the sequences of primers used for PCR. The putative polyadenylation signal is double underlined. (B) Amino acid sequence alignment of C. reinhardtii Fer1 with ferritins from other organisms. The alignment was generated by using the ClustalW algorithm and BioEdit software (34). Residues that are similar or identical in a majority of sequences are shaded gray and black, respectively. The conserved amino acids required for ferroxidase activity are indicated by arrowheads above the alignment. The conserved RExxE motif is indicated by a line below the alignment. GenBank accession numbers: A. thaliana, AF229850; G. max, U31648; H. sapiens ferritin light chain, P02792; H. sapiens ferritin heavy chain, XP_043419.
Fer1 contained the highly conserved iron-binding motif RExxE found in other ferritins and, like other plant ferritins, contained the conserved amino acids required for ferroxidase activity (Fig. 2). Note that His77 (C. reinhardtii numbering) is conserved between the C. reinhardtii and human sequences but is replaced by Ala in the plant sequences, where it was shown previously to have a role in the ferroxidase acitivity of the plant proteins (109). We conclude that Chlamydomonas contains at least one plastid-localized ferritin.
The abundance of Fer1 transcripts increases in iron deficiency, up to 10-fold, rather than under conditions of iron excess (up to 200 μM as Fe-EDTA tested [data not shown]), suggesting that the plastid Fer1 gene product may be important for iron buffering (see Discussion). Nevertheless, the protein abundance was not changed noticeably in −Fe relative to +Fe cells. It is well known that ferritin abundance is controlled by translational mechanisms in animals (104), and such mechanisms may operate also in C. reinhardtii. A search of the Chlamydomonas dbEST (with Arabidopsis Fer1 as input) database reveals more than a dozen ESTs (July 2002), all of which appear to arise from a single gene. Comparisons between the sequenced clone LC007f05 and the ESTs also suggest that all the Fer1 sequences represent a single gene or highly related gene sequences.
Iron permease (Ftr1).
The cDNA clone (CL42d10) corresponding to EST AV395492 was obtained from the Kazusa DNA Research Institute. The 2.9-kb cDNA insert, probably corresponding to the full-length mRNA based on comparison with the size of the mRNA transcript (Fig. 1), was sequenced completely (GenBank accession no. AF478411), and found to contain an ORF encoding 541 amino acids flanked by 264 bp of the 5′ UTR and 1,059 bp of the 3′ UTR with a canonical polyadenylation signal, TGTAA, at the expected position relative to the poly(A) tail (Fig. 3). A BLAST search of the nonredundant sequence database with the complete Ftr1 reading frame revealed its relationship to S. cerevisiae, S. pombe, and Candida albicans Ftr1p and Ftr1p homologues, each of which has a demonstrated role in iron metabolism (7, 92, 100), with scores ranging from 1e−12 to 3e−08. All other sequences showing weak similarities and carrying RExxE motifs, including Synechocystis sp. strain PCC 6803 (BAA16870, ORF slr0964, 0.001), were of prokaryotic origin. A search of the dbEST database revealed several sequences in Physcomitrella patens (moss) that contained two RExxE motifs (expected value, 10−9 to 10−8). These could represent a plant Ftr1 homologue. A multiple alignment of C. reinhardtii Ftr1 with S. cerevisiae Ftr1p and Fth1p, S. pombe Fip1p, Candida albicans CaFtr1 and CaFtr2, and Synechocystis sp. strain PCC 6803 ORF slr0964 is shown in Fig. 4. Although the C. reinhardtii and Synechocystis sequences display less than 20% amino acid identity overall, the latter is included in the alignment because of the relevance of cyanobacterial metabolism to chloroplast biology. Like the fungal sequences, C. reinhardtii Ftr1 is predicted to have a cleavable N-terminal leader sequence (SignalP V1.1 [80]). Six transmembrane domains at positions that correspond to those shown previously for S. cerevisiae Ftr1p are also predicted (58). Two RExxE motifs were present within all sequences: the first one, within the putative N-terminal leader sequence, is less conserved; the other, within the hypothesized third transmembrane domain, is more conserved. In all of the fungal sequences, this motif is REGLE, whereas in the C. reinhardtii and Synechocystis sequences, the L was replaced by I and F, respectively. ExxE has been identified as a potential iron-binding motif in Ftr1p and CaFTR1 and CaFTR2 (92, 100); two such motifs are found in the C-terminal region. Although the predicted C. reinhardtii transmembrane regions corresponded closely with those predicted for the S. cerevisiae Ftr1p sequence, there was a region of 151 amino acids between transmembrane domains 2 and 3 that was unique to C. reinhardtii. This region was hydrophobic and rich in proline residues and perhaps may represent a highly folded structure with a hydrophobic interior.
FIG. 3.
Analsyis of the Ftr1 cDNA. The nucleotide sequence of the 2.9-kb cDNA from clone CL42d10 corresponding to EST AV395492 (Kazusa DNA Research Institute) is shown with the deduced amino acid sequence of the longest ORF given below the nucleotide sequence. The numbers on the left refer to the nucleotide sequence, which is numbered +1 from the first nucleotide of the GenBank entry. The numbers on the right indicate the positions of the amino acids in the reading frame. The polyadenylation signal is double underlined. The putative N-terminal signal peptide is underlined. Boxes denote sequences corresponding to putative transmembrane regions. The iron-binding RExxE motifs are given in boldface and shaded gray. The ExxE motifs are double underlined. Half-arrows denote the sequences of primers used for PCR.
FIG. 4.
Alignment of C. reinhardtii Ftr1 with Ftr1 homologues from other organisms. The alignment was generated by using the ClustalW algorithm and BioEdit software (34). Residues that are similar or identical in a majority (four) of sequences are shaded gray and black, respectively. A line above the alignment indicates the conserved RExxE motifs. GenBank accession numbers: C. reinhardtii Ftr1, AF478411; Synechocystis, BAA16870, S. cerevisiae Ftr1p, NP_011072; S. cerevisiae Fth1p, AF177330; S. pombe Fip1, AF177330; C. albicans CaFTR1, AF195775; and C. albicans CaFTR2, AF195776.
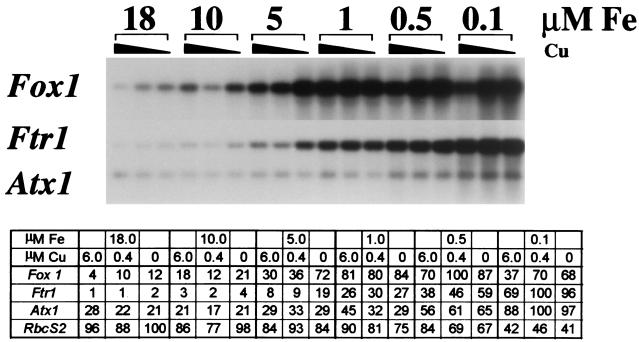
RNA blot analysis had indicated that Ftr1 mRNA accumulated to higher levels in cells displaying iron-deficiency chlorosis than in cells not displaying iron-deficiency chlorosis (Fig. 1). To investigate the pattern of iron-dependent expression of Ftr1, total RNA from C. reinhardtii grown in medium supplemented with various amounts of copper and iron (from 0 to 6 μM Cu and from 0.1 to 18 μM Fe) was analyzed by hybridization. Ftr1 mRNA accumulation increased steadily as the medium iron concentration decreased from 18 μM (concentration in standard TAP medium [36]) to 0.1 μM (concentration where cell growth was inhibited and chlorosis was evident [see below and Fig. 12]) (Fig. 5). The magnitude of Ftr1 induction relative to total RNA between the maximum and minimum medium iron concentrations tested was at least 102-fold. The effect of copper deficiency, at most a twofold increase in copper-deficient cells (no added copper) over that in copper-supplemented cells (6 μM supplemental copper), was not consistent, nor did it display a reproducible pattern with respect to copper concentration (Fig. 5). The extent of copper deficiency of the culture was verified routinely by analyzing the expression of the Cyc6 gene (89). When medium iron concentration was increased about 10-fold to 200 μM, Ftr1 mRNA abundance was decreased further (data not shown). Based on the overall sequence relationship between C. reinhardtii Ftr1 and Ftr1 homologues, the presence of two conserved RExxE motifs, a predicted membrane topology similar to that of S. cerevisiae Ftr1p, and its iron-regulated expression, we conclude that Ftr1 encodes an iron permease with a function related to S. cerevisiae FTR1. Hence, the name Ftr1, for ferric transporter, was adopted. The C. reinhardtii dbEST database contains only five clones (six ESTs) representing this Ftr1 gene, and these appear to be the only Ftr1-related sequences in the dbEST database (July 2002).
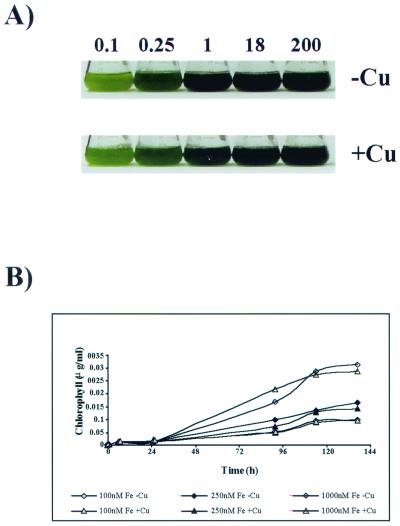
FIG. 12.
Effect of iron concentration on growth of C. reinhardtii. (A) C. reinhardtii was grown to exponential phase in TAP medium and then subcultured into TAP medium containing the indicated iron supplement and either 6 μM copper (+Cu) or no copper (−Cu), and growth was monitored over a period of 6 days. (B) C. reinhardtii was cultured in TAP medium containing iron concentrations that ranged from 100 nM (−Fe) to 1 μM (+Fe) and either 0 μM (−Cu) or 6 μM copper (+Cu). The cell number of each culture was monitored over a period of 6 days, and chlorophyll content as a measure of iron sufficiency was monitored over the same time period by removing 10 μl of the culture into 1.0 ml of 80% acetone-20% methanol. All measurements were carried out in duplicate.
FIG. 5.
Increased abundance of Fox1, Ftr1, and Atx1 transcipts as medium iron is reduced. A 100-ml culture of C. reinhardtii was adapted to low iron (0.1 μM in copper-free TAP medium) at late log phase and was used to inoculate cultures with the indicated iron concentrations and copper to either 6 μM (normal copper-supplemented TAP medium), 0.4 μM (saturating for Cyc6 repression and plastocyanin biosynthesis [76]), or 0 μM. RNA was harvested the following day when the cultures were at mid-log phase (2 × 106 to 3 × 106 cells ml−1) and analyzed by RNA hybridization. Parallel samples were probed for RbcS2 expression for quantitation of the data (tabulated). The relative intensities were obtained after object average background correction. The values in the table represent the signal for each sample relative to the maximum intensity for each probe, which was arbitrarily set at 100.
Ferroxidase (Fox1).
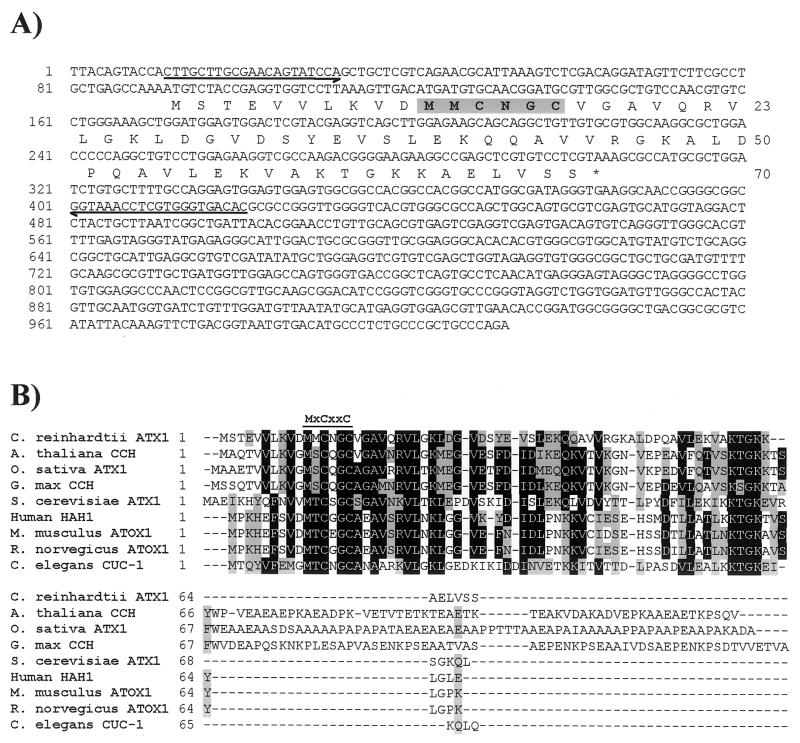
In the fungi, Ftr1p and its homologues function in obligate partnership with an MCO with ferroxidase activity, Fet3p and its homologues (7, 21, 100). Together the two proteins accomplish high-affinity, highly selective transport of iron versus other metals (6). A search of the Chlamydomonas EST database (4) with Fet3p did not reveal any candidate sequences of interest, but when other members of the MCO family such as laccase and ascorbate oxidase were used as the input sequence, several ESTs (accession no. AV395796, AV394462, and AV394010) showing significant similarity (1e−13) to human ceruloplasmin, a serum MCO, were identified. The cDNA clone (CL48f10) corresponding to EST AV395796 was obtained (Kazusa DNA Research Institute), and the 4.1-kb cDNA insert was sequenced completely to reveal that the accessions probably represented a single Fox1 sequence. Based on the size of the Fox1 transcript (Fig. 1) and alignment with mammalian ceruloplasmins and hephaestin, we concluded that the cDNA in clone CL48f10 lacked approximately 1 kb at the 5′ end including the initiator methionine. The missing region was cloned by 5′-RACE (see Materials and Methods), sequenced, and assembled to generate a sequence of 4,908 nucleotides (GenBank accession no. AF450137), including an ORF of 3,429 bases flanked by 297 bases of 5′ UTR and 1,182 bases of 3′ UTR (Fig. 6). A long poly(A) tail with a canonical TGTAA polyadenylation signal was identified within the clone. The predicted ORF encoded a protein of 1,142 amino acids. However, based on signal peptide predictions and the relationship of Fox1 with other MCOs, the second methionine residue at position 41 is likely to represent the initiator methionine, yielding an encoded protein of 1,102 amino acids.
A putative N-terminal signal peptide was identified (80), which if cleaved around position 59 would yield a mature protein with a mass of 117 × 103 Da. A potential C-terminal transmembrane domain was predicted by the algorithm of Kyte and Doolittle (58) and also by TMpred (49) and TopPred2 (110). Five potential N-glycosylation sites also were identified. A BLAST search of the nonredundant database with the entire ORF validated its identification as a likely ferroxidase, because the sequence displayed a relationship (27% identity and 40% overall similarity) to the mammalian MCOs hephaestin and ceruloplasmin with high probability, e−131 and e−115, respectively, and to other MCOs such as ascorbate oxidase and laccase, or to blood coagulation factor VIII, with weaker scores. The length of the polypeptide was also most similar to those of mammalian ceruloplasmins and hephaestin, which are 1,048 to 1,119 amino acids in length, rather than to laccases and ascorbate oxidases, which are approximately half that length. The gene was designated Fox1. Over 20 entries corresponding to this Fox1 sequence were found in the C. reinhardtii dbEST database (July 2002). Another MCO was not evident in the dbEST database, and Southern analysis results are consistent with a single-copy gene.
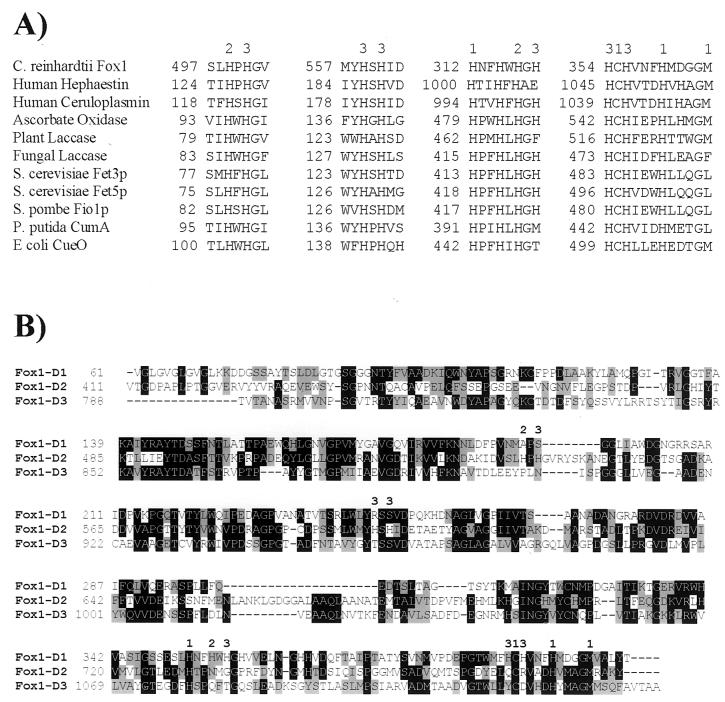
Prosite analysis revealed the presence of three sequences that corresponded to MCO signature 1 and one sequence that corresponded to MCO signature 2 (Fig. 6). These patterns were derived from a region within ascorbate oxidase, laccase, ceruloplasmin, and Pseudomonas syringae CopA that contains five residues known to be involved in the binding of copper. Although the Fox1 sequence aligned poorly with the other sequences, all of the ligands that form the type I copper binding sites in ceruloplasmin (His, Cys, His, and Met) were conserved in the predicted Fox1 protein. In addition, there were several His-Xaa-His motifs within Fox1 which, by comparison with the other MCO sequences, may represent the ligands that form the type II and type III copper binding sites (Fig. 7A). However, the position of these putative copper ligands within the C. reinhardtii Fox1 sequence was different from their relative locations within the other sequences.
FIG. 7.
Domain-like structure of the Fox1 product with candidate copper binding ligands. The potential ligands for type I, type II, and type III copper binding sites are designated 1, 2, and 3, respectively. (A) Alignment of amino acid sequences of the putative copper binding sites in Fox1 with those of other MCOs. GenBank accession numbers: C. reinhardtii Fox1, AF450137; human hephaestin, AF148860; human ceruloplasmin, XM011006; ascorbate oxidase, A51027; plant laccase, U12757; fungal laccase, 17943174; S. cerevisiae Fet3p, P38993; S. cerevisiae Fet5p, P38993; S. pombe Fio1p, CAA91955; P. putida CumA, CAA91955; and E. coli CueO (yacK), P36649. (B) Alignment of Fox1 domains. The putative ligands for type I, II, and III copper binding sites within each domain are shown above the alignment. Each domain contained the four ligands (His, Cys, His, and Met) that characterize a type I Cu binding site. The numbers on the left indicate the amino acid position within the sequence.
The amino acid sequence of human ceruloplasmin can be divided into three contiguous similar units of approximately 350 residues (77). If the predicted Fox1 mature sequence was similarly divided into units of approximately 360 residues and the units were aligned with each other, a high degree of sequence identity was revealed (26 to 29%) (Fig. 7B). Some regions were very highly conserved among the three domain-like units. Each putative domain contained the four copper ligands (His, Cys, His, and Met) that form the type I Cu binding site, and the position and spacing of these ligands were perfectly conserved within each domain (Fig. 7B). We concluded that Fox1 is likely to represent an MCO that is capable of binding at least three type I, one type II, and one type III copper ion.
Regulation of ferroxidase abundance by iron.
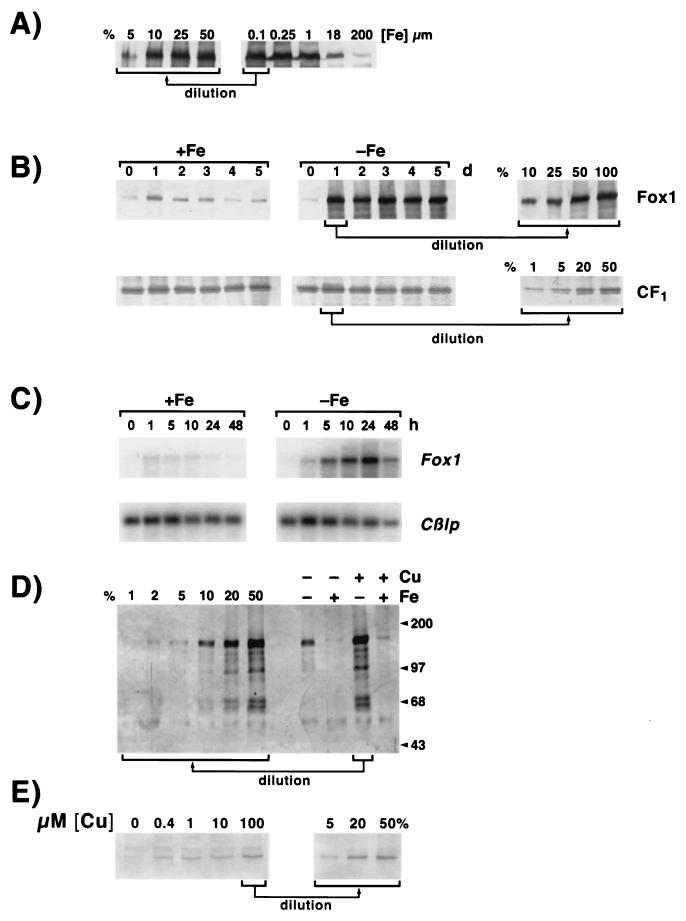
To address the hypothesis that Fox1 encodes an MCO involved in iron metabolism, we examined Fox1 mRNA as a function of iron nutrition (Fig. 5). As expected for a component of iron assimilation, Fox1 mRNA accumulation is increased as iron in the medium is decreased. The pattern of expression appears to be coordinate with that for Ftr1, with most of the mRNA increase evident at a 1 μM medium iron concentration when the physiological symptoms of iron deficiency are not yet strongly evident. The extent of regulation depends strongly on the cell density. At 2 × 106 to 3 × 106 cells/ml (Fig. 5), there is a 10- to 20-fold increase (relative to total RNA loaded) in the abundance of Fox1 mRNA at low iron versus 18 μM iron in the medium. At a higher cell density, the relative difference can be as high as 1 × 102- to 4 × 102-fold (data not shown). As for Ftr1 regulation, copper did not have a significant effect on Fox1 mRNA abundance, a two- to threefold change at most and without a reproducible pattern. The coordinate iron-dependent expression of Ftr1 and Fox1 is consistent with the hypothesis that a permease-oxidase complex is involved in iron assimilation in Chlamydomonas.
Antibodies raised against one of the three domains of Fox1 (see Materials and Methods) were used to monitor the location and abundance of Fox1 in Chlamydomonas. Immunoblot analysis identified a prominent signal corresponding to migration at 138 kDa, which compares well with the migration of ceruloplasmin at 132 kDa (77) (Fig. 8).
FIG. 8.
Fox1 abundance in iron-deficient Chlamydomonas. Protein extracts from C. reinhardtii cells grown in TAP medium containing various concentrations of copper and iron, and collected at a density of 107 cells ml−1, were prepared as described in Materials and Methods. Extracts were analyzed after separation by denaturing gel electrophoresis (7.5% acrylamide), transfer to nitrocellulose (1 h, 100 V, in 25 mM Tris-192 mM glycine-0.04% [wt/vol] SDS-20% [vol/vol] methanol), and incubation with anti-Fox1 antiserum (1:300 dilution). Bound antibody was visualized colorimetrically after incubation with alkaline phosphatase-conjugated secondary antibody. (A) The Fox1 gene product accumulates in −Fe cells. Cells grown in medium containing 200 μM added Fe-EDTA were transferred to fresh medium supplemented with 0.1, 0.25, 1, 18, or 200 μM Fe-EDTA and sampled after 5 days of growth. Protein extracts from 2.5 × 106 cells were analyzed as described above. Samples were normalized for loading on the basis of equal cell numbers and verified for accumulation of CF1, which is iron independent (see Materials and Methods). Percentages on the left are the fractional amounts of the “0.1 μM Fe” sample that were loaded. (B) Time course of Fox1 induction in −Fe cells. Cells grown with 200 μM added Fe-EDTA were transferred to fresh medium lacking iron (−Fe) or containing 200 μM added Fe-EDTA (+Fe). Cultures were sampled each day for 5 days (d), and ferroxidase abundance was analyzed as described above. Percentages shown on the right arethe amounts of the day 1 sample that were loaded. The accumulation of the α and β subunits of CF1 is shown as a loading control. (C) Time course of Fox1 mRNA accumulation upon transfer from Fe-replete (200 μM) to Fe-deficent (0 μM) medium. Cultures were sampled at the indicated time after transfer for RNA isolations. The RNAs were analyzed by blot hybridization. The Cβlp mRNA was used as an internal control. Its behavior as a function of cell growth and iron nutrition is typical of many other RNAs. (D) Cu is required for accumulation of Fox1. C. reinhardtii was cultured in copper-supplemented (6 μM, +Cu) or copper-deficient (no added copper, −Cu), iron-supplemented (18 μM, +Fe) or iron-deficient (no added iron, −Fe) TAP medium. One microliter of protein extract from 4 × 108 cells was analyzed as described above. Percentages on the left are the fraction of the −Fe-+Cu sample that was applied for quantitation. Molecular masses of markers (Gibco BRL) are indicated in kilodaltons. (E) Cu-dependent accumulation of Fox1 in iron-replete cells. Copper-deficient, iron-replete (18 μM) CC125 cells were transferred to fresh (0 μM supplemental iron) medium containing the indicated amounts of copper and sampled for immunoblot analysis of Fox1 accumulation after 3 days. Each lane was loaded with material from 2 × 107 cells (equivalent to approximately 5 μg of chlorophyll). Percentages on the right are the fraction of the sample from medium containing 100 μM supplemental copper.
The discrepancy between the apparent and the predicted sizes of Fox1 may be due to glycosylation, as is the case with other MCOs (26, 77, 81). Based on its presence in the pellet rather than the soluble fraction, we conclude that Fox1 is probably membrane bound. The signals with higher mobility correspond possibly to degradation products because their presence is completely correlated with the intensity of the 138-kDa signal. To test whether the protein is induced in iron-deficient cells, cells cultured in media containing 200 μM added Fe-EDTA were transferred to fresh media supplemented with 0.1, 0.25, 1, 18, or 200 μM Fe-EDTA; sampled after 5 days of growth; and examined by immunoblotting for Fox1 accumulation. Fox1 accumulation as a function of medium iron nutrition clearly mirrors the pattern noted for its mRNA (Fig. 8A). Fox1 abundance was substantially increased as the medium iron concentration was reduced from 18 μM (normal TAP medium) and was already increased maximally in medium containing 1 μM iron. When the normal iron supplement was increased about 10-fold to 200 μM, the amount of Fox1 was reduced. To monitor the kinetics of the response to iron deficiency, cells grown in medium containing 200 μM iron were transferred to fresh-iron-supplemented (200 μM) or iron-depleted (0 μM) medium and sampled each day for 5 days. The response to iron depletion was rapid. Fox1 abundance was maximal within 24 h (Fig. 8B) even though cell division had not occurred during this time (data not shown). The change in RNA, although transient, was even more rapid and was noticeable at 5 h (Fig. 8C). The rapidity of the response suggests either that the cells can measure external iron or that an internal signaling pool is depleted rapidly when the external iron supply is reduced.
Copper-dependent accumulation of the ferroxidase.
Since copper is an essential cofactor for the ferroxidase, we wondered whether copper nutritional status might affect its accumulation, by analogy with the effect of copper status on plastocyanin accumulation in Chlamydomonas cells (73). The copper dependency of ferroxidase accumulation was tested in both iron-deficient and iron-supplemented media to assess whether the outcome was influenced by biosynthetic demand for copper. Regardless of iron nutritional status and the extent of Fox1 mRNA increase, ferroxidase accumulation was strongly dependent on copper availability (Fig. 8D). The amount in −Cu iron-deficient cells was only 10% of that in +Cu iron-deficient cells. The effect was noted even for iron-supplemented cells that have a smaller demand for biosynthetic copper because of the lower level of Fox1 expression (Fig. 8E). We conclude that copper does not affect the iron-responsive regulation of Fox1 but is required instead for accumulation of the protein under both iron-deficient and -sufficient conditions. This effect is executed at the level of protein accumulation as evident from the finding that copper deficiency does not reduce the abundance of Fox1 transcripts. As noted above (Fig. 5), any effect of copper is minimal and, in any case, occurs in the opposite direction of the change in protein abundance.
Other components of the iron assimilation pathway.
The involvement of an MCO in iron assimilation prompted us to search for copper-metabolizing components analogous to Atx1p and Ccc2p of S. cerevisiae (66, 116). S. cerevisiae Atx1p is the metallochaperone required for delivery of copper to apo-Fet3p via Ccc2p-dependent transport of copper in a post-Golgi vesicle (66, 116). This copper-requiring step is essential for the complete maturation of the Fet3p/Ftr1p complex and its passage to the plasma membrane (100). Therefore, loss of Atx1p function in S. cerevisiae results in loss of high-affinity iron uptake and failure to grow on iron-deficient medium.
Copper chaperone (Atx1).
Oligonucleotide primers were designed and used in reverse transcription-PCR to amplify a 411-bp fragment encoding C. reinhardtii Atx1. The amplified product was cloned into the NotI site of the multicopy yeast expression vector pFL61, three independent clones were sequenced to validate the accuracy of the amplification, and the confirmed sequence for Atx1 was deposited under accession no. AF280056. Subsequently, clone CM017g07 (Kazusa DNA Research Institute), containing also the 5′ and 3′ UTRs, was also obtained and sequenced, and the sequence was deposited under accession no. AY120936 (Fig. 9A). A poly(A) tail and polyadenylation signal were not identified within the sequenced insert. The dbEST database contains about a dozen accessions representing this Atx1 gene. The predicted C. reinhardtii Atx1 amino acid sequence was aligned with that of S. cerevisiae Atx1p and homologues from A. thaliana (CCH), rice (Oryza sativa ATX1), soybean (G. max CCH), yeast (S. cerevisiae ATX1), human (Homo sapiens HAH1), mouse (Mus musculus Atox1), rat (Rattus norvegicus Atox1), and Caenorhabditis elegans (CUC-1) (Fig. 9B). An extended C terminus of approximately 50 to 60 amino acids was present in homologues from the three photosynthetic species employed for comparison in the alignment (Arabidopsis, rice, and soybean) but not in the C. reinhardtii Atx1, which was comparable in length (70 amino acids) with the yeast, mammalian, and C. elegans homologues (68 to 73 amino acids). C. reinhardtii Atx1 shared the greatest sequence identity with S. cerevisiae ATX1 (36%) and less but still significant identity with the other sequences (29 to 32%). Overall similarity between Atx1 and the other sequences ranged from 38 to 40%. All Atx1p homologues contained the highly conserved motif MxCxxC (where x is any amino acid), which has been shown elsewhere to bind copper (88). This motif, with a glycine residue immediately preceding the methionine, is found also in the N-terminal domain of the copper-transporting ATPases, often in multiple copies. Among the Atx1 homologues, A. thaliana CCH, rice ATX1, soybean CCH, and C. elegans CUC-1 all possess the glycine at this position, whereas the motif in C. reinhardtii Atx1 is more similar to the mammalian homologues, which have an aspartic acid residue in this position in place of glycine. The conserved C-terminal lysine-rich region also was present in the C. reinhardtii sequence, although the arrangement of the lysines was most similar to the homologues from the photosynthetic species (KTGKK) rather than to those from the nonphotosynthetic ones (KKTGK). Southern analysis reveals a single hybridizing fragment in the genome of Chlamydomonas.
FIG. 9.
Analysis of the Atx1 cDNA. (A) The nucleotide sequence of the 1.0-kb Atx1 cDNA is shown. The sequence was determined for clone CM017g07 (Kazusa DNA Research Institute) by Qiagen Genomics (this study; GenBank accession no. AY120936). A poly(A) tail was not present within the sequenced insert from clone CM017g07 but was identified within EST BE441651 at a position corresponding to 1007 of the sequence shown. The numbers on the left refer to the nucleotide sequence, which is numbered +1 from the first nucleotide of ESTs AV631680, AV638507, and BE441652. The deduced amino acid sequence of the longest ORF is given below the nucleotide sequence. The numbers on the right indicate the positions of the amino acids in the reading frame. The putative copper binding motif is shown in boldface and gray shading. Half-arrows denote the sequences of primers used for PCR. (B) Amino acid sequence alignment of C. reinhardtii Atx1 with homologues from other organisms. The alignment was generated by using the ClustalW algorithm and BioEdit software (34). Residues that are similar or identical in a majority (five) of sequences are shaded gray and black, respectively. The conserved MxCxxC motif is indicated by a line above the alignment. GenBank accession numbers: C. reinhardtii Atx1, AF280056; A. thaliana CCH, U88711; O. sativa ATX1, AF198626; G. max CCH, T50778; S. cerevisiae ATX1, T50778; H. sapiens HAH1, U70660; M. musculus ATOX1, AF004591; R. norvegicus ATOX1, NM_053359; C. elegans CUC-1, AB017201.
Copper delivery.
A role for C. reinhardtii Atx1 in copper delivery to a copper-containing ferroxidase is supported by RNA blot analysis, which indicates an approximately fivefold increase in Atx1 mRNA accumulation relative to total RNA in cells exhibiting symptoms of iron deficiency (0.1 μM supplemental iron) compared to iron-replete cells (18 μM). Medium copper content, on the other hand, did not affect Atx1 mRNA abundance (Fig. 5).
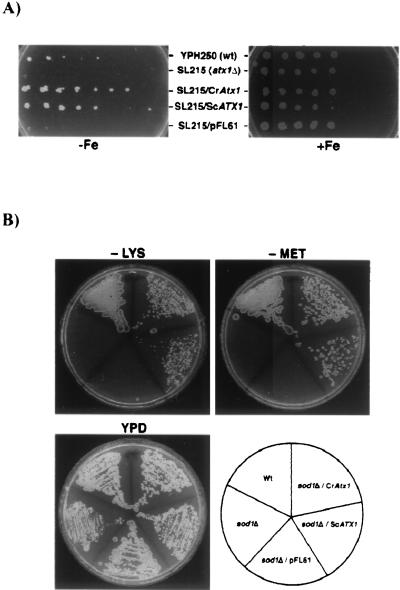
C. reinhardtii Atx1 function in copper delivery was also tested by functional complementation of an S. cerevisiae atx1Δ strain. Atx1p is essential for high-affinity iron uptake in S. cerevisiae. Accordingly, a strain with the ATX1 gene deleted (atx1Δ, SL215) is unable to grow on iron-depleted medium (66). CCH (Arabidopsis), HAH1 (human), and CUC-1 (C. elegans) have each been shown to restore the growth of an S. cerevisiae atx1Δ strain on iron-depleted medium, indicating that the heterologous proteins can function in S. cerevisiae (45, 55, 66, 112). In this study, S. cerevisiae strains YPH250 (wt) and SL215 (atx1Δ) were transformed with either C. reinhardtii Atx1 (CrAtx1) or S. cerevisiae ATX1 (ScATX1) under the control of the PGK1 promoter in pFL61 (78). Transformants were plated on SD complete medium containing the iron chelator ferrozine (1.5 mM) in the absence (−Fe) or presence (+Fe) of 350 μM ferrous ammonium sulfate. Only the wt strain and transformants expressing Atx1 of C. reinhardtii or S. cerevisiae were able to grow on the iron-deficient medium (Fig. 10A). Complementation of the mutant phenotype was plasmid dependent, and similar results were obtained when the experiment was repeated with independent transformants expressing CrAtx1 or ScATX1 (data not shown). The vector by itself did not alter the iron-dependent growth of the atx1Δ strain. C. reinhardtii Atx1 was comparable to S. cerevisiae ATX1 in the context of this experiment. Differences in growth between the wt strain and the transformants reflect perhaps the difference between ATX1 function at a chromosomal location and that on a plasmid. We conclude that C. reinhardtii Atx1 can function to deliver copper to a copper transporter in the secretory pathway.
FIG. 10.
Chlamydomonas Atx1 rescued S. cerevisiae atx1 and sod1 mutants. (A) Complementation of iron-deficient growth in S. cerevisiae atx1Δ mutant strain SL215. S. cerevisiae strains YPH250 (wt), SL215 (atxΔ), and SL215 transformed with CrAtx1, ScATX1, or control vector pFL61 were serially diluted and grown on SD complete medium containing the iron chelator ferrozine (1.5 mM) in the absence (−Fe) or presence (+Fe) of 350 μM ferrous ammonium sulfate. (B) Complementation of aerobic lysine and methionine auxotrophy of S. cerevisiae sod1Δ mutant. wt, sod1Δ mutant, and sod1Δ strains transformed with CrAtx1, ScATX1, or control vector pFL61 were grown on YPD or SD complete medium lacking lysine or methionine.
Antioxidant function.
S. cerevisiae ATX1 was isolated originally for its ability to protect the sod1Δ sod2Δ mutant strain against oxygen toxicity, hence the name ATX1 for antioxidant (65). The sod1Δ mutant strain exhibits auxotrophy for lysine and methionine, since the biosynthetic pathways for these amino acids contain steps that are sensitive to reactive oxygen. Overexpression of ATX1 in a sod1Δ mutant can rescue the lysine and methionine auxotrophy of this strain (65) via the antioxidant activity of Atx1p, which is attributed to its copper binding function. To test whether C. reinhardtii Atx1 has an analogous function, wt, sod1Δ mutant, and sod1Δ strains transformed with C. reinhardtii Atx1 (CrAtx1), S. cerevisiae ATX1 (ScATX1), or pFL61 alone were compared for growth on medium lacking lysine or methionine. C. reinhardtii Atx1 was indeed able to restore aerobic growth of the sod1Δ strain on medium lacking lysine (−Lys) or methionine (−Met) (Fig. 10B), confirming yet another aspect of its function and the validity of the designation Atx1.
Copper ATPase.
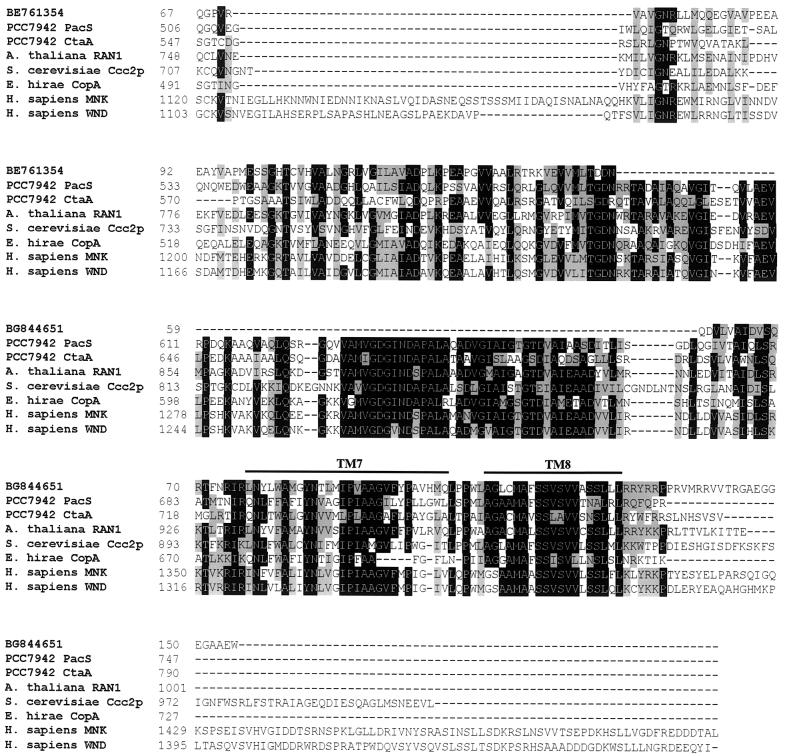
The functionality of C. reinhardtii Atx1 argues in favour of a Ccc2- or MNK/WND-like copper-transporting P-type ATPase in the ferroxidase assembly pathway. A BLAST search of the C. reinhardtii EST database (31) with the sequences of A. thaliana copper ATPases as the input revealed two C. reinhardtii ESTs represented by accessions BE761354 and BG844651 with good probability scores, 2e−13 and 2e−18, respectively. When each is used as the input to search the nonredundant protein database, sequences encoding copper-transporting ATPases are retrieved with the best probability relative to other ATPases. The 3′ ends of the two clones from which the ESTs were derived are identical, indicating that the two ESTs represent a single gene, which increases the likelihood of our assignment of this sequence as a copper-transporting ATPase. The amino acid sequences deduced from the two C. reinhardtii ESTs were aligned with those of copper-transporting ATPases from Synechococcus strain PCC 7942 (PacS and CtaA), A. thaliana (RAN1), S. cerevisiae (Ccc2p), Enterococcus hirae (CopA), and the human Menkes (MNK) and Wilson (WND) proteins. The protein sequence derived from accession BG844651 showed similarity to a highly conserved region that spans transmembrane domain 7 of the MNK and WND proteins and is close to the C terminus of the copper ATPases. The protein sequence derived from accession BE761354 showed similarity to a less well conserved region between the phosphorylation site and the ATP binding site of MNK and WND that is located within the cytoplasmic loop that lies between transmembrane domains 6 and 7 (Fig. 11). A fragment corresponding to the EST represented by accession BE761354 was amplified and cloned (see Materials and Methods) and used as a probe in RNA blot analysis with the objective of testing its expression in response to iron deficiency. Unfortunately, a signal was not detected, and this question could not be addressed.
FIG. 11.
Sequence comparison between C. reinhardtii ESTs and copper ATPases. The predicted amino acid sequences encoded by C. reinhardtii ESTs (GenBank accession no. BE761354 and BG844651) were aligned with the relevant regions of copper-transporting ATPases from other organisms. The alignment was generated by using the ClustalW algorithm and BioEdit software (34). The numbers indicate the positions of the amino acids in each sequence. The first sequence in the alignment represents the amino acids encoded by the two C. reinhardtii ESTs, which align with different regions of the copper ATPases but are derived from a single gene. The alignment of the ESTs was based on BLAST output. Residues that are similar or identical in a majority (five) of sequences are shaded gray and black, respectively. A line above the alignment indicates the transmembrane regions. GenBank accession numbers: Synechococcus PacS, P37279; Synechococcus CtaA, AAB82020; A. thaliana RAN1, AF082565; S. cerevisiae Ccc2p, L36317; E. hirae CopA, L13292; H. sapiens MNK, NM_000052; H. sapiens WND, NM_000053.
Is copper-deficient C. reinhardtii also iron deficient?
In yeast and mammals, the requirement of multicopper-containing ferroxidases for iron metabolism results in an obligate link between copper nutritional status and iron metabolism, such that copper deficiency leads to impairment of ferroxidase function and hence secondary iron deficiency. Previously, we have demonstrated that this is not the case for C. reinhardtii based on the distinct, nonoverlapping pattern of responses to copper deficiencies versus those to iron deficiencies (42). In this work, we provide substantial evidence for the involvement of an MCO in iron metabolism (Fig. 5 and 8), including a pathway for assembly of its copper binding site (Fig. 9 to 11). We also show that the induced accumulation of the ferroxidase in iron deficiency is greatly inhibited in copper deficiency. The lack of effect of copper nutritional status on iron metabolism as suggested previously (42) appeared contrary. We considered, therefore, whether the operation of low-affinity iron assimilation pathways might mask the effect of copper deficiency on iron uptake. If so, one might expect a greater impact of copper deficiency when iron supply is limiting to growth. The effect of copper-deficient growth conditions on the iron status of C. reinhardtii cells was therefore reassessed on wt C. reinhardtii as a function of iron concentration from 0.1 to 200 μM (Fig. 12A). Growth was monitored over a period of 6 days by cell counts, and iron sufficiency was monitored over the same time by chlorophyll content (Fig. 12B). Iron-deficient cells became chlorotic due to degradation of chlorophyll proteins (J. Moseley, S. Merchant, and M. Hippler, unpublished results), and as deficiency became more severe, cell growth was inhibited so that the final cell density of iron-deficient cells was lower than that for iron-replete cells. The medium iron concentration at which these changes were evident was independent of medium copper concentration (Fig. 12), indicating that copper deficiency and loss of ferroxidase function do not affect cellular iron status in C. reinhardtii. By analogy with the occurrence of a copper-independent pathway for photosynthetic electron transfer, we suggest that (i) there must be a backup copper-independent pathway for iron assimilation and (ii) the expression of this pathway must be increased in copper deficiency to compensate for the loss of Fox1 function.
DISCUSSION
The molecular components of iron metabolism in green algae are largely unidentified. Physiological experiments implied the involvement of reductases in iron assimilation because this activity is induced in iron-deficient cells (22, 42, 67, 114). The question of whether these reductases are iron specific or whether they function also in copper assimilation is unexplored yet because the identity of the enzymes is unknown. The nature of iron transporters is also largely unknown, although molecules like transferrin have been implicated in iron assimilation in a halotolerant alga, Dunaliella species (27, 28) (Dunaliella tertiolecta accession no. AAF72064). With the objective of defining the molecular components of iron assimilation and homeostasis in Chlamydomonas, which is an excellent model for the investigation of metal metabolism, we searched the EST database for candidate homologues of known components of iron metabolism in plants, animals, and fungi. Sequences representing four molecules were analyzed in this work: ferritin (Fer1), an iron permease (Ftr1), an MCO (Fox1), and a copper chaperone (Atx1). ESTs representing a copper transporter that probably interacts with the chaperone also were identified, but these have not yet been analyzed. Homologues of the Saccharomyces FRE genes or the Arabidopsis FRO genes were not identified (February 2002); instead, ESTs representing at least two different genes for cytochrome b5-reductase type proteins were identified (3e−36 and 3e−40) that are related to maize NADM-dependent Fe3+-chelate reductase, which are implicated in iron metabolism (8). As for the candidate copper transporter, the relevance of the NADM-dependent Fe3+-chelate reductase-like sequences in iron metabolism needs to be tested by expression analyses.
Ferritin.
The ESTs representing a plastid-targeted ferritin appeared to represent a single gene. The discovery of ferritin was not surprising, because algal ferritin has been described previously (50, 87). Ferritin has a storage function and is a key enzyme in maintaining iron homeostasis. Plant ferritin is localized in the plastids in the stroma (10, 102). It is found primarily in roots and leaves of young plants, with much lower levels occurring in mature plants (10). Incorrect expression of ferritin in mature plants results in iron-deficiency chlorosis, confirming that ferritin sequesters iron and supporting the model that ferritin serves as an iron reservoir for iron supply to the photosynthetic apparatus in developing leaves (108). Since ferritins probably function also for transient iron storage, for example, during senescence or other situations where iron proteins are degraded (10), this might explain the anomalous finding that mRNAs encoding ferritin are up-regulated during iron deficiency (9, 84); in iron-deficiency-induced chlorosis, iron might be released as photosystem I is degraded. In Chlamydomonas, ferritin mRNA is increased about fourfold in iron deficiency relative to total RNA (Fig. 1). This iron-deficiency increase in the mRNA may be part of a mechanism to anticipate iron overload. Two models are offered to explain C. reinhardtii Fer1 gene induction under iron deficiency. First, increased ferritin might allow the cell to handle transient overload when iron is resupplied to the deficient cells following the induction of the uptake pathway, and this is consistent with the observation that the increased mRNA does not result in increased protein (M. Hippler, personal communication). A second possibility is that ferritin may serve to buffer iron as it is released from degrading photosystem I and recycled to other compartments (such as the mitochondrion). This model is supported by the finding that iron deficiency impacts the chloroplast more severely than it does respiration. It also is compatible with the increase in Fer1 gene expression in degreening nitrogen-starved Chlorella cells (50).
MCOs.
MCOs have not previously been implicated in iron metabolism in plants although plants contain several abundant MCOs whose in vivo functions remain to be elucidated. The discovery of a multicopper ferroxidase in Chlamydomonas is, therefore, of interest as the first one identified in a photosynthetic species. MCOs constitute a family of copper-containing proteins that catalyze the four-electron reduction of molecular oxygen to water coupled to the one-electron oxidation of the substrate (77). Members of this family include mammalian ceruloplasmin, hephaestin and blood coagulation factor, factor VIII, S. cerevisiae Fet3p, ascorbate oxidase found in plants, plant and fungal laccase, and several bacterial proteins involved in copper resistance (5, 11, 54, 60, 77, 83, 99, 111). Thus, MCOs occur in a wide variety of organisms and have a range of cellular functions, which is reflected in the variety of organic substrates that can be oxidized (77). A distinguishing feature of these proteins is the presence of copper ions that are classified according to their spectroscopic properties and are referred to as type I or “blue,” type II or “normal,” and type III or “binuclear” copper ions, the latter comprising a pair of copper ions (77). The type I copper is bound as a mononuclear species, while the copper ions of the type II and type III sites form a trinuclear cluster. With the exception of ceruloplasmin, which has three type I sites, in general there is one type I site associated with a trinuclear cluster. Typical type I sites are formed by a set of four copper-binding ligands, His, Cys, His, and Met, although one of the three type I sites in ceruloplasmin and the type I sites of fungal laccase and Fet3p have Leu in place of the Met. The trinuclear cluster has eight histidine ligands arranged as four His-Xaa-His sequences along the polypeptide chain (77).
We suggest that Fox1 may be a type I, C-terminally anchored, membrane glycoprotein by analogy to Fet3p (99) and hephaestin (111). Its amino-terminal domain would be extracellular or within an extracytosolic compartment, and only four putative N-glycosylation sites that precede the transmembrane domain would be glycosylated. In sequence, length, and conservation of type I, II, and III copper binding sites, Fox1 was most similar to the mammalian ferroxidases. Till the recent discovery of hephaestin, a peculiarity of ceruloplasmin was the presence of three type I copper sites, whereas other MCOs have only one. With three potential type I copper sites, Fox1 represents the third member of the ceruloplasmin-hephaestin family. This observation together with its three-domain structure makes Fox1 very similar to ceruloplasmin. Given that the MCOs function in specific partnerships with individual iron transporters (7), it seemed unlikely that the Chlamydomonas protein would rescue a fet3 mutant, and hence this experiment was not attempted. If the topological prediction of Fox1 were correct, then one of the three type I sites would have an intracellular location. This raises the possibility for intracellular recognition of the occupancy state of the copper binding sites and perhaps subsequent activation of a degradation pathway. The other MCO sites would occur extracellularly, where they could function in iron uptake together with Ftr1. Another novelty of Fox1 is the position of the type I sites relative to the type II and type III sites. In the other MCOs the HxH motifs precede the HxxHxH and HCHxxxH motifs, whereas in Fox1 the latter two motifs precede the two HxH motifs. In addition, these four motifs occur within the first half of Fox1, while in the other MCOs, the HxH motifs are close to the N terminus and the HxxHxH and HCHxxxH motifs are close to the C terminus (Fig. 7A). The functional significance of these features of Fox1 can be tested with respect to its structure, copper binding properties, and regulation by copper nutrition.
Ftr1.
The Ftr1-like permease had features consistent with a role in the transmembrane transport of iron. If Ftr1 functions in a complex with Fox1 as in S. cerevisiae (100), then it is predicted to be located in the plasma membrane, where Fox1 is located (39). In this case it would function in assimilation rather than intracellular distribution. The pattern of expression of Ftr1 argues in favor of a function in iron assimilation. The activation of Ftr1 and Fox1 mRNA accumulation occurs at nutritionally relevant concentrations and precedes the appearance of iron-deficiency phenotypes such as chlorosis (compare Fig. 5 and Fig. 12). Ftr1 contains two copies of the highly conserved RExxE motif found in ferritin and Ftr1p. In ferritin the glutamates interact with iron (64, 107), and in Ftr1p they are necessary for iron transport (100). As with the other Ftr1 homologues, one of the two RExxE motifs occurs within the putative signal sequence of C. reinhardtii Ftr1, which raises the possibility of a function in iron-dependent trafficking. A further possibility is that the N-terminal RExxE may be involved in the maturation of the protein and iron-dependent posttranslational control of the level of functional protein in the membrane, in a manner somewhat analogous to the copper-dependent posttranslational control of S. cerevisiae Ctr1p (82). Alternatively, since this motif is also embedded within a hydrophobic region, it may simply serve as an additional binding site for iron as it traverses the membrane. Two ExxE motifs that were proposed to be involved in iron binding in Ftr1p of S. cerevisiae also were found in the Chlamydomonas sequence. We can propose two topological models for C. reinhardtii Ftr1. If the N-terminal hydrophobic region serves as a signal sequence and is cleaved, then the N and C termini would be extracellular, whereas cleavage of the N terminus would yield a protein with a cytoplasmic N terminus and a C terminus that is extracellular.
Copper metabolism.
Molecules that might function in the biosynthesis of Fox1, such as homologues of the Ccc2p transporter and the metallochaperone Atx1p, also are found in Chlamydomonas. The latter occurs as a single-copy gene (data not shown) and is induced under iron-deficient conditions, albeit not as strongly as Fox1 and Ftr1, supporting its assignment in copper delivery to the secretory pathway. The accumulation of Cox17 mRNA (encoding a different copper chaperone involved in copper supply to the mitochondrion) is not affected by iron, suggesting that the effect of iron deficiency on Atx1 mRNA is specific and significant. Ccc2 and Atx1 homologues probably function to load a variety of copper-containing enzymes in the secretory pathway in Chlamydomonas besides Fox1, which would account for the smaller fold difference in regulation. For instance, in plants, RAN1, a Ccc2p homologue, is required for loading copper into the active site of the ethylene receptor (46). In Synechocystis strain PCC 6803 a copper metallochaperone, also designated Atx1, was identified and shown to interact with the CtaA copper importer to acquire and then deliver copper to the PacS copper ATPase which in turn provides copper for proteins involved in photosynthetic and respiratory electron transport within the thylakoid membranes (106).
Functional assignment of C. reinhardtii Atx1 is also supported by its ability to complement S. cerevisiae atx1Δ and sod1Δ mutant strains, the former reflecting its ability to interact with the copper binding domains of Ccc2p and the latter reflecting its ability to bind copper. Atx1 has the conserved metal binding motif MxCxxC present in all metallochaperones identified to date and in the copper-transporting ATPases. The Thr residue immediately adjacent to the Met is conserved among the yeast, C. elegans, and mammalian Atx1 homologues as well as the copper ATPases and is conservatively replaced by Ser in the plant sequences. Based on structural studies of ATX1 and HAH1, a suggested function for this residue is to modulate, via hydrogen bonding, the interaction between the chaperone and its copper ATPase partner (51). The significance of the Met residue at the equivalent position in the C. reinhardtii sequence is unclear. Several lysine residues form a positively charged surface on the ATX1 molecule and were hypothesized previously to be involved in electrostatic interactions with a corresponding acidic face of the copper ATPase target protein (85, 94). Three of these Lys residues (corresponding to S. cerevisiae Lys28, Lys62, and Lys65) are conserved in all of the Atx1 homologues, including C. reinhardtii Atx1. Lys65 is required for the copper-dependent interaction and possibly metal transfer between ATX1 and Ccc2p and for the antioxidant role of ATX1 and HAH1 (51, 52, 85, 94), whereas Lys28 and Lys62 as well as Lys24 and Lys61 of ATX1 are required only for copper delivery to Ccc2p (85).
C. reinhardtii Atx1 had two features in common with the mammalian homologues. One of these was the aspartic acid residue immediately preceding the methionine of the MxCxxC motif instead of the glycine residue that is conserved in the copper ATPases, A. thaliana CCH, rice ATX1, soybean CCH, and C. elegans CUC-1. Structural studies will clarify the significance of this substitution. The other was the absence of the C-terminal extension that is present in the plant homologues. In A. thaliana this region was postulated to interact with other molecules (45). A. thaliana CCH is down-regulated by copper and up-regulated during leaf senescence, possibly for copper sequestration during this process (45). In constrast to A. thaliana CCH but similar to S. cerevisiae ATX1, C. reinhardtii Atx1 is not regulated by copper but is regulated by iron and therefore is likely to share more structural and functional similarities with the mammalian and yeast homologues than with the plant homologues.
Copper versus iron nutritional status.
Previously, we had argued that iron metabolism in Chlamydomonas was independent of copper because copper-deficient cells did not display any symptoms of iron deficiency, such as chlorosis. Also, the pattern of gene expression in −Cu versus −Fe cells was distinct (42). The analysis of iron-deficiency-induced transcripts in this study confirms the previous work (Fig. 5). Transcripts that increase up to several-hundredfold in iron-deficient cells are not affected by copper deficiency. The discovery of a ceruloplasmin-hephaestin-like MCO in Chlamydomonas was unexpected (Fig. 6 to 8). Its involvement in iron metabolism and, more specifically, iron assimilation is supported on the basis of its coordinated expression with Ftr1, a putative iron permease, in iron-deficient cells (Fig. 5); its membrane localization (Fig. 8); and its extracellular ferroxidase activity (39) (http://lin2.biologie.hu-berlin.de/∼botanik/deutsch/flp.html). The lackof impact of copper nutritional status on iron metabolism presented a conundrum.
We considered two possibilities: first, that ferroxidase function was not compromised in copper-deficient cells because of preferential allocation of residual copper to iron assimilation versus photosynthesis, and second, that the effect of copper nutritional status would be apparent only in iron-limited cells. To address these hypotheses, ferroxidase abundance was monitored as a function of copper concentration (Fig. 8). Regardless of cellular iron status, copper deficiency (this work) or copper removal (39) had a drastic impact on ferroxidase accumulation or activity, respectively. We suggest that assembly of the holoprotein is defective when copper is limiting, leading perhaps to degradation of the apoprotein. No change in the abundance of lower- or higher-molecular-weight immunoreactive bands, indicative of enhanced protease susceptibility or ubiquitination, was noted, but this does not argue against degradation, because such intermediates may be short lived. An effect of copper on translation also cannot be ruled out. Interestingly, ferroxidase abundance does increase in iron-deficient cells compared to iron-replete cells even without copper in the medium. This may result from increased allocation of intracellular copper to the ferroxidase biosynthetic pathway in iron-deficient versus iron-sufficient cells through increased Atx1 expression (Fig. 5). Alternatively, it may merely reflect a new steady state resulting from increased synthesis as a consequence of an increase in the Fox1 mRNA pool. The second hypothesis, that copper deficiency might impact only iron-limited cells, was tested by examining the effect of copper nutrition on marker gene expression (Fig. 5) and chlorophyll accumulation and growth (Fig. 12) at a range of medium iron concentrations. Clearly copper nutritional status does not affect the response to iron nutrition despite its impact on ferroxidase accumulation.
A third hypothesis that can be tested genetically is that there may be a copper-independent pathway for iron assimilation. We propose that the expression of such a pathway would be regulated by copper so that intracellular iron abundance is controlled tightly. It would serve only as a backup for the Fox1-dependent pathway, just as cytochrome c6 serves as a backup for plastocyanin (71, 74).
The similarity in catalytic ability between copper and iron enzymes prompts speculation that the replacement of a copper enzyme with a backup iron version might represent a general metabolic adaptation to copper deficiency. There are several examples where copper- and iron-containing proteins represent alternatives that carry out the same function, and these include plastocyanin/cytochrome c6, cytochrome oxidase/alternative oxidase, and CuZn-SOD/Fe-SOD, among others. In Chlamydomonas, clearly there must exist a copper-independent pathway for iron uptake, and whether there are components of such a pathway that serve as true backup proteins for the copper proteins involved in copper-dependent iron uptake remains to be determined. A genetic screen for C. reinhardtii iron metabolism mutants will help to identify such components.
Acknowledgments
We thank Michael Allen, Patrice Hamel, Dan Kosman, and Matthew Portnoy for helpful discussions and advice; Joan Valentine and Edie Gralla, UCLA, California, for S. cerevisiae strains DBY746 (wt) and EG118 (sod1Δ); Valeria Culotta, Johns Hopkins University, Maryland, for S. cerevisiae strains YPH250 (wt) and SL215 (atxΔ); Francois Lacroute, Centre de Génétique Moléculaire, France, for the yeast expression vector pFL61; and the Kazusa DNA Research Institute, Chiba, Japan, for the EST clones.
This work is supported by grants from the National Institutes of Health (GM42143) (S.M.); the Council on Research of the Los Angeles Division of the Academic Senate, UCLA (S.M.); and the Australian Research Council International Researcher Exchange scheme (X00001622) (S.L. and S.M.) and by a National Health and Medical Research Council R. Douglas Wright Fellowship (S.L.).
REFERENCES
- 1.Abboud, S., and D. J. Haile. 2000. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J. Biol. Chem. 275:19906-19912. [DOI] [PubMed] [Google Scholar]
- 2.Aisen, P., M. Wessling-Resnick, and E. A. Leibold. 1999. Iron metabolism. Curr. Opin. Chem. Biol. 3:200-206. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C. 1999. The iron transporter DMT1. Int. J. Biochem. Cell Biol. 31:991-994. [DOI] [PubMed] [Google Scholar]
- 4.Asamizu, E., Y. Nakamura, S. Sato, H. Fukuzawa, and S. Tabata. 1999. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6:369-373. [DOI] [PubMed] [Google Scholar]
- 5.Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. [DOI] [PubMed] [Google Scholar]
- 6.Askwith, C., and J. Kaplan. 1998. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23:135-138. [DOI] [PubMed] [Google Scholar]
- 7.Askwith, C., and J. Kaplan. 1997. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272:401-405. [DOI] [PubMed] [Google Scholar]
- 8.Bagnaresi, P., S. Thoiron, M. Mansion, M. Rossignol, P. Pupillo, and J. F. Briat. 1999. Cloning and characterization of a maize cytochrome-b5 reductase with Fe3+-chelate reduction capability. Biochem. J. 338:499-505. [PMC free article] [PubMed] [Google Scholar]
- 9.Briat, J.-F., and S. Lobreaux. 1997. Iron transport and storage in plants. Trends Plant Sci. 2:187-193. [Google Scholar]
- 10.Briat, J. F., S. Lobreaux, N. Grignon, and G. Vansuyt. 1999. Regulation of plant ferritin synthesis: how and why. Cell. Mol. Life Sci. 56:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouwers, G.-J., J. P. M. de Vrind, P. L. A. M. Corstjens, P. Cornelis, C. Baysse, and E. W. de Vrind-de Jong. 1999. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 65:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruce, B. D. 2000. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 10:440-447. [DOI] [PubMed] [Google Scholar]
- 13.Bucher, P., and A. Bairoch. 1994. A generalized profile syntax for biomolecular sequences motifs and its function in automatic sequence interpretation, p. 53-61. In R. Altman, D. Brutlag, P. Karp, R. Lanthrop, and D. Searls (ed.), ISMB-94. Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 14.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Cohen, A., H. Nelson, and N. Nelson. 2000. The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 275:33388-33394. [DOI] [PubMed] [Google Scholar]
- 16.Crichton, R. R., and J.-L. Pierre. 2001. Old iron, young copper: from Mars to Venus. Biometals 14:99-112. [DOI] [PubMed] [Google Scholar]
- 17.Curie, C., J. M. Alonso, M. L. Jean, J. R. Ecker, and J. F. Briat. 2000. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem. J. 347:749-755. [PMC free article] [PubMed] [Google Scholar]
- 18.Curie, C., Z. Panaviene, C. Loulergue, S. L. Dellaporta, J. F. Briat, and E. L. Walker. 2001. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346-349. [DOI] [PubMed] [Google Scholar]
- 19.Dix, D., J. Bridgham, M. Broderius, and D. Eide. 1997. Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J. Biol. Chem. 272:11770-11777. [DOI] [PubMed] [Google Scholar]
- 20.Donovan, A., A. Brownlie, Y. Zhou, J. Shepard, S. J. Pratt, J. Moynihan, B. H. Paw, A. Drejer, B. Barut, A. Zapata, T. C. Law, C. Brugnara, S. E. Lux, G. S. Pinkus, J. L. Pinkus, P. D. Kingsley, J. Palis, M. D. Fleming, N. C. Andrews, and L. I. Zon. 2000. Positional cloning of zebrafish ferroportin 1 identifies a conserved vertebrate iron exporter. Nature 403:776-781. [DOI] [PubMed] [Google Scholar]
- 21.Eck, R., S. Hundt, A. Hartl, E. Roemer, and W. Kunkel. 1999. A multicopper oxidase gene from Candida albicans: cloning characterization and disruption. Microbiology 145:2415-2422. [DOI] [PubMed] [Google Scholar]
- 22.Eckhardt, U., and T. J. Buckout. 1998. Iron assimilation in Chlamydomonas reinhardtii involves ferric reduction and is similar to Strategy I higher plants. J. Exp. Bot. 49:1219-1226. [Google Scholar]
- 23.Eide, D., M. Broderius, J. Fett, and M. L. Guerinot. 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93:5624-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenstein, R. S. 2000. Discovery of the ceruloplasmin homologue hephaestin: new insight into the copper/iron connection. Nutr. Rev. 58:22-26. [DOI] [PubMed] [Google Scholar]
- 25.Engelke, D. R., A. Krikos, M. E. Bruck, and D. Ginsburg. 1990. Purification of Thermus aquaticus DNA polymerase expressed in Escherichia coli. Anal. Biochem. 191:396-400. [DOI] [PubMed] [Google Scholar]
- 26.Esaka, M., J. Imagi, K. Suzuki, and K. Kubota. 1988. Formation of ascorbate oxidase in cultured pumpkin cells. Plant Cell Physiol. 29:231-235. [Google Scholar]
- 27.Fisher, M., I. Gokhman, U. Pick, and A. Zamir. 1997. A structurally novel transferrin-like protein accumulates in the plasma membrane of the unicellular green alga Dunaliella salina grown in high salinities. J. Biol. Chem. 272:1565-1570. [DOI] [PubMed] [Google Scholar]
- 28.Fisher, M., A. Zamir, and U. Pick. 1998. Iron uptake by the halotolerant alga Dunaliella is mediated by a plasma membrane transferrin. J. Biol. Chem. 273:17553-17558. [DOI] [PubMed] [Google Scholar]
- 29.Fox, T. C., and M. L. Guerinot. 1998. Molecular biology of cation transport in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:669-696. [DOI] [PubMed] [Google Scholar]
- 30.Gietz, R. D., and R. A. Woods. 1994. High efficiency transformation in yeast, p. 121-134. In J. A. Johnston (ed.), Molecular genetics of yeast: practical approaches. Oxford University Press, Oxford, United Kingdom.
- 31.Grossman, A., J. Davies, N. Federspiel, E. Harris, P. Lefebvre, J. P. McDermott, C. Silflow, D. Stern, and R. Surzycki. 2000. Analyses of the Chlamydomonas reinhardtii genome: a model unicellular system for analyzing gene function and regulation in vascular plants; project phase 2.
- 32.Guerinot, M. L. 2000. The ZIP family of metal transporters. Biochim. Biophys. Acta 1465:190-198. [DOI] [PubMed] [Google Scholar]
- 33.Guerinot, M. L., N. Grotz, S. Hibbard, E. Rogers, E. Connolly, and D. J. Eide. 2001. Molecular characterization of the uptake of iron and other divalent cations in Arabidopsis. J. Exp. Bot. 52(Suppl.):62. [Google Scholar]
- 34.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 35.Hammacott, J. E., P. H. Williams, and A. M. Cashmore. 2000. Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant. Microbiology 146:869-876. [DOI] [PubMed] [Google Scholar]
- 36.Harris, E. H. 1989. The chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 37.Hassett, R., D. R. Dix, D. J. Eide, and D. J. Kosman. 2000. The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 351:477-484. [PMC free article] [PubMed] [Google Scholar]
- 38.Hentze, M. W., T. A. Rouault, S. W. Caughman, A. Dancis, J. B. Harford, and R. D. Klausner. 1987. A cis-acting element is necessary and sufficient for translational regulation of human ferritin expression in response to iron. Proc. Natl. Acad. Sci. USA 84:6730-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herbik, A., and T. J. Buckhout. 2001. Is a ferroxidase involved in the high-affinity iron uptake in Chlamydomonas? J. Exp. Bot. 52(Suppl.):80. [Google Scholar]
- 40.Heymann, P., J. F. Ernst, and G. Winkelmann. 2000. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186:221-227. [DOI] [PubMed] [Google Scholar]
- 41.Heymann, P., J. F. Ernst, and G. Winkelmann. 1999. Identification of a fungal triacetylfusarinine C siderophore transport gene (TAF1) in Saccharomyces cerevisiae as a member of the major facilitator superfamily. Biometals 12:301-306. [DOI] [PubMed] [Google Scholar]
- 42.Hill, K. L., R. Hassett, D. Kosman, and S. Merchant. 1996. Regulated copper uptake in Chlamydomonas reinhardtii in response to copper availability. Plant Physiol. 112:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill, K. L., H. H. Li, J. Singer, and S. Merchant. 1991. Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266:15060-15067. [PubMed] [Google Scholar]
- 44.Himelblau, E., and R. M. Amasino. 2000. Delivering copper within plant cells. Curr. Opin. Plant Biol. 3:205-210. [PubMed] [Google Scholar]
- 45.Himelblau, E., H. Mira, S. J. Lin, V. C. Culotta, L. Penarrubia, and R. M. Amasino. 1998. Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 117:1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirayama, T., and J. M. Alonso. 2000. Ethylene captures a metal! Metal ions are involved in ethylene perception and signal transduction. Plant Cell Physiol. 41:548-555. [DOI] [PubMed] [Google Scholar]
- 47.Hirayama, T., J. J. Kieber, N. Hirayama, M. Kogan, P. Guzman, S. Nourizadeh, J. M. Alonso, W. P. Dailey, A. Dancis, and J. R. Ecker. 1999. RESPONSE TO ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97:383-393. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database; its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 50.Hortensteiner, S., J. Chinner, P. Matile, H. Thomas, and I. S. Donnison. 2000. Chlorophyll breakdown in Chlorella protothecoides: characterization of degreening and cloning of degreening-related genes. Plant Mol. Biol. 42:439-450. [DOI] [PubMed] [Google Scholar]
- 51.Huffman, D. L., and T. V. O'Halloran. 2001. Function, structure and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 70:677-701. [DOI] [PubMed] [Google Scholar]
- 52.Hung, I. H., R. L. B. Casareno, G. Labesse, F. S. Mathews, and J. D. Gitlin. 1998. HAH1 is a copper-binding protein with distinct amino acid residues mediating copper homeostasis and antioxidant defense. J. Biol. Chem. 273:1749-1754. [DOI] [PubMed] [Google Scholar]
- 53.Kaplan, J., and J. P. Kushner. 2000. Mining the genome for iron. Nature 403:712-713. [DOI] [PubMed] [Google Scholar]
- 54.Kim, C., W. W. Lorenz, J. T. Hoopes, and J. F. D. Dean. 2001. Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J. Bacteriol. 183:4866-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klomp, L. W., S.-J. Lin, D. S. Yuan, R. D. Klausner, V. C. Culotta, and J. D. Gitlin. 1997. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J. Biol. Chem. 272:9221-9226. [DOI] [PubMed] [Google Scholar]
- 56.Knight, S. A. B., E. Lesuisse, R. Stearman, R. D. Klausner, and A. Dancis. 2002. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 148:29-40. [DOI] [PubMed] [Google Scholar]
- 57.Korshunova, Y., D. Eide, W. G. Clark, M. L. Guerinot, and H. B. Pakrasi. 1999. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40:37-44. [DOI] [PubMed] [Google Scholar]
- 58.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 157:105-142. [DOI] [PubMed] [Google Scholar]
- 59.Laulhere, J. P., A. M. Laboure, and J. F. Briat. 1989. Mechanism of the transition from plant ferritin to phytosiderin. J. Biol. Chem. 264:3629-3635. [PubMed] [Google Scholar]
- 60.Lee, Y.-A., M. Hendson, N. J. Panopoulos, and M. N. Schroth. 1994. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Xanthomonas campestris pv. juglandis: homology with small blue copper proteins and multicopper oxidase. J. Bacteriol. 176:173-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lescure, A. M., D. Proudhon, H. Pesey, M. Ragland, E. C. Theil, and J. F. Briat. 1991. Ferritin gene transcription is regulated by iron in soybean cell cultures. Proc. Natl. Acad. Sci. USA 88:8222-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lesuisse, E., P. L. Blaiseau, A. Dancis, and J. M. Camadro. 2001. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology 147:289-298. [DOI] [PubMed] [Google Scholar]
- 63.Lesuisse, E., M. Simon-Casteras, and P. Labbe. 1998. Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144:3455-3462. [DOI] [PubMed] [Google Scholar]
- 64.Levi, S., P. Santambrogio, A. Cozzi, E. Rovida, B. Corsi, E. Tamborini, S. Spada, A. Albertini, and P. Arosio. 1994. The role of the L-chain in ferritin iron incorporation. Studies of homo and heteropolymers. J. Mol. Biol. 238:649-654. [DOI] [PubMed] [Google Scholar]
- 65.Lin, S.-J., and V. C. Culotta. 1995. The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc. Natl. Acad. Sci. USA 92:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin, S.-J., R. A. Pufahl, A. Dancis, T. V. O'Halloran, and V. C. Culotta. 1997. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem. 272:9215-9220. [PubMed] [Google Scholar]
- 67.Lynnes, J. A., T. L. M. Derzaph, and H. G. Weger. 1998. Iron limitation results in induction of ferricyanide reductase and ferric chelate reductase activities in Chlamydomonas reinhardtii. Planta 204:360-365. [Google Scholar]
- 68.Martins, L. J., L. T. Jensen, J. R. Simons, G. L. Keller, and D. R. Winge. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 69.McKie, A. T., D. Barrow, G. O. Latunde-Dada, A. Rolfs, G. Sager, E. Mudaly, M. Mudaly, C. Richardson, D. Barlow, A. Bomford, T. J. Peters, K. B. Raja, S. Shirali, M. A. Hediger, F. Farzaneh, and R. J. Simpson. 2001. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291:1755-1759. [DOI] [PubMed] [Google Scholar]
- 70.McKie, A. T., P. Marciani, A. Rolfs, K. Brennan, K. Wehr, D. Barrow, S. Miret, A. Bomford, T. J. Peters, F. Farzaneh, M. A. Hediger, M. W. Hentze, and R. J. Simpson. 2000. A novel duodenal iron-regulated membrane protein (Ireg1) implicated in basolateral transfer of iron to the circulation. Mol. Cell 5:299-309. [DOI] [PubMed] [Google Scholar]
- 71.Merchant, S. 1997. Reciprocal, copper-responsive accumulation of plastocyanin and cytochrome c6 in algae and cyanobacteria: a model for metalloregulation of metalloprotein synthesis, p. 450-467. In S. Silver and W. Walden (ed.), Metal ions in gene regulation. Chapman Hall, New York, N.Y.
- 72.Merchant, S. 1998. Synthesis of metalloproteins involved in photosynthesis: plastocyanin and cytochromes, p. 597-611. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), Molecular biology of Chlamydomonas: chloroplasts and mitochondria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 73.Merchant, S., and L. Bogorad. 1986. Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J. Biol. Chem. 261:15850-15853. [PubMed] [Google Scholar]
- 74.Merchant, S., and L. Bogorad. 1986. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol. Cell. Biol. 6:462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Merchant, S., and B. W. Dreyfuss. 1998. Posttranslational assembly of photosynthetic metalloproteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:25-51. [DOI] [PubMed] [Google Scholar]
- 76.Merchant, S., K. Hill, and G. Howe. 1991. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 10:1383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Messerschmidt, A., and R. Huber. 1990. The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur. J. Biochem. 187:341-352. [DOI] [PubMed] [Google Scholar]
- 78.Minet, M., M.-E. Dufour, and F. Lacroute. 1992. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2:417-422. [DOI] [PubMed] [Google Scholar]
- 79.Moseley, J., J. Quinn, M. Eriksson, and S. Merchant. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen, H., J. Engelbrecht, S. Brunak, and G. Von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 81.Ohkawa, J., N. Okada, A. Shinmyo, and M. Takano. 1989. Primary structure of cucumber (Cucumis sativus) ascorbate oxidase deduced from cDNA sequence: homology with blue copper proteins and tissue-specific expression. Proc. Natl. Acad. Sci. USA 86:1239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ooi, C. E., E. Rabinovich, A. Dancis, J. S. Bonifacino, and R. D. Klausner. 1996. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15:3515-3523. [PMC free article] [PubMed] [Google Scholar]
- 83.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 84.Petit, J.-M., J.-F. Briat, and S. Lobreaux. 2001. Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J. 359:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Portnoy, M. E., A. C. Rosenzweig, T. Rae, D. L. Huffman, T. V. O'Halloran, and V. C. Culotta. 1999. Structure-function analyses of the ATX1 metallochaperone. J. Biol. Chem. 274:15041-15045. [DOI] [PubMed] [Google Scholar]
- 86.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 87.Pueschel, C. M., and K. M. Cole. 1980. Phytoferritin in the red alga Constantinea (Cryptonemiales). J. Ultrastruct. Res. 73:282-287. [DOI] [PubMed] [Google Scholar]
- 88.Pufahl, R. A., C. P. Singer, K. L. Peariso, S.-L. Lin, P. J. Schmidt, V. Fahrni, V. C. Culotta, J. E. Penner-Hahn, and T. V. O'Halloran. 1997. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278:853-856. [DOI] [PubMed] [Google Scholar]
- 89.Quinn, J. M., and S. Merchant. 1998. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297:263-279. [DOI] [PubMed] [Google Scholar]
- 90.Quinn, J. M., S. S. Nakamoto, and S. Merchant. 1999. Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper-response elements and increased translation from a copper-deficiency-specific form of the transcript. J. Biol. Chem. 274:14444-14454. [DOI] [PubMed] [Google Scholar]
- 91.Radisky, D., and J. Kaplan. 1999. Regulation of transition metal transport across the yeast plasma membrane. J. Biol. Chem. 274:4481-4484. [DOI] [PubMed] [Google Scholar]
- 92.Ramanan, N., and Y. Wang. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 288:1062-1064. [DOI] [PubMed] [Google Scholar]
- 93.Robinson, N. J., C. M. Procter, E. L. Connolly, and M.-L. Guerinot. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397:694-697. [DOI] [PubMed] [Google Scholar]
- 94.Rosenzweig, A. C., D. L. Huffman, M. Y. Hou, A. K. Wernimont, R. A. Pufahl, and T. V. O'Halloran. 1999. Crystal structure of the Atx1 metallochaperone protein at 1.02 Å resolution. Structure 7:605-617. [DOI] [PubMed] [Google Scholar]
- 95.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 96.Schloss, J. A. 1990. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221:443-452. [DOI] [PubMed] [Google Scholar]
- 97.Schneider, B. D., and E. A. Leibold. 2000. Regulation of mammalian iron homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 3:267-273. [DOI] [PubMed] [Google Scholar]
- 98.Sikorski, R. S., and P. Heiter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Silva, D. D., C. C. Askwith, D. Eide, and J. Kaplan. 1995. The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270:1098-1101. [DOI] [PubMed] [Google Scholar]
- 100.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 101.Tandy, S., M. Williams, A. Leggett, M. Lopez-Jimenez, M. Dedes, B. Ramesh, S. K. Srai, and P. Sharp. 2000. Nramp2 expression is associated with pH-dependent iron uptake across the apical membrane of human intestinal Caco-2 cells. J. Biol. Chem. 275:1023-1029. [DOI] [PubMed] [Google Scholar]
- 102.Theil, E. C. 1987. Ferritin: structure, gene regulation, and cellular function in animals, plants and microorganisms. Annu. Rev. Biochem. 56:289-315. [DOI] [PubMed] [Google Scholar]
- 103.Thomine, S., R. Wang, J. M. Ward, N. M. Crawford, and J. I. Schroeder. 2000. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA 97:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thomson, A. M., J. T. Rogers, and P. J. Leedman. 1999. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 31:1139-1152. [DOI] [PubMed] [Google Scholar]
- 105.Tottey, S., P. R. Rich, S. A. M. Rondet, and N. J. Robinson. 2001. Two Menkes-type ATPases supply copper for photosynthesis in Synechocystis PCC6803. J. Biol. Chem. 276:19999-20004. [DOI] [PubMed] [Google Scholar]
- 106.Tottey, S., S. A. M. Rondet, G. P. M. Borrelly, P. J. Robinson, P. R. Rich, and N. J. Robinson. 2002. A copper metallochaperone for photosynthesis and respiration reveals metal-specific targets, interaction with an importer and alternative sites for copper acquisition. J. Biol. Chem. 277:5490-5497. [DOI] [PubMed] [Google Scholar]
- 107.Trikha, J., E. C. Theil, and N. M. Allewell. 1995. High resolution crystal structures of amphibian red-cell L ferritin: potential roles for structural plasticity and solvation in function. J. Mol. Biol. 248:949-967. [DOI] [PubMed] [Google Scholar]
- 108.van Wuytswinkel, O., G. Vansuyt, N. Grignon, P. Fourcroy, and J. F. Briat. 1999. Iron homeostasis alteration in transgenic tobacco overexpressing ferritin. Plant J. 17:93-97. [DOI] [PubMed] [Google Scholar]
- 109.van Wuytswinkel, O., and J. F. Briat. 1995. Conformational changes and in vitro core-formation modifications induced by site-directed mutagensis of the specific N-terminus of pea seed ferritin. Biochem. J. 305:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.von Heijne, G. 1992. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 111.Vulpe, C. D., Y. M. Kuo, T. L. Murphy, L. Cowley, C. Askwith, N. Libina, J. Gitschier, and G. J. Anderson. 1999. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21:195-199. [DOI] [PubMed] [Google Scholar]
- 112.Wakabayashi, T., N. Nakamura, Y. Sambongi, Y. Wada, T. Oka, and M. Futai. 1998. Identification of the copper chaperone, CUC-1, in Caenorhabditis elegans: tissue specific co-expression with the copper transporting ATPase, CUA-1. FEBS Lett. 440:141-146. [DOI] [PubMed] [Google Scholar]
- 113.Wardrop, A. J., R. E. Wicks, and B. Entsch. 1999. Occurrence and expression of members of the ferritin gene family in cowpeas. Biochem. J. 337:523-530. [PMC free article] [PubMed] [Google Scholar]
- 114.Weger, H. G. 1999. Ferric and cupric reductase activities in the green alga Chlamydomonas reinhardtii: experiments using iron-limited chemostats. Planta 207:377-384. [Google Scholar]
- 115.Yi, Y., and M. L. Guerinot. 1996. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10:835-844. [DOI] [PubMed] [Google Scholar]
- 116.Yuan, D. S., A. Dancis, and R. D. Klausner. 1997. Restriction of copper export in Saccharomyces cerevisiae to a late Golgi or post-Golgi compartment in the secretory pathway. J. Biol. Chem. 272:25787-25793. [DOI] [PubMed] [Google Scholar]
- 117.Yuan, D. S., R. Stearman, A. Dancis, T. Dunn, T. Beeler, and R. D. Klausner. 1995. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. USA 92:2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yun, C.-W., T. Ferea, J. Rashford, O. Ardon, P. O. Brown, D. Botstein, J. Kaplan, and C. C. Philpott. 2000. Desferrioxamine-mediated iron uptake in Saccharomyces cereivisiae. Evidence for two pathways of iron uptake. J. Biol. Chem. 275:10709-10715. [DOI] [PubMed] [Google Scholar]
- 119.Yun, C.-W., J. S. Tiedeman, R. E. Moore, and C. C. Philpott. 2000. Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J. Biol. Chem. 275:16354-16359. [DOI] [PubMed] [Google Scholar]