Abstract
Candida albicans is an important pathogen of immunocompromised patients which grows with true hyphal, pseudohyphal, and yeast morphologies. The dynamics of cell cycle progression are markedly different in true hyphal relative to pseudohyphal and yeast cells, including nuclear movement and septin ring positioning. In Saccharomyces cerevisiae, two forkhead transcription factors (ScFKH1 and ScFKH2) regulate the expression of B-cyclin genes. In both S. cerevisiae and Schizosaccharomyces pombe, forkhead transcription factors also influence morphogenesis. To explore the molecular mechanisms that connect C. albicans morphogenesis with cell cycle progression, we analyzed CaFKH2, the single homolog of S. cerevisiae FKH1/FKH2. C. albicans cells lacking CaFkh2p formed constitutive pseudohyphae under all yeast and hyphal growth conditions tested. Under hyphal growth conditions levels of hyphae-specific mRNAs were reduced, and under yeast growth conditions levels of several genes encoding proteins likely to be important for cell wall separation were reduced. Together these results imply that Fkh2p is required for the morphogenesis of true hyphal as well as yeast cells. Efg1p and Cph1p, two transcription factors that contribute to C. albicans hyphal growth, were not required for the pseudohyphal morphology of fkh2 mutants, implying that Fkh2p acts in pathways downstream of and/or parallel to Efg1p and Cph1p. In addition, cells lacking Fkh2p were unable to damage human epithelial or endothelial cells in vitro, suggesting that Fkh2p contributes to C. albicans virulence.
Candida albicans, an important human pathogen, is multimorphic and is able to grow in yeast, pseudohyphal, and true hyphal forms. Yeast are round-to-ovoid cells that usually undergo cell separation following cytokinesis; pseudohyphae form chains of ellipsoid daughter cells that remain attached for several divisions. Both yeast and pseudohyphae septate at a constriction between the mother and daughter cells. In contrast, true hyphal cells form long narrow filaments with parallel sides that do not separate from one another and that exhibit little, if any, constriction at the septa (41). Furthermore, in hyphal cells the first site of septation is located distal to the mother-daughter junction (53, 56). The transition between yeast, pseudohyphal, and true hyphal growth is influenced by environmental factors, such as temperature, pH, carbon source, nitrogen source, the presence of a quorum sensor, and physical contact with surfaces, and is thought to be an important contributor to virulence (reviewed in references 14, 36, and 40). Therefore, understanding the molecular mechanisms that contribute to these morphogenetic transitions is vital to understanding C. albicans pathogenicity.
At least eight different transcription factors (Cph1p, Efg1p, Rbf1p, Tup1p, Czf1p, Nrg1p, Cph2p, and Rim101p) contribute to hyphal morphogenesis under different environmental conditions (reviewed in references 15 and 30) by affecting the levels of expression of specific subsets of downstream genes, such as those that encode the hyphae-specific cell wall proteins (28, 39). Cph1p contributes to filamentous growth on starvation media and under embedded conditions. Efg1p is critical for hyphal growth in response to inducers, such as serum and N-acetylglucosamine, and is a negative regulator of filamentation in response to embedded conditions. Double mutants lacking both Efg1p and Cph1p form virtually no hyphae in response to serum, starvation, N-acetylglucosamine, or high pH at 37°C, although they do form hyphae under some in vitro and in vivo conditions (reviewed in reference 15).
In C. albicans, the position and dynamics of fundamental structures required for cell cycle progression are very different in true hyphae relative to yeast and pseudohyphae (56). For example, in yeast and pseudohyphae (as in Saccharomyces cerevisiae), the septin filament ring forms at the incipient bud site prior to bud emergence, and septation occurs later at this mother-bud junction (17, 37, 53, 56). In contrast, in true hyphae, as in Schizosaccharomyces pombe and Aspergillus nidulans (reviewed in references 20 and 33), the first septum forms at a position distal to the mother cell, well after evagination of the growing germ tube (56). Thus, yeast and pseudohyphal growth are characterized by septin ring formation and bud emergence occurring in rapid succession, while in hyphae, daughter cell evagination occurs well before determination of the first site of septation. Accordingly, in yeast and pseudohyphae the dividing nucleus and mitotic spindle span the mother-bud neck, while in true hyphae the mitotic spindle is positioned distal to the mother cell and the nucleus migrates to this position prior to mitosis (21, 56).
C. albicans yeast, pseudohyphae, and true hyphae also differ in their cell wall properties. Hyphal cell walls are more adherent than yeast cell walls to human endothelial and epithelial cells (25, 46, 57) and contain slightly more chitin (7). In addition, true hyphae express a specific constellation of genes, including HWP1 (51, 55), ECE1 (2), HYR1 (1), RBT1, -4, and -5, and WAP1 (3), which all encode predicted glycosylphosphatidylinositol (GPI)-modified cell wall proteins. In contrast, yeast cell walls contain more alkali-extractable proteins (thought to reflect the internal-repeat cell wall proteins) than hyphae (reviewed in reference 27). Furthermore, genes such as CHT2 (encoding a homolog of the ScChs1p chitinase, which is required for the separation of daughter yeast cells after cytokinesis [10]), are specifically transcribed at higher levels in yeast relative to hyphae (5, 34).
In most organisms, morphogenesis is tightly coordinated with cell cycle progression and is regulated by the cyclin-dependent kinase (CDK). In the filamentous fungus A. nidulans, hyphal differentiation appears to be regulated by a protein kinase cascade that regulates septation and mitotic exit and that, in turn, is regulated by CDK activity (reviewed in reference 20). Similarly, in S. cerevisiae pseudohyphal growth is the result of signal transduction pathways and transcriptional outputs from them that ultimately regulate the activity of Clb2/Cdc28p, the mitotic B-cyclin CDK (reviewed in reference 47). Clb2/Cdc28p antagonizes polarized growth by affecting the localization of actin and the secretory vesicles that export cell wall components and enzymes necessary for growth at the appropriate cortical sites.
In S. cerevisiae two partially redundant forkhead domain-containing transcription factors, Fkh1p and Fkh2p, regulate the periodic, cell-cycle-dependent expression of genes whose transcription peaks in early M phase (61). This CLB2 cluster includes 33 genes, among them CLB1 and CLB2, which encode the mitotic B-cyclins, and SWI5 and ACE2, which encode two transcription factors required for expression of the SIC1 cluster, a gene cluster expressed in late mitosis and early G1 (54). Genes of the SIC1 cluster include CTS1 and SCW11, both important for cell separation following cytokinesis. In mutants lacking Fkh1p and Fkh2p, the pattern of cell-cycle-regulated transcription of CLB2 cluster genes is lost, which also indirectly affects the SIC1 cluster (61). In addition, fkh1 fkh2 mutants display constitutive pseudohyphal growth (22, 61). S. pombe mutants lacking the forkhead transcription factor Sep1p also grow with a filamentous morphology and are defective in cell separation (45). Thus, Fkh-like proteins appear to play a key role in regulating the cell cycle machinery, which in turn affects cell morphology.
To explore the molecular mechanisms that connect cell cycle progression and morphogenesis in C. albicans, we analyzed the role of CaFKH2, the single C. albicans homolog of S. cerevisiae FKH1/FKH2, in morphogenesis and gene expression. We found that CaFKH2 mutants formed constitutive pseudohyphae under both yeast and hyphal growth conditions. Furthermore, fkh2 mutants exhibited reduced expression of hyphae-specific mRNAs in the presence of serum and exhibited reduced expression of several genes likely to be important for yeast cell separation under yeast growth conditions. In addition, fkh2 mutants were defective in the ability to damage human epithelial cells in vitro. Our results suggest that Fkh2p plays a crucial role in the morphogenetic transition between cell types in C. albicans and is likely to be important for C. albicans virulence.
MATERIALS AND METHODS
Plasmids.
To place FKH2 under the control of the MET3 promoter, the 5′ end of FKH2 was amplified from genomic DNA of S. cerevisiae strain SC5314 (19) by using primer 559, engineered to include a BamHI site (5′-GGCGGATCCATGTCAGCACAATTTATCAC-3′), and primer 560, engineered to include a PstI site (5′-GGGCCTGCAGCGCATAAGAATAGGGAGGCT-3′). The PCR product was digested with BamHI and PstI and was ligated into BamHI/PstI-cut pBluescript II SK(+) (Stratagene, La Jolla, Calif.) to generate plasmid pBSK-CaFKH2-5′. The PCR-derived FKH2 fragment was sequenced and compared to the sequence at the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida/index.html). A discrepancy was found in multiple, independent PCR products at codon 126. The reported sequence at this position is ATA (ile), while we consistently found the sequence at this position to read TAT (leu). The BamHI/PstI fragment from pBSK-CaFKH2-5′ was subcloned into BamHI/PstI pCaDis, adjacent to the MET3 promoter (12), to form pDis-FKH2-5′. Full-length C. albicans FKH2 was PCR amplified from genomic DNA isolated from strain SC5314 by using primer 559 and primer 581, which contains a PstI site on its 5′ end (5′-GGGCCTGCAGGGACATTTCGTGCAACTGTGT-3′) and was inserted into pCaEXP (12) cut with BamHI/PstI to form pEXP-FKH2. pRSET-FKH2 contains the BamHI/PstI FKH2 fragment from pEXP-FKH2 ligated into BamHI/PstI-digested pRSET-A (Invitrogen, Carlsbad, Calif.). For heterologous expression of CaFKH2 in S. cerevisiae, a BamHI/XhoI fragment containing the C. albicans FKH2 open reading frame (ORF) from pRSET-FKH2 was subcloned into BamHI/XhoI-digested pYES2 (Invitrogen) to form pYES-CaFKH2.
Strains.
C. albicans and S. cerevisiae strains used in this study are described in Table 1. All strains were checked for correct genome integration by PCR and/or Southern analysis (data not shown). To construct disruption strains we used PCR-mediated gene disruption as previously described (59). A fkh2::HIS1 PCR product was amplified with the primers 561 (5′-CGATTATTTACAAATGTCAGCACAATTTATCACACCGAAAAAGCGTCCCCACTCACCACTAGATAGGTTTTCCCAGTCACGACGTT-3′) and 562 (5′-CACGCCAATTTTGTGGACTATTAATTATTTAAAATACATACCTGTGCCTTTAGCCATTTTTGGTGTGTTGTGTGGAATTGTGAGCGGATA-3′), with pGEM-HIS1 (59) as the DNA template. The fkh2::HIS1 product was transformed into BWP17 (59) to generate multiple independent heterozygous strains, including YJB4744 and YJB4745. YJB4744 and YJB4745 were transformed with an fkh2::ARG4 disruption fragment by using primers 561 and 562 with pRS-ARGΔSpeI (59) as the DNA template to generate strains YJB5387 and YJB4899, respectively.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype or description | Source |
|---|---|---|
| C. albicans | ||
| SC5314 | Prototrophic | 19 |
| CAF2 | ura3Δ::λimm434/URA3 | 16 |
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 59 |
| HLC52 | ura3Δ::λimm434/ura3Δ::λimm434 efg1::hisG/efg1::hisG-URA3-hisG | 32 |
| HLC54 | ura3Δ::λimm434/ura3Δ::λimm434 cph1::hisG/cph1::hisG efg1::hisG/efg1::hisG-URA3-hisG | 32 |
| HLC67 | ura3Δ::λimm434/ura3Δ::λimm434 efg1::hisG/efg1::hisG | 32 |
| HLC69 | ura3Δ::λimm434/ura3Δ::λimm434 cph1::hisG/cph1::hisG efg1::hisG/efg1::hisG | 32 |
| JKC18 | ura3Δ::λimm434/ura3Δ::λimm434 cph1::hisG/cph1::hisG | 32 |
| JKC19 | ura3Δ::λimm434/ura3Δ::λimm434 cph1::hisG/cph1::hisG-URA3-hisG | 32 |
| YJB4744 | BWP17 fkh2::HIS1/FKH2 | This study |
| YJB4745 | BWP17 fkh2::HIS1/FKH2 | This study |
| YJB4826 | BWP17 fkh2::HIS1/PMET3-FKH2-URA3::fkh2 | This study |
| YJB4899 | BWP17 fkh2::HIS1/fkh2::ARG4 | This study |
| YJB5387 | BWP17 fkh2::HIS1/fkh2::ARG4 | This study |
| YJB5535 | BWP17 FKH2-GFP:URA3/FKH2 | This study |
| YJB5653 | JKC18 fkh2::URA3-dpl200/FKH2 | This study |
| YJB5655 | HLC69 fkh2::URA3-dpl200/FKH2 | This study |
| YJB5658 | HLC67 fkh2::URA3-dpl200/FKH2 | This study |
| YJB5707 | HLC69 fkh2::dpl200/FKH2 | This study |
| YJB5709 | JKC18 fkh2::dpl200/FKH2 | This study |
| YJB5800 | HLC67 fkh2::dpl200/FKH2 | This study |
| YJB5802 | HLC69 fkh2::dpl200/fkh2::URA3-dpl200 | This study |
| YJB6403 | BWP17 fkh2::HIS1/fkh2::ARG4 PMET3-FKH2-URA3::RP10/RP10 | This study |
| YJB6413 | HLC67 fkh2::dpl200/fkh2::URA3-dpl200 | This study |
| YJB6441 | JKC18 fkh2::dpl200/fkh2::URA3dpl200 | This study |
| YJB6284 | ura3Δ::λimm434/ura3Δ::λimm434 HIS1::his1::hisG/his1::hisG ARG4-URA3::arg4::hisG/arg4::hisG | This study |
| YJB6288 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG ARG4-URA3::arg4::hisG/arg4::hisG fkh2::HIS1/FKH2 | This study |
| YJB6290 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG ARG4-URA3::arg4::hisG/arg4::hisG fkh2::HIS1/FKH2 | This study |
| YJB6292 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG ARG4-URA3::arg4::hisG/arg4::hisG fkh2::HIS1/fkh2::ARG4 | This study |
| S. cerevisiae | ||
| CFY147 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 HMR-SSΔIa lys2Δ fkh1Δ::TRP1 fkh2::HIS3 | 22 |
| YJB6004 | CFY147 + pYES2 (PGALI URA3, 2μ) | This study |
| YJB6005 | CFY147 + pYES-CaFKH2 (PGALI-CaFKH2, URA3,2μ) | This study |
To place FKH2 under the control of the MET3 promoter at the RP10 locus, pEXP-FKH2 was linearized with StuI and transformed into strain YJB4899 to generate strain YJB6403. To place FKH2 under the control of the MET3 promoter at the FKH2 locus, pDis-FKH2-5′ was linearized with ApaI and transformed into strain YJB4744 to generate strain YJB4826.
Fkh2p was tagged at the C terminus with green fluorescent protein (GFP) by PCR-mediated gene modification (18) with primers 595 (5′-CCAAAAAGAAATCCATCTATTTCTAACAACACACCAAAGATGGCTAAAGGCACAGGTATGTATTTTAAAGGTGGTGGTTCTAAAGGTGAAGAATTATT-3′) and 596 (5′-CTTCTCGAATGGCTTTCTGTTGATACTAATTCATAATATATTCGTTAGTAGCTGTTATTTCACGCCTCTAGAAGGACCACCTTTGATTG-3′) with pGFP-URA3 (18) as the DNA template. This FKH2-GFP cassette was transformed into strain BWP17 to generate strain YJB5535.
For epistasis studies, FKH2 was deleted by using a PCR-derived recyclable URA3 cassette (58). Primers 561 and 562 were used to PCR amplify an fkh2::URA3-dpl200 fragment from plasmid pDDB57 (58). This fragment was transformed into JKC18 (32) to generate strain JBY5653, into HLC67 (32) to generate YJB5658, and into strain HLC69 (32) to generate strain YJB5655. Ura− derivatives of YJB5653, YJB5658, and YJB5655 were selected by plating onto 5-fluoroorotic acid to generate strains YJB5709, YJB5800, and YJB5707, respectively. These Ura− strains were subjected to another round of transformation with the fkh2::URA3-dpl200 disruption fragment to generate YJB6441, YJB6413, and YJB5802, respectively.
C. albicans strains were made prototrophic by transforming them with pGEM-HIS1 linearized with NruI and/or pRS-ARG-URA-BN (13) linearized with NotI. BWP17 was transformed with the above plasmids to generate a Ura+, Arg+, His+ prototroph, YJB6284. YJB4744 and YJB4745 were transformed with NotI-digested pRS-ARG-URA-BN to generate the FKH2/FKH2::HIS1 Ura+ Arg+ strains YJB6288 and YJB6290, respectively. YJB4899 was transformed with NotI-digested pRS-ARG4-URA-BN to generate an fkh2::HIS1/fkh2::ARG4 Ura+ strain, YJB6292.
Media and growth conditions.
Rich media (YPAD), synthetic complete media (SDC), and synthetic minimal medium lacking specific nutrients have been described previously (52). M199 medium (Life Technologies, Rockville, Md.) was buffered with 150 mM HEPES and adjusted to pH 4.0 or 8.0. To induce hyphal growth, cells were grown for the indicated times and temperatures in YPAD plus 10% bovine calf serum (Sigma, St. Louis, Mo.), YPA (YPAD without glucose) plus 4 mM N-acetyl-d-glucosamine (GlcNAc), solid Spider medium (31), milk-Tween agar (24), or under embedded conditions in YPS (YPA containing 2% sucrose [9]). All media contained 80 mg of uridine/liter except when Ura+ prototrophs were being selected. Strains grown on Spider and milk-Tween agar plates were incubated in petri dish sleeves with damp paper towels to maintain a humid environment.
Morphological observations.
Calcofluor White (Sigma) staining was performed by incubating formaldehyde-fixed cells in 0.1 mg of Calcofluor/ml for 10 min at room temperature and washing them with phosphate-buffered saline. To localize Fkh2p-GFP and nuclear DNA (see Fig. 8), strain YJB5535 was grown overnight in SDC containing 1 μg of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI)/ml. Cells were diluted into SDC containing 1 μg of DAPI/ml and grown to mid-logarithmic phase. Differential interference contrast (DIC) and epifluorescence microscopy were performed with the use of a Nikon Eclipse E600 photomicroscope equipped with a standard UV filter set and an Endow GFP bandpass emission filter set (Chroma Technology Corporation, Brattleboro, Vt.). Digital images were collected with a DVC-1310 digital camera (Digital Video Camera Company, Austin, Tex.), captured to a Pentium III 550-MHz computer by using C-View V2.1 software (Digital Video Camera Company), and processed with the use of Image J (National Institutes of Health, Bethesda, Md.) and Adobe Photoshop (Adobe Systems, San Jose, Calif.). C. albicans cells were prepared for electron microscopy by using the PIPES-KMO4 protocol detailed by Wright (60).
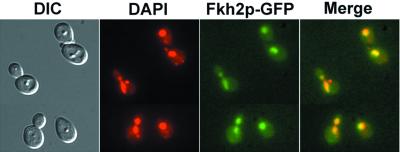
FIG. 8.
Fkh2p-GFP localizes to the nucleus. A strain expressing GFP-tagged Fkh2p (YJB5535) was grown overnight in SDC containing 1 μg of DAPI/ml. Cells were diluted into SDC containing 1 μg of DAPI/ml and were grown at 30°C to mid-logarithmic phase. DIC and fluorescent micrographs were taken of unfixed cells. DAPI images were false-colored red, and Fkh2p-GFP images were false-colored green for merging.
RNA isolation.
Cells (10- to 25-ml cultures) were rapidly harvested by centrifugation, washed in H20, and flash frozen in liquid nitrogen. Pellets were thawed in 0.5 ml of extraction buffer (100 mM Tris-HCl [pH 7.6], 100 mM LiCl, 1% sodium dodecyl sulfate [SDS], 50 mM EDTA) and lysed by vortexing (15 min, 4°C) with 1 ml of 0.45-μ acid-washed glass beads and 0.8 ml of phenol:chloroform:isoamyl alcohol (25:24:1). The aqueous phase was phenol extracted, ethanol precipitated, and resuspended in H20.
Microarray construction.
Primer pairs were used to amplify portions of 319 C. albicans ORFs (see supplementary material, designated Table 2A, that is located at http://www.cbs.umn.edu/labs/berman/eu.cell.sup.htm) from SC5314 genomic DNA. The PCR fragments were purified on MultiScreen-PCR 96-well filtration units (Millipore, Bedford, Mass.) and were eluted in spotting buffer (3× SSC [(1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] plus 0.01% SDS). Purified PCR products were spotted in triplicate on poly-l-lysine-coated slides with a MicroGrid II (BioRobotics, Cambridge, United Kingdom). Slides were postprocessed with succinic anhydride in 1-methyl-2-pyrrolidinone (http://www.microarrays.org/protocols.html).
Microarray analysis.
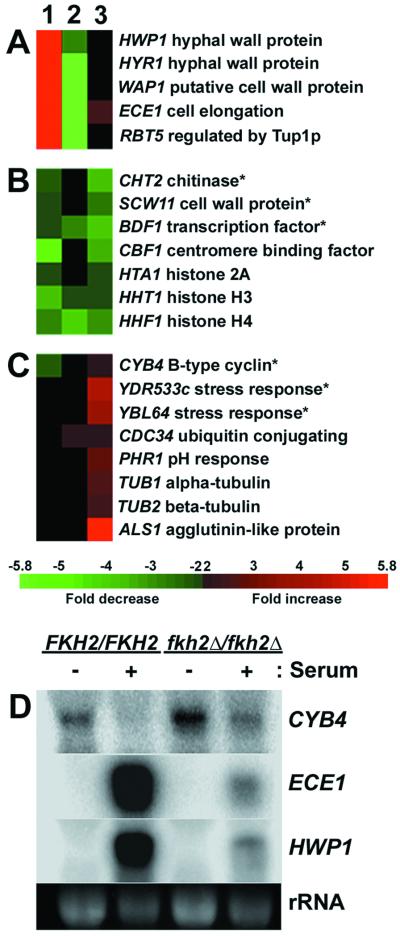
Microarray experiments were performed by using modified protocols from J. DeRisi (http://www.microarrays.org/protocols.html). For detailed protocols see the supplementary material at http://www.cbs.umn.edu/labs/berman/eu.cell.sup.htm. In brief, 10 μg of total RNA was annealed with oligo(dT) primer (Invitrogen) followed by reverse transcription in the presence of 5-(3-aminoallyl)-2′-deoxyuridine 5′-triphosphate (aa-dUTP). Samples were coupled with Cy3 or Cy5 monoreactive dye (Amersham Biosciences, Piscataway, N.J.) and were mixed. Labeled probe was resuspended in hybridization buffer (DIG Easy Hyb [Roche, Indianapolis, Ind.] containing 0.5-mg sheared salmon sperm DNA [Ambion Inc., Austin, Tex.]), heated at 65°C for 2 min, applied to the microarray, and incubated at 37°C for 12 to 16 h. Arrays were visualized on a ScanArray 5000 microarray scanner (Packard Biosci, Meriden, Conn.). Scanned images were quantitated with QuantArray software (Packard Biosci), normalizing each signal for total fluorescence. Fold induction and repression values were colorized with Array File Maker 4.0 software (8). In Fig. 9, one of two experiments is shown for experiment 1, one of two experiments is shown for experiment 2, and one of seven experiments is shown for experiment 3. Genes that displayed a consistent increase or decrease in expression that was independent of the fluor label used are shown.
FIG. 9.
Fkh2p affects transcription of a subset of genes. (A to C) A total of 319 ORFs on a C. albicans cDNA microarray were competitively hybridized with Cy3- and Cy5-labeled cDNA derived from the following strains: (1) YJB6284 (FKH2/FKH2) grown for 3 h in YPAD plus 10% serum at 37°C and YJB6284 grown in YPAD at 30°C; (2) YJB6292 (fkh2Δ/fkh2Δ) grown for 3 h in YPAD plus 10% serum at 37°C and YJB6284 grown for 3 h in YPAD plus 10% serum at 37°C; and (3) YJB6292 grown in YPAD at 30°C and YJB6284 grown in YPAD at 30°C. Fold induction or repression is indicated by the color scale where red indicates an increase and green indicates a decrease in expression. Black indicates a less than twofold change. (A) Genes with increased expression in FKH2/FKH2 but not fkh2Δ/fkh2Δ cells when grown under hyphae-inducing conditions. (B) Genes with decreased expression in fkh2Δ/fkh2Δ cells compared to that of FKH2/FKH2 cells grown under yeast growth conditions. (C) Genes with increased expression in fkh2Δ/fkh2Δ cells compared to that of FKH2/FKH2 cells grown under yeast growth conditions. Asterisks indicate C. albicans genes similar to S. cerevisiae genes that display altered expression in S. cerevisiae fkh1 fkh2 mutants. (D) Northern analysis is consistent with microarray data. Northern analysis of RNA from YJB6284 and YJB6292 grown in the absence (−) or presence (+) of serum, as described above, is depicted. CYB4, ECE1, and HWP1 probes were prepared as described in Materials and Methods. rRNA was used as a loading control.
Northern analysis.
Twenty micrograms of total RNA was resolved by electrophoresis through a formaldehyde-containing agarose gel (48). Separated RNA was transferred to GeneScreen Plus (NEN Life Science, Boston, Mass.). Probes were made from PCR-amplified fragments by using SC5314 genomic DNA as template and the following primer pairs: HWP1F (5′-TCAATTGGGGCCACTGTC-3′) and HWP1R (5′-TGGAATCCAATCGGTTGG-3′), ECE1F (5′-AGATGTTGCTCCAGCTGC-3′) and ECE1R (5′-AACAGTTTCCAGGACGCC-3′), and CYB4F (5′-ATTGCCAATTTGAGCAGC-3′) and CYB4R (5′-CCACATTGGTTGCATGGG-3′). Fragments were radiolabeled with [γ-32P]dCTP by using the Random Primed DNA Labeling kit (Roche). Blots were prehybridized for 4 h at 50°C in hybridization solution (DIG Easy Hyb with 100 μg of sheared salmon sperm DNA/ml) and then incubated in fresh hybridization solution containing radiolabeled probe for 16 h at 50°C. Blots were washed twice in solution 1 (2× SSC, 0.1% SDS) for 15 min at 23°C, twice in solution 2 (0.1× SSC, 0.1% SDS) for 15 min at 23°C, and once in solution 2 for 60 min at 50°C. Bands were visualized by phosphorimage analysis.
Epithelial and endothelial cell damage assays.
The ability of the various mutants to damage the HOK-16B-BaP-T1 oral epithelial cell line (42) or human umbilical vein endothelial cells in a 96-well tissue culture plate was determined by using the chromium release assay described previously (43). Both cell types were loaded with 51Cr and were infected for 3 h with prototrophic FKH2/FKH2, fkh2Δ/FKH2, or fkh2Δ/fkh2Δ strains in RPMI 1640 medium. The inoculum for the epithelial cells was 105 cells per well, and the inoculum for the endothelial cells was 4 × 104 cells per well.
RESULTS
C. albicans has a single FKH homolog.
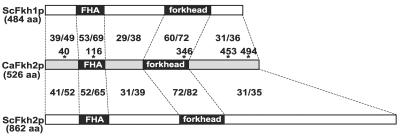
In a search of the C. albicans genomic sequence data generated by the Stanford Technology Group (http://www-sequence.stanford.edu/group/candida/index.html), we identified one ORF, orf6.8625, that was closely related to S. cerevisiae FKH1 and FKH2. Like ScFkh1p and ScFkh2p, CaFkh2p contains a forkhead-associated domain which is thought to participate in the binding of phosphoproteins (reviewed in reference 29). While full-length orf6.8625 is equally similar to both ScFkh1p and ScFkh2p, it is significantly more similar to ScFkh2p within its forkhead DNA-binding domain (72% identical and 82% similar) than it is to that of ScFkh1p (60% identical and 72% similar) (Fig. 1). Thus, we refer to orf6.8625 as FKH2.
FIG. 1.
Protein sequence comparison of C. albicans Fkh2p (CaFkh2p) and S. cerevisiae Fkh1 and Fkh2p (ScFkh1p, ScFkh2p). Percent identity/percent similarity values are shown for regions between dashed lines. Values were obtained by using the GAP program from the Wisconsin Package, version 10.2 (Genetics Computer Group, Madison, Wis.). Forkhead DNA-binding domains (forkhead) and forkhead-associated domains (FHA) are shown in black boxes. Asterisks represent positions of CUG codons in the CaFkh2p sequence. aa, amino acids.
While searching the C. albicans genome for Fkhp homologs, we identified two additional ORFs coding for putative forkhead domain-containing proteins, orf6.7358 and orf6.1892. These two ORFs display higher identity to S. cerevisiae Hcm1p and Fhl1p, respectively, than to either ScFkh protein: orf6.7358 is most similar to Hcm1p (31% identical and 37% similar), while orf6.1892 is most similar to Fhl1p (27% identical and 36% similar). Thus, it appears that the C. albicans genome includes a single ScFKH1- or ScFKH2-like ORF.
CaFKH2 complements a Scfkh1 Scfkh2 strain.
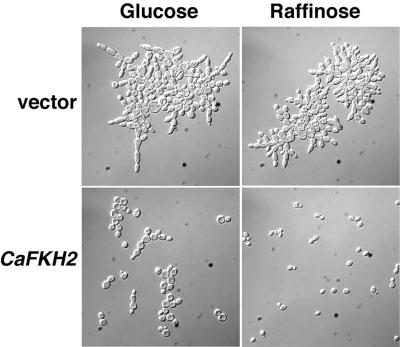
To ask if the C. albicans FKH2 gene encodes a protein with functions like those of S. cerevisiae Fkh1p and FKh2p, we expressed CaFKH2 from the ScGAL1 promoter in an S. cerevisiae fkh1Δfkh2Δ strain (22). The GAL1 promoter directs high levels of transcription when cells are grown in galactose, low-to-moderate levels of transcription in raffinose, and only very low levels of transcription when cells are grown in glucose. The S. cerevisiae fkh1Δfkh2Δ strain carrying only the GAL1 vector in the presence of either glucose, raffinose, or galactose grew with a pseudohyphal morphology, forming elongated cells that remain attached and that were generally larger than wild-type cells (Fig. 2 and data not shown). In contrast, in the presence of raffinose PGAL1-CaFKH2 restored Scfkh1Δfkh2Δ cells to the ovoid morphology, cell size, and separation characteristics of wild-type yeast cells (Fig. 2). Even in the presence of glucose, Scfkh1Δfkh2Δ cells carrying PGAL1-CaFKH2 were rounder, less enlarged, and less attached than cells carrying only vector (Fig. 2). Because C. albicans reads the CUG codon as serine rather than leucine (49), these results indicate that the substitution of five leucines for serines in CaFKH2 (Fig. 1) did not obviate the ability of the protein to functionally complement the lack of S. cerevisiae Fkh proteins. However, S. cerevisiae cells expressing PGAL1-CaFKH2 in the presence of galactose arrested as large-budded yeast cells (data not shown). The inability of S. cerevisiae cells to tolerate very high levels of CaFKH2 transcripts may be due to the high levels of CaFKH2 expression from the GAL1 promoter, since extra copies of ScFKH2 expressed from its own promoter are tolerated in both the w303 and Σ strain backgrounds (22, 61). Alternatively, the substitution of leucine for serine at some of the conserved positions may confer negative properties on CaFkh2p when it is overexpressed in S. cerevisiae.
FIG. 2.
CaFKH2 suppresses the cell morphology defect of an S. cerevisiae fkh1Δfkh2Δ strain. Strain CFY147 (fkh1Δfkh2Δ) was transformed with pYES2 (vector) or pYES-CaFKH2 (PGAL1-CaFKH2) and grown in the presence of 2% glucose or 2% raffinose liquid media. Exponentially growing cells were viewed with DIC optics.
C. albicans strains lacking Fkh2p do not form round yeast cells.
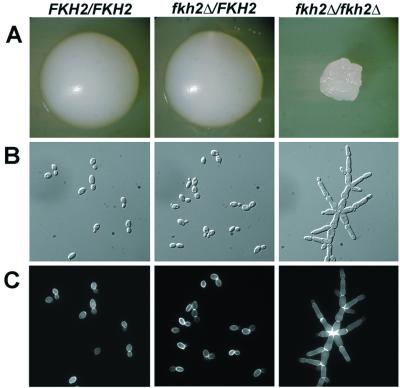
To analyze the phenotypes of C. albicans strains lacking Fkh2p, we generated several independent strains (prototrophic for all nutritional markers) in which both copies of the FKH2 ORF were deleted (see Materials and Methods for details). Colonies of fkh2Δ/fkh2Δ strains were irregularly shaped, wrinkled, dry, and crunchy (in contrast to the round, smooth, moist, soft wild-type colonies) (Fig. 3A). It was difficult to restreak such colonies in a manner that assured that new colonies arose from a single progenitor cell. The growth rate of the fkh2Δ/fkh2Δ strains, determined from colony size and time to generate saturated cultures, was also reduced relative to the growth of wild-type cells. Accurate quantitation of growth rates in liquid cultures was impeded by the fact that fkh2Δ/fkh2Δ cells tended to flocculate extensively.
FIG. 3.
fkh2Δ/fkh2Δ cells form irregular colonies and pseudohyphal cells on yeast medium. (A) Colony morphology of fkh2Δ/fkh2Δ cells. YJB6284 (FKH2/FKH2), YJB6290 (fkh2Δ/FKH2), and YJB6292 (fkh2Δ/fkh2Δ) cells were grown on YPAD agar medium for 2 days at 30°C. (B and C) Cellular morphology of fkh2Δ/fkh2Δ cells. Strains depicted in panel A were grown in YPAD liquid medium to mid-logarithmic phase and were stained with Calcofluor White to visualize chitin distribution. DIC images (B) and fluorescence micrographs (C) are shown.
Individual fkh2Δ/fkh2Δ cells were elongated, were attached to one another in large clumps, and were, on average, twice as long as wild-type cells grown under similar yeast growth conditions (Fig. 3B). These elongated cells had constricted septa (as detected with Calcofluor White; Fig. 3C), which are diagnostic of pseudohyphal rather than true hyphal cells. The morphology of heterozygous fkh2Δ/FKH2 strains was indistinguishable from that of the parental FKH2/FKH2 strain (Fig. 3). Deleting both copies of the putative C. albicans HCM1 gene did not affect colony or cellular morphology (data not shown). Therefore, consistent with sequence data, C. albicans contains a single Fkh-like protein affecting cellular morphology.
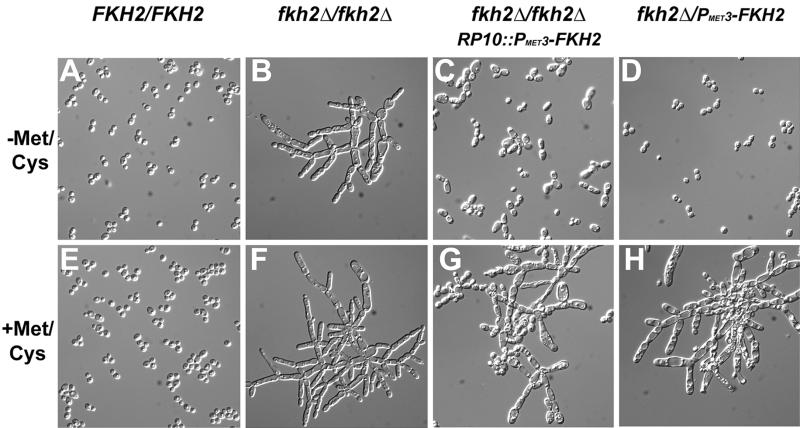
FKH2 was reintroduced into the fkh2Δ/fkh2Δ strains by insertion of FKH2 under control of the conditional MET3 promoter at the RP10 locus (12). When expression was induced (in the absence of methionine and cysteine), cells grew with a yeast-like morphology (Fig. 4C) and colonies were smooth and indistinguishable from wild-type colonies (data not shown). Repression of this ectopic copy of PMET3-FKH2, by the addition of methionine and cysteine, resulted in a pseudohyphal morphology like that of fkh2Δ/fkh2Δ strains (Fig. 4G) and a return of the wrinkled, crunchy colony phenotype (data not shown). In addition, we constructed a strain carrying PMET3-FKH2 at the FKH2 locus. This strain also grew with a yeast morphology and smooth colony phenotype when expression was induced (Fig. 4D) and exhibited a pseudohyphal morphology and wrinkled colony phenotype when expression was repressed (Fig. 4H). The presence or absence of methionine and cysteine did not affect the morphology of either wild-type or fkh2Δ/fkh2Δ strains (Fig. 4A, B, E, and F). The similar phenotypes of cells lacking both copies of FKH2 (Fig. 3) and cells not expressing FKH2 (Fig. 4G and H), as well as restoration of yeast-form growth when FKH2 was reintroduced at two different loci (Fig. 4C and D), indicates that the pseudohyphal morphology and wrinkled colony phenotypes were caused by a lack of Fkh2p and not by some other mutation in the strains. Furthermore, these results imply that FKH2 is required for the formation of ovoid yeast cells.
FIG. 4.
PMET3-FKH2 restores wild-type cellular morphology to fkh2 mutant cells. YJB6284 (FKH2/FKH2), YJB6292 (fkh2Δ/fkh2Δ), YJB6403 (fkh2Δ/fkh2ΔRP10::PMET3-FKH2), and YJB4828 (fkh2Δ/PMET3-FKH2) cells were grown to saturation in SDC-methionine-cysteine-uridine at 30°C. Cells were diluted into either SDC-methionine-cysteine-uridine (−Met/Cys) or SDC plus 10 mM methionine plus 2 mM cysteine-uridine (+Met/Cys) and were grown overnight at 30°C. DIC images are shown.
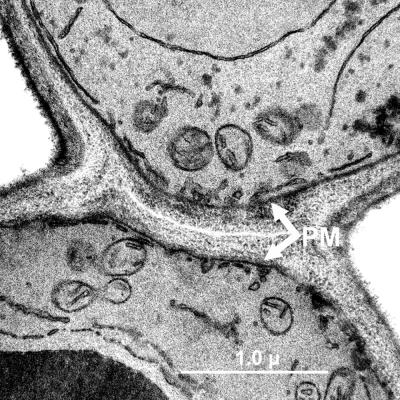
Cells lacking Fkh2p appear to have intact septa, as indicated by Calcofluor staining (Fig. 3C), although they remain attached, forming large clusters of cells that did not readily disperse after mild agitation (vortexing). Electron microscopy of fkh2Δ/fkh2Δ strains reveals that fkh2Δ/fkh2Δ cells, like wild-type pseudohyphal cells, remain attached by cell wall material while there is no plasma membrane connection between the mother and daughter cells (Fig. 5). Thus, fkh2 mutant strains undergo cytokinesis and septation but not cell separation.
FIG. 5.
fkh2Δ/fkh2Δ cells are defective in cell separation. Transmission electron micrograph of YJB6292 (fkh2Δ/fkh2Δ) cells grown in YPAD liquid medium to mid-logarithmic phase at 30°C. White arrows indicate plasma membrane (PM).
C. albicans strains lacking Fkh2p do not form true hyphae.
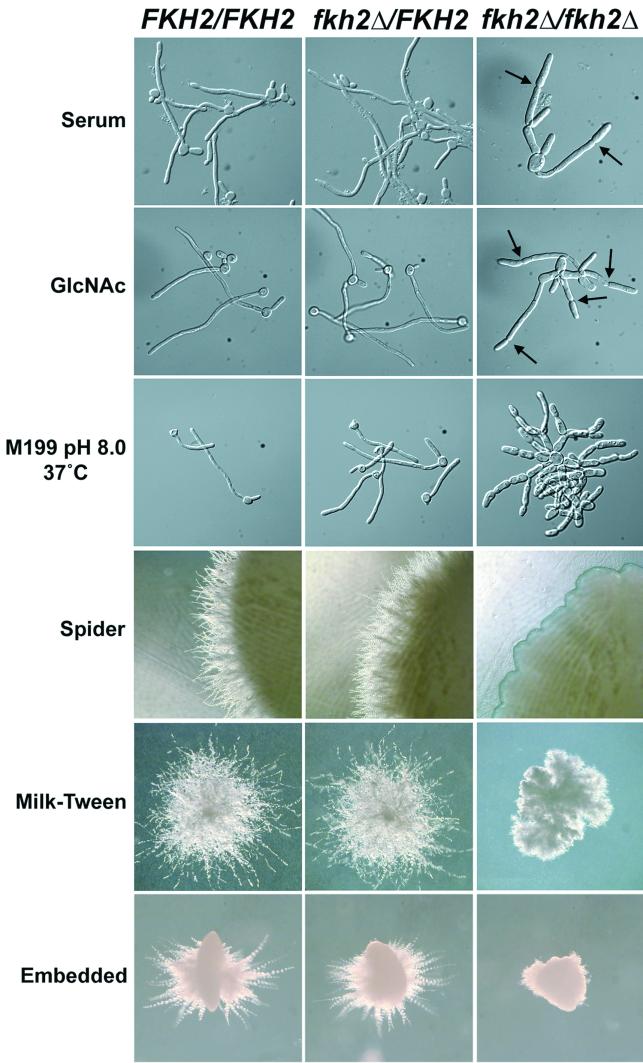
We grew fkh2Δ/fkh2Δ strains under several different conditions that stimulate hyphal growth. First, we used liquid medium containing serum or GlcNAc, strong inducers of true hyphal growth. In the presence of either inducer, wild-type cells and fkh2Δ/FKH2 heterozygotes formed true hyphae, while fkh2Δ/fkh2Δ strains formed elongated cells with a pseudohyphal morphology. The fkh2Δ/fkh2Δ cells were wider than hyphal cells and included distinct septal constrictions (Fig. 6, arrows). On solid media, such as Spider and milk-Tween agar, wild-type and fkh2Δ/FKH2 heterozygotes formed highly filamentous colonies, while the colonies formed by fkh2Δ/fkh2Δ strains were much less filamentous (Fig. 6). These media are thought to induce hyphal growth in response to low levels of nitrogen or other critical nutrients (15). When cells were embedded in agar media, a condition that induces hyphae in response to contact with matrix (9), fkh2Δ/fkh2Δ cells displayed significantly fewer filaments emanating from the embedded colony than did wild-type cells (Fig. 6). Furthermore, wild-type cells were able to form true hyphae under embedded conditions while fkh2Δ/fkh2Δ cells grew as chains of budding cells (data not shown). Thus, C. albicans FKH2 is required for the formation of true hyphal cells under many different hyphal induction conditions.
FIG. 6.
Fkh2p affects the formation of true hyphae. YJB6284 (FKH2/FKH2), YJB6290 (fkh2Δ/FKH2), and YJB6292 (fkh2Δ/fkh2Δ) cells were grown under different hyphae-inducing conditions. For the panels labeled serum and GlcNAc, cells were grown to saturation in YPAD at 30°C, diluted into YPAD plus 10% serum or YPA plus 4 mM GlcNAc, and incubated for 4 h. For the panel labeled M199, cells were grown to saturation in M199 (pH 4.0 at 37°C), diluted into M199 (pH 8.0), and incubated at 37°C for 4 h. For the panel labeled Spider, cells were grown to saturation in YPAD at 30°C, plated onto solid Spider medium, and incubated for 7 days at 30°C. For the panel labeled milk-Tween, cells were grown to saturation in YPAD at 30°C, plated onto milk-Tween agar plates, and incubated for 7 days at 37°C. For the panel labeled embedded, cells were grown to saturation in YPAD at 30°C, diluted into cooled, molten YPS agar, and incubated for 3 days at 28°C. Arrows indicate septal constrictions.
Interestingly, when strong inducers of hyphal growth such as serum or GlcNAc were used, fkh2Δ/fkh2Δ pseudohyphal cells were more elongated than when they were grown on yeast media. The average cell length was 15.75 ± 8.27 μm (n = 58) and 19.11 ± 12.59 μm (n = 50) on rich medium with serum or GlcNAc, respectively, compared to 11.35 ± 1.66 μm (n = 61) on rich medium alone. The high standard deviations for cell length in serum or GlcNAc-treated cells reflects the observation that ∼50% of the cells were similar in size to cells grown on rich medium alone, while the other ∼50% of the cells were up to four times longer than the rich-medium-grown cells. This indicates that about half of the fkh2Δ/fkh2Δ cells responded to signals from serum or GlcNAc that mediate polarized growth, while they did not respond to the signals that mediate cell cycle changes, such as altered septum placement, nuclear migration, and cell width properties of true hyphal cells.
In contrast, when weaker hyphal induction conditions were used no increased polarized growth of fkh2 strains was observed. In liquid M199 medium, which induces true hyphae in response to pH and temperature (pH 8.0 at 37°C), fkh2Δ/fkh2Δ cells grew as pseudohyphae with dimensions (average cell length, 11.52 ± 3.73 μm [n = 54]) similar to those of fkh2Δ/fkh2Δ cells grown on M199 under yeast growth conditions (pH 4.0 at 37°C; average length, 10.87 ± 1.92 μm [n = 53]) (Fig. 6 and data not shown). Furthermore, cells removed from colonies of fkh2Δ/fkh2Δ strains grown on milk-Tween agar or Spider medium or under embedded conditions did not show increased cell length (data not shown). Thus, unlike the strong hyphal inducers, weaker inducers do not stimulate an FKH2-independent polarized growth response.
Pseudohyphal growth of fkh2Δ/fkh2Δ cells does not require Efg1p or Cph1p.
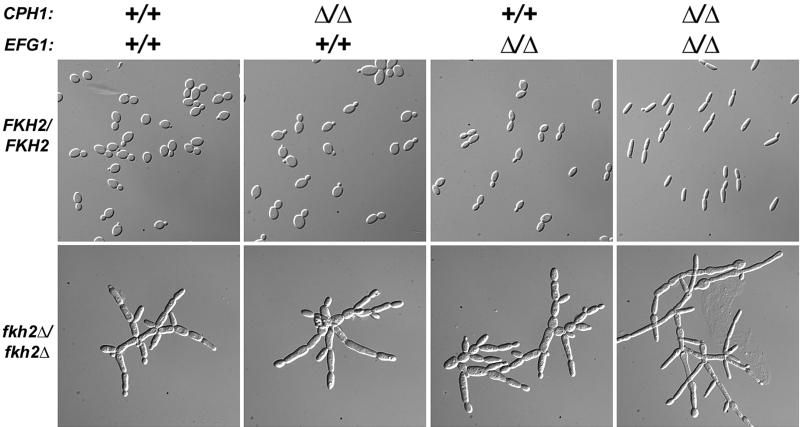
To determine the relationship between Fkh2p and the Efg1p and Cph1p transcription factors, we constructed mutants lacking FKH2 as well as EFG1 and/or CPH1 and grew them under conditions that promote yeast form growth. The fkh2Δ/fkh2Δcph1Δ/cph1Δ, fkh2Δ/fkh2Δefg1Δ/efg1Δ, and fkh2Δ/fkh2Δcph1Δ/cph1Δefg1Δ/efg1Δ mutant strains formed pseudohyphal cells that resembled fkh2Δ/fkh2Δ single mutants: they were generally elongated and remained attached to one another in a branching pattern (Fig. 7). These results indicate that the role of Fkh2p in morphogenesis is not dependent upon either Efg1p or Cph1p. Rather, Fkh2p acts downstream of and/or parallel to these transcription factors which regulate morphogenesis in response to specific environmental stimuli.
FIG. 7.
Epistasis analysis of fkh2. CAF2 (FKH2/FKH2 CPH1/CPH1 EFG1/EFG1), JKC19 (cph1Δ/cph1Δ), HLC52 (efg1Δ/efg1Δ), HLC54 (cph1Δ/cph1Δefg1Δ/efg1Δ), YJB6292 (fkh2Δ/fkh2Δ), YJB6441 (fkh2Δ/fkh2Δcph1Δ/cph1Δ), YJB6413 (fkh2Δ/fkh2Δefg1Δ/efg1Δ), and YJB5802 (fkh2Δ/fkh2Δefg1Δ/efg1Δcph1Δ/cph1) cells were grown to mid-logarithmic phase at 30°C in YPAD. DIC micrographs are shown.
Fkh2p-GFP localizes to the nucleus throughout the cell cycle.
If Fkh2p is a transcription factor, it is expected to localize to the nucleus for at least some portion of the cell cycle. We analyzed the localization of C. albicans Fkh2p by generating a Fkh2p-GFP fusion protein at the FKH2 locus in C. albicans strain BWP17. This fusion protein is fully functional as determined by the smooth colony and round yeast cell morphology of a FKH2-GFP/fkh2Δ strain. Fkh2p-GFP was detected as a single major fluorescent spot in each cell that was stretched across the mother-bud neck in cells undergoing mitosis (Fig. 8). The Fkh2p-GFP fluorescence colocalized with the major region of DAPI-stained DNA (Fig. 8). (DAPI stains mitochondrial DNA as well as nuclear DNA. Fkh2p-GFP did not colocalize with the mitochondrial DNA.) Thus, Fkh2p, like S. cerevisiae Fkh1p and Fkh2p and S. pombe Sep1p (44, 62), is a nuclear protein.
Fkh2p is required for the expression of several hyphae-specific and yeast-specific genes.
In S. cerevisiae, the Fkh1 and Fkh2 proteins are required for the periodic transcription of the CLB2 cluster of cell-cycle-regulated genes. To begin to study the role of C. albicans Fkh2p in transcriptional regulation, we constructed a DNA array composed of 319 PCR products amplified from C. albicans genomic DNA with gene-specific primers (see Materials and Methods for details). Among the genes on this miniarray were genes reported to affect filamentous growth as well as genes that were the apparent homologs of S. cerevisiae genes whose transcription patterns were altered in Scfkh1 and Scfkh2 mutants. (A complete list of genes used on the miniarray is provided in Table 2A of the online supplementary material at http://www.cbs.umn.edu/labs/berman/eu.cell.sup.htm). RNA was isolated from asynchronous wild-type and fkh2Δ/fkh2Δ cultures under yeast (YPAD, 30°C) or hyphal (YPAD plus 10% serum, 37°C) growth conditions, reverse transcribed, labeled with Cy3 or Cy5 fluors, and hybridized in pair-wise combinations to the miniarrays. Representative results for mRNAs that exhibited consistent changes in their expression levels in all of the experiments performed are shown in Fig. 9.
In wild-type cells, RNAs whose expression levels increased dramatically upon hyphal induction (Fig. 9A, column 1) encode proteins, such as Hwp1p, Hyr1p, Wap1p, Ece1p, and Rbt5p, which are all predicted to be GPI-modified cell wall proteins. Northern analysis of several of these mRNAs confirmed the array results (Fig. 9D). These results are consistent with previous reports from others that use microarrays or conventional Northern blots (5, 28, 38, 39).
Interestingly, under serum induction conditions these hyphae-specific genes were expressed at much lower levels in fkh2 mutants than in the wild-type parent (Fig. 9A, column 2). This indicates that serum treatment of fkh2 mutants did not result in the dramatic induction of genes that, in wild-type cells, are associated with hyphal growth and cell elongation.
Under yeast growth conditions, several C. albicans RNAs encoding apparent homologs of S. cerevisiae mRNAs that exhibit altered transcription levels in S. cerevisiae fkh1 and fkh2 mutants were also affected in CaFKH2 mutants (Fig. 9B and C, column 3). Those whose transcript levels increased in fkh2 mutants include homologs of S. cerevisiae YDR533 (orf6.1991) and PRX1/YBL064 (orf6.6956), which have roles in stress response, and CYB4 (orf6.8086), which encodes a B-cyclin. Those whose transcript levels decreased in fkh2 mutants relative to wild-type strains include homologs of S. cerevisiae SCW11/YGL028 (orf6.2346), CTS1 (CHT2/orf6.2344), and BDF1 (orf6.8227), which are predicted to encode a cell wall protein, a chitinase, and a transcription factor, respectively.
The increased level of CYB4 mRNA in the fkh2 mutant strains was confirmed on Northern blots (Fig. 9D) and suggests that Fkh2p, directly or indirectly, is a negative regulator of CYB4 transcription in these asynchronous cultures. Other genes expressed at higher levels in fkh2 strains than in wild-type control cultures were CDC34 (orf6.7631), PHR1 (orf6.7524), TUB1, TUB2, and one or more of the ALS1 to -5 genes (the ALS1 probe used on this array is expected to cross-react with ALS family members 1 through 5; Lois Hoyer, personal communication).
Several genes were expressed at lower levels in fkh2 mutant cells. Among these were CBF1/CP1 (orf6.4385), an apparent homolog of the S. cerevisiae CBF1 centromere-binding protein, and genes encoding histones H2A (orf6.7310), H3 (orf6.8840), and H4 (orf6.4930). Reduced levels of these genes may reflect the slower growth of fkh2 mutant cells. Similarly, increased levels of PHR1, a gene repressed under acidic conditions (50), may reflect the slower growth and reduced acidification of the medium by fkh2 mutant strains.
Fkh2p is required for damage to human tissue culture cells.
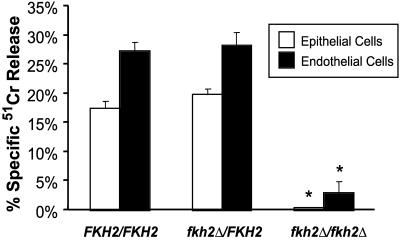
Because cells lacking Fkh2p grow slower than wild-type cells, we analyzed the ability of fkh2 strains to damage cells in a tissue culture assay for cell damage that is not dependent upon cell growth. The results are shown in Fig. 10. Both the parental and the heterozygous strains caused significant damage to oral epithelial and vascular endothelial cells. In contrast, the fkh2Δ/fkh2Δ strain caused virtually no damage to these human host cells (Fig. 10). Thus, Fkh2p is required for C. albicans cells to damage human epithelial and endothelial cells in vitro.
FIG. 10.
Fkh2p is required for epithelial and endothelial cell injury. The HOK-16B-BaP-T1 oral epithelial cell line and human umbilical vein endothelial cells were incubated with YJB6284 (FKH2/FKH2), YJB6288 (fkh2Δ/FKH2), and YJB6292 (fkh2Δ/fkh2Δ) cells. The extent of epithelial cell damage was measured with a chromium release assay. Results are the averages ± standard deviations of triplicate measurements. Asterisks indicate a P value of <0.001 compared to that for cells infected with the FKH2/FKH2 strain by analysis of variance.
DISCUSSION
We have identified a single C. albicans Fkh-like protein that participates in cellular morphogenesis. Depleting cells of Fkh2p resulted in constitutive pseudohyphal growth, suggesting that it functions in the formation of both true hyphal as well as yeast cells. The pseudohyphal growth of fkh2 mutants did not require either Efg1p or Cph1p, suggesting that Fkh2p acts in a pathway that is downstream and/or parallel to these transcription factors. Consistent with its proposed action as a transcription factor, Fkh2p localized to the nucleus. Furthermore, the transcription of a subset of genes, including CYB4 (which encodes a B-type cyclin), genes required for efficient cell separation, and hyphae-specific genes, was altered in fkh2 mutants. Among the Fkh2p-regulated genes were homologs of genes regulated in a cell-cycle-specific manner in S. cerevisiae. These results indicate that Fkh2p is important for C. albicans morphogenesis, potentially through its role as a transcription factor regulating genes in a cell-cycle-specific manner.
Fkh2p contributes to the ability of C. albicans cells to damage human epithelial and endothelial cells in vitro. Since cells lacking Fkh2p grow slowly in vitro, in vivo virulence experiments using fkh2Δ/fkh2Δ strains are unlikely to produce meaningful results. We therefore performed in vitro cell damage assays that are not dependent upon growth rates. The reduction in cell damage by fkh2 mutant cells is likely due to multiple effects of Fkh2p. The fkh2Δ/fkh2Δ strain grew as pseudohyphae on the epithelial and endothelial cells, whereas the FKH2/FKH2 and fkh2Δ12/FKH2 strains formed true hyphae. A tup1 mutant, which also grows only as pseudohyphae, is similarly defective in its ability to damage endothelial cells (43). Thus, pseudohyphae appear to have a significantly reduced capacity to damage host cells. It is also possible that the cell cycle defects of fkh2 mutant strains contribute to their inability to cause host cell damage.
The absence of CaFkh2p resulted in constitutive pseudohyphal growth and changes in the transcription of a group of genes involved in cell cycle progression. Specifically, CaFkh2p appears to be a transcriptional regulator of Cyb4p, a B-cyclin most similar to S. cerevisiae Clb3p and Clb4p. Interestingly, strains lacking Cyb4p, like strains lacking Fkh2p, exhibit a constitutive pseudohyphal morphology (E. S. Bensen and J. Berman, unpublished data). The increase in CYB4 mRNA observed in asynchronous cultures of fkh2Δ/fkh2Δ cells suggests that Fkh2p is a negative regulator of CYB4 transcription. Future studies comparing genome-wide transcription profiles following cell cycle progression in synchronous wild-type and fkh2Δ/fkh2Δ cells will be needed to address the mechanisms by which Fkh2p regulates CYB4 transcript levels. Expression levels of CYB1 (orf6.7127), which encodes a B-type cyclin most similar to S. cerevisiae Clb1p and Clb2p, were not consistently detected by either microarray or Northern analysis.
In addition to CYB4 mRNA, a gene closely related to ScCDC34 which encodes the ubiquitin conjugating component of the SCF complex is negatively regulated by Fkh2p under both yeast and hyphal conditions. The SCF complex is required for the degradation of many proteins at specific cell cycle stages. ScCDC34 transcript levels are not regulated in a cell-cycle-dependent manner, nor are they regulated by ScFkh1 and ScFkh2p in synchronous or asynchronous cells. Perhaps C. albicans, unlike S. cerevisiae, regulates SCF function at the transcriptional level. Consistent with this idea, Murad and coworkers found that CaCDC34 mRNA levels increased 2.17-, 3.62-, and 3.32-fold in mig1, nrg1, and tup1 mutant strains, respectively, relative to the isogenic wild-type parental strains (38, 39).
Electron microscopy studies indicate that fkh2Δ/fkh2Δ cells fail to efficiently separate after cytokinesis, retaining cell wall material between the mother-daughter cells. Consistent with this observation, we found that CaFkh2p positively regulates several genes important for cell separation, such as CaCht2p, an endochitinase important for cell separation in C. albicans yeast form cells (26) that is expressed at higher levels in yeast than in hyphal cells (34). CaScw11p/YGL028p is most similar to ScScw11p, a protein with similarity to glucanases that is important for cell separation in S. cerevisiae (11). ScCts1p and ScScw11p, the S. cerevisiae proteins most closely related to Cht2p and Scw11p, are members of the SIC1 cluster, which is indirectly regulated by ScFkh1 and ScFkh2p (54). Thus, it is likely that the cell separation defect seen in fkh2Δ/fkh2Δ cells is due to reduced expression of CHT2 and SCW11. Furthermore, our results confirm the previous report that CHT2 transcript levels are elevated in yeast cells relative to hyphal cells (34) and extend this observation to SCW11 as well.
Importantly, CaFkh2p is required for the formation of true hyphae and for high levels of hyphae-specific gene expression. Unlike many of the characterized genes that affect hyphal morphogenesis under a subset of induction conditions, FKH2 is required for true hyphal morphogenesis under all conditions tested, including strong inducers (serum and GlcNAc), high pH at 37°C (M199), starvation media (milk-Tween, Spider), or embedding conditions. These results support the hypothesis that Fkh2p is required to execute cell cycle events such as delayed septin ring formation and decreased cell width, events that distinguish true hyphal cells from pseudohyphal and yeast cells (56).
Interestingly, fkh2 mutants highlight differences between different hyphal induction stimuli. In response to strong hyphal inducers such as serum and GlcNAc, fkh2 mutant cells were more elongated than when they were grown under yeast growth conditions (Fig. 3 and 6), yet they exhibited the cell cycle characteristics of pseudohyphae. In contrast, in response to weaker inducers, such as high pH at 37°C, starvation, or embedded conditions, the length of fkh2 mutant cells did not increase relative to fkh2 mutant cells grown in conditions of low pH at 30°C. Thus, strong inducers activate an Fkh2p-independent polarized growth activity that is not seen when weaker inducers are used, and the Fkh2p-independent polarized growth response is separable from hyphae-specific, Fkh2p-dependent cell cycle events, such as altered septin positioning. This is consistent with the observation that hyphal evagination in response to serum occurs prior to other measurable cell cycle events (21). Furthermore, it demonstrates that serum induces a complex set of responses, including cell-cycle-independent events, Fkh2p-independent polarized growth, cell-cycle-dependent events, and Fkh2p-dependent events.
Fkh2p is required for high-level expression of hyphae-specific genes (e.g., HWP1) that previous works placed downstream of transcription factors such as Efg1p, Cph1p, Tup1p, and Cph2p (3, 15, 28, 30, 38, 39). Furthermore, our epistasis analysis suggests that Fkh2p is either downstream of and/or in a parallel pathway for morphogenesis with both Efg1p and Cph1p. Our results are consistent with those of a model in which Fkh2p is downstream of the Efg1p and Cph1p transcription factors and is upstream of several of the genes regulated by them. Since combinatorial control of many of these genes appears likely, we cannot yet determine if Fkh2p regulates these genes by binding to their promoters directly or if indirect mechanisms are involved.
An alternative model is that Fkh2p is a negative regulator of pseudohyphal growth and that this negative regulation is required for the formation of both yeast and true hyphal cells. While pseudohyphae were thought to be a cell type that is an intermediate between yeast and true hyphae (35), more recent studies indicate that true hyphae have cell cycle dynamics that distinguish them from pseudohyphal and yeast cells (56). Nonetheless, pseudohyphal growth may be a default pathway that must be repressed in order for yeast or true hyphal growth to proceed. This is consistent with the observation that strains lacking other transcriptional repressors (e.g., Tup1p, Nrg1p, and Rbf1p), like strains lacking Fkh2p, exhibit primarily pseudohyphal rather than true hyphal morphology when grown on yeast medium (4, 6, 23, 39). It will be interesting to determine the overlapping set of genes regulated by all four of these transcription factors. Furthermore, it will be important to determine if it is the altered expression of cell-cycle-regulated genes that causes the pseudohyphal morphology of cells lacking these factors.
In summary, C. albicans, like other fungi, uses a forkhead transcription factor to regulate morphogenesis. CaFkh2p appears to regulate the expression of at least one of the two C. albicans B-cyclins. Future studies will aim to determine the role of CaFkh2p in regulating the B-cyclins and other coregulated genes as yeast, pseudohyphal, and true hyphal cells progress through the cell cycle.
Acknowledgments
We thank Bridget Jacques-Fricke, Jolene Ruf, Maryam Gerami-Nejad, Michelle Dessrouseaux, and Mark McClellan for technical assistance, Ann Palmer and Gilbert Ahlstrand at the University of Minnesota Imaging Center for the preparation of electron micrograph samples, Jeff Lande and Kevin Roberg-Perez for help with bioinformatics, Angela Sanchez for performing the cell damage assays, and Vivek Kapur and Mike Paustian from the University of Minnesota Applied Genetics Analysis Center for assistance with preparation and scanning of the DNA miniarrays. We thank Catherine Fox and Gerald Fink for strains and Ted White for primers. We thank Catherine Fox, Bruce Futcher, and Peter Sudbery for providing data prior to publication and helpful discussions, and we thank Dana Davis, Cheryl Gale, Lynn Glowczewski, David Kirkpatrick, and Duncan Clarke for critical review of the manuscript. We are indebted to the Stanford Genome Technology Center for providing C. albicans genome sequence data.
This work was supported by a Burroughs Wellcome Senior Scholar Award 0677 and NIH-RO1-DE14666 to JB, NIH R01-DE13974 to SGF, and by NIH fellowships T32-AI07421 and F32-AI10647 to ESB.
REFERENCES
- 1.Bailey, D. A., P. J. Feldmann, M. Bovey, N. A. Gow, and A. J. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birse, C. E., M. Y. Irwin, W. A. Fonzi, and P. S. Sypherd. 1993. Cloning and characterization of ECE1 a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 5.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, P. C., and R. A. Calderone. 1978. Chitin synthesis in Candida albicans: comparison of yeast and hyphal forms. J. Bacteriol. 133:1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitkreutz, B. J., P. Jorgensen, A. Breitkreutz, and M. Tyers. 2001. AFM 4.0: a toolbox for DNA microarray analysis. Genome Biol. 2:0001.1-0001.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, D. H., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 10.Cabib, E., S. J. Silverman, and J. A. Shaw. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:97-102. [DOI] [PubMed] [Google Scholar]
- 11.Cappellaro, C., V. Mrsa, and W. Tanner. 1998. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180:5030-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 13.Davis, D., J. E. Edwards, Jr., A. P. Mitchell, and A. S. Ibrahim. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst, J. F. 2000. Regulation of dimorphism in Candida albicans. Contrib. Microbiol. 5:98-111. [DOI] [PubMed] [Google Scholar]
- 15.Ernst, J. F. 2000. Transcription factors in Candida albicans-environmental control of morphogenesis. Microbiology UK 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 16.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale, C. A., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell. 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 19.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 20.Harris, S. D. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4:736-739. [DOI] [PubMed] [Google Scholar]
- 21.Hazan, I., M. Sepulveda-Becerra, and H. Liu. 2002. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell 13:134-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollenhorst, P. C., M. E. Bose, M. R. Mielke, U. Muller, and C. A. Fox. 2000. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154:1533-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii, N., M. Yamamoto, F. Yoshihara, M. Arisawa, and Y. Aoki. 1997. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology 143:429-435. [DOI] [PubMed] [Google Scholar]
- 24.Jitsurong, S., S. Kiamsiri, and N. Pattararangrong. 1993. New milk medium for germ tube and chlamydoconidia production by Candida albicans. Mycopathologia 123:95-98. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, L. H., and N. N. Pearsall. 1980. Relationship between germination of Candida albicans and increased adherence to human buccal epithelial cells. Infect. Immun. 28:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, L., and G. Butler. 1998. Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr. Genet. 34:183-191. [DOI] [PubMed] [Google Scholar]
- 27.Klis, F. M., P. De Groot, and K. Hellingwerf. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39:1-8. [PubMed] [Google Scholar]
- 28.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., G. I. Lee, S. R. Van Doren, and J. C. Walker. 2000. The FHA domain mediates phosphoprotein interactions. J. Cell Sci. 113:4143-4149. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 31.Liu, H., J. Köhler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 32.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 33.McCollum, D., and K. L. Gould. 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11:89-95. [DOI] [PubMed] [Google Scholar]
- 34.McCreath, K. J., C. A. Specht, and P. W. Robbins. 1995. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc. Natl. Acad. Sci. USA 92:2544-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merson-Davies, L. A., and F. C. Odds. 1989. A morphology index for characterization of cell shape in Candida albicans. J. Gen. Microbiol. 135:3143-3152. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell, A. P. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687-692. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell, L. H., and D. R. Soll. 1979. Temporal and spatial differences in septation during synchronous mycelium and bud formation by Candida albicans. Exp. Mycol. 3:298-309. [Google Scholar]
- 38.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 39.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro-Garcia, F., M. Sanchez, C. Nombela, and J. Pla. 2001. Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol. Rev. 25:245-268. [DOI] [PubMed] [Google Scholar]
- 41.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Baillière Tindall, London, United Kingdom.
- 42.Park, N. H., B. M. Min, S. L. Li, M. Z. Huang, H. M. Cherick, and J. Doniger. 1991. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis 12:1627-1631. [DOI] [PubMed] [Google Scholar]
- 43.Phan, Q. T., P. H. Belanger, and S. G. Filler. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pic, A., F. L. Lim, S. J. Ross, E. A. Veal, A. L. Johnson, M. R. Sultan, A. G. West, L. H. Johnston, A. D. Sharrocks, and B. A. Morgan. 2000. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19:3750-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribar, B., A. Grallert, E. Olah, and Z. Szallasi. 1999. Deletion of the sep1(+) forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 263:465-474. [DOI] [PubMed] [Google Scholar]
- 46.Rotrosen, D., J. E. Edwards, Jr., T. R. Gibson, J. C. Moore, A. H. Cohen, and I. Green. 1985. Adherence of Candida to cultured vascular endothelial cells: mechanisms of attachment and endothelial cell penetration. J. Infect. Dis. 152:1264-1274. [DOI] [PubMed] [Google Scholar]
- 47.Rua, D., B. T. Tobe, and S. J. Kron. 2001. Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4:720-727. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Santos, M. A., and M. F. Tuite. 1995. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 23:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman, F. 1991. Getting started with yeast, p. 3-20. In C. Guthrie and G. R. Fink (ed.), Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif.
- 53.Soll, D. R., and L. H. Mitchell. 1983. Filament ring formation in the dimorphic yeast Candida albicans. J. Cell Biol. 96:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staab, J. F., C. A. Ferrer, and P. Sundstrom. 1996. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271:6298-6305. [DOI] [PubMed] [Google Scholar]
- 56.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 57.Sundstrom, P. 1999. Adhesins in Candida albicans. Curr. Opin. Microbiol. 2:353-357. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 59.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright, R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496-510. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]
- 62.Zilahi, E., E. Salimova, V. Simanis, and M. Sipiczki. 2000. The S. pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett. 481:105-108. [DOI] [PubMed] [Google Scholar]