Abstract
The c-Jun-like transcriptional activator Gcn4p controls biosynthesis of translational precursors in the yeast Saccharomyces cerevisiae. Protein stability is dependent on amino acid limitation and cis signals within Gcn4p which are recognized by cyclin-dependent protein kinases, including Pho85p. The Gcn4p population within unstarved yeast consists of a small relatively stable cytoplasmic fraction and a larger less stable nuclear fraction. Gcn4p contains two nuclear localization signals (NLS) which function independently of the presence or absence of amino acids. Expression of NLS-truncated Gcn4p results in an increased cytoplasmic fraction and an overall stabilization of the protein. The same effect is achieved for the entire Gcn4p in a yrb1 yeast mutant strain impaired in the nuclear import machinery. In the presence of amino acids, controlled destabilization of Gcn4p is triggered by the phosphorylation activity of Pho85p. A pho85Δ mutation stabilizes Gcn4p without affecting nuclear import. Pho85p is localized within the nucleus in the presence or absence of amino acids. Therefore, there is a strict spatial separation of protein synthesis and degradation of Gcn4p in yeast. Control of protein stabilization which antagonizes Gcn4p function is restricted to the nucleus.
In the yeast Saccharomyces cerevisiae, a large number of genes encoding enzymes of different biosynthetic pathways are coregulated by a genetic network known as the general control system of amino acid biosynthesis (23). Starvation for a single amino acid results in an increased expression and stability of the transcriptional activator Gcn4p, which subsequently upregulates the transcription of multiple target genes in various biosynthetic pathways for translational precursors. Like mammalian c-Jun, Gcn4p belongs to the bZIP family of transcription factors. Gcn4p expression is regulated at the level of translation initiation and protein stability. Four short upstream open reading frames (ORFs) prevent an efficient translation of the GCN4 mRNA under nonstarvation conditions (14). In addition, GCN4 mRNA translation is repressed by nitrogen starvation (10). When cells are starved for amino acids, uncharged tRNA molecules bind to the tRNA synthetase domain of the kinase Gcn2p. As a consequence, the kinase becomes activated and phosphorylates the α-subunit of eukaryotic initiation factor eIF-2 (8, 34). Phosphorylation inhibits eIF-2B, which normally exchanges eIF-2-bound GDP for GTP. This results in a reduced amount of active eIF-2 that is available for translation initiation (19, 33). The diminished overall translation efficiency is counteracted by increased expression of Gcn4p, because the modified translational apparatus allows the utilization of the GCN4 start codon. A strain lacking the kinase Gcn2p is not able to turn on the general control of amino acid biosynthesis in response to amino acid limitation.
Whereas translational regulation of the GCN4 mRNA has been studied for many years, the regulation of Gcn4p stability is a more recent research field. Gcn4p is a highly unstable protein with a half-life of about 5 min. Starvation for specific amino acids increases the half-life of the protein (17). Rapid degradation of Gcn4p depends on phosphorylation by cyclin-dependent protein kinases such as Pho85p. Amino acid residue Thr165 has been identified as one of the crucial phosphorylation sites. Another cyclin-dependent protein kinase which was recently identified to be involved in Gcn4p stability is Srb10p (5). Correspondingly, a pho85Δ or a srb10Δ mutation results in Gcn4p stabilization (18, 5). Phosphorylated Gcn4p subsequently serves as substrate for ubiquitinylation by the SCFCdc4 ubiquitin ligase complex.
S. cerevisiae cells are subdivided into the typical eukaryotic compartments which separate different cellular processes. The Gcn4 protein is synthesized in the cytoplasm and has to be transported into the nucleus to fulfill its transcriptional activation function. To analyze whether Gcn4p stability is regulated at the level of its subcellular localization, we investigated the localization of Gcn4p and Pho85p in living yeast cells under various conditions. Gcn4p is predominantly localized in the nucleus in the presence or absence of amino acid limitation due to two nuclear localization signals (NLS). Nuclear localization of Gcn4p does not require a functional general control system. Pho85p, which triggers Gcn4p degradation by initial phosphorylation, is also predominantly localized within the nucleus, independently of the availability of amino acids. Neither functional Pho85p nor a functional Srb10p are required for Gcn4p transportation into the nucleus. Gcn4p stability is regulated within the nucleus in response to the amount of available amino acids. Correspondingly, Gcn4p can be stabilized by preventing its entering into the nucleus. Our results show that the regulation of Gcn4p synthesis and the regulation of Gcn4p stability are two independent compartment-specific processes. Amino acid limitation as initial stimulus increases the synthesis of Gcn4p in the cytoplasm and increases the stability of the protein within the nucleus.
MATERIALS AND METHODS
S. cerevisiae strains and growth conditions.
Yeast strains used in this study are either congenic to S. cerevisiae S288c (RH1347, RH1376, and RH1408) or the W303 genetic background. Details of the yeast strains used in this study are given in Table 1. Standard methods for genetic crosses and transformation were used and standard yeast culture yeast extract-peptone-dextrose (YPD) and yeast nitrogen base (YNB) media were prepared as described elsewhere (11).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| RH1347 | MATaaro7 ura3-52 | Our collection |
| RH1376 | MATaura3-52 | Our collection |
| RH1408 | MATagcn4-103 ura3-52 gal2 | Our collection |
| RH1479 | MATα gcn2 ura3-52 | Our collection |
| W303 | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 | 22 |
| EY0140 | W303 pho85Δ::LEU2 | 24 |
| RH2663 | W303 yrb1-51 | 1 |
| RH2713 | W303 srb10Δ::KANr | Our collection |
Plasmids.
The plasmids used in this study are described in Table 2. Plasmids pME2126, pME2127, pME2128, pME2129, pME2134, and pME2135 expressing different green fluorescent protein (GFP)-Gcn4p derivatives from the MET25 promoter were obtained by amplifying the different GCN4 fragments with Pfu polymerase and introducing them via SmaI/HindIII into p426MET25 (21) or a low-copy-number GFP-N-Fus vector (23). A BglII site was introduced in front of the coding region for insertion of a 750-bp BglII fragment encoding the GFP-uv variant of GFP that was amplified from plasmid pBAD-GFPuv (Clontech, Heidelberg, Germany).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| p426MET25 | pRS426 containing MET25 promoter and CYC1 terminator | 20 |
| pME2232 | pGFP-N-Fus | 23 |
| pME2231 | MET25prom-GFP-GCN4 fusion in pGFP-N-Fus | This study |
| pME2126 | MET25prom-GFP-GCN4 fusion in p426MET25 | This study |
| pME2127 | MET25prom-GFP-GCN4aa1-249 fusion in p426MET25 | This study |
| pME2128 | MET25prom-GFP-GCN4aa1-221 fusion in p426MET25 | This study |
| pME2129 | MET25prom-GFP-GCN4aa1-169 fusion in p426MET25 | This study |
| pME2130 | MET25prom-GFP-ARO7 fusion in p426MET25 | This study |
| pME2131 | MET25prom-GFP-ARO7-GCN4aa167-200 fusion in p426MET25 | This study |
| pME2132 | MET25prom-GFP-ARO7-GCN4aa221-281 fusion in p426MET25 | This study |
| pME2133 | MET25prom-GFP-ARO7-GCN4aa231-249 fusion in p426MET25 | This study |
| pME2134 | MET25prom-GFP-GCN4S214A, S218A, S224A fusion in p426MET25 | This study |
| pME2135 | MET25prom-GFP-GCN4S214D, S218D, S224D fusion in p426MET25 | This study |
| pME2136 | MET25prom-GFP-ARO7-GCN4aa167-249 fusion in p426MET25 | This study |
| pME2137 | Like pME2136 but using GCN4S214A, S218A, S224A | This study |
| pME2138 | Like pME2136 but using GCN4S214D, S218D, S224D | This study |
| pME2139 | MET25prom-GFP-PHO85 fusion in p426MET25 | This study |
| KB294 | GAL1prom-myc3-GCN4 fusion in URA3-marked2μm vector | D. Kornitzer |
| pME2140 | GAL1prom-myc3-GCN4aa1-169 fusion in URA3-marked2μm vector | This study |
| pME2316 | GAL1prom-GFP-GCN4aa1-169 fusion in URA3-marked2μm vector | This study |
| pME2317 | GAL1prom-GFP-NLSaa215-249-GCN4aa1-169 fusion in URA3-marked2μm vector | This study |
Plasmids pME2130, pME2131, pME2132, pME2133, pME2136, pME2137, and pME2138 expressing GFP-Aro7p and GFP-Aro7p fused with different Gcn4p fragments driven from the MET25 promoter were constructed similarly to the GFP-Gcn4p plasmids. GCN4 fragments were fused via EcoRI/ClaI to the 3′ end of ARO7. PHO85 was introduced as a SmaI/ClaI fragment into p426MET25, and GFP-uv was inserted as a BglII fragment at the PHO85 5′ end.
Plasmids KB294 and pME2140, expressing a triple myc epitope-tagged version of either wild-type Gcn4 or Gcn4aa1-169 under the control of the GAL1 promoter, were obtained by insertion of a 120-bp BamHI fragment carrying the triple myc epitope (myc3) into a BglII restriction site after the fourth amino acid. Plasmids pME2316 and pME2317 express a GFP-tagged GCN4aa1-169 SmaI/HindIII fragment in a high-copy vector from a GAL1 promoter without and with the inserted NLS2aa215-249 between GFP and GCN4aa1-169, respectively.
GFP fluorescence microscopy.
Yeast strains harboring plasmids encoding GFP fusion proteins were grown to exponential phase in selective minimal medium. Cells from 1 ml of the cultures were harvested by centrifugation and immediately viewed in vivo on a Zeiss Axiovert microscope by either differential interference contrast (DIC) microscopy or fluorescence microscopy using a GFP filter set (AHF Analysentechnik AG, Tübingen, Germany). 4′,6-Diamidino-2-phenylindole (DAPI) staining was used for visualization of nuclei using standard DAPI filter sets. Cells were photographed using a Xillix microimager digital camera and the Improvision Openlab software (Improvision, Coventry, United Kingdom).
Protein analysis.
Preparation of whole yeast cell extracts were performed as described previously (31). Routinely, 10 μg of crude protein extracts was separated. After separation on sodium dodecyl sulfate gels, proteins were transferred to nitrocellulose membranes. Proteins were visualized using ECL technology (Amersham) after incubation of membranes with polyclonal mouse anti-Myc or mouse anti-GFP antibodies and a peroxidase-coupled goat anti-mouse secondary antibody.
RESULTS
The transcription factor Gcn4p is targeted to the yeast nucleus in the presence or absence of amino acids.
The transcriptional activator Gcn4p has to be imported into the nucleus to fulfill its function. Gcn4p is an unstable protein which can be stabilized in response to amino acid limitation (17). In the presence of sufficient amounts of amino acids, only small amounts of the Gcn4 protein are synthesized. Amino acid limitation results in increased translation of GCN4 mRNA. We asked whether Gcn4p stability correlates with its subcellular localization. The localization of Gcn4p was monitored in vivo by expressing the coding region of the GFP variant GFP-uv (7), which was fused to the 5′ end of the GCN4 ORF. The chimeric gene was analyzed in yeast strain RH1408 (ura3 gcn4Δ) by fluorescence microscopy. The N-terminal half of Gcn4p carries the transcriptional activation domain, whereas the C-terminal part includes the bZIP region for DNA binding and dimerization. The GFP-GCN4 hybrid ORF on a low- and also on a high-copy vector was driven from the MET25 promoter, which allows for downregulation by adding methionine to the medium.
Expression of GFP-Gcn4p was verified by Western analysis of S. cerevisiae cell extracts using polyclonal anti-GFP antibodies. GFP signals of the expected size could be visualized in cells expressing GFP-Gcn4p (Fig. 1) and were compared to the expression levels of the unregulated housekeeping gene ARO7 (29), which was used as a control. The GCN4-deficient yeast strain RH1408 is unable to grow under amino acid starvation conditions because the amino acid biosynthetic genetic network cannot be induced by the transcriptional activator Gcn4p. The GFP-Gcn4p fusion protein was able to complement the gcn4Δ phenotype. Transcription factor function was tested in vivo by inducing amino acid starvation conditions using the analogue 3-aminotriazole (3AT), which acts as a competitive inhibitor of the HIS3 gene product (16) and therefore prevents growth without functional Gcn4p-induced gene expression.
FIG. 1.
Expression of the GFP-Gcn4p fusion protein in S. cerevisiae. For Western hybridization analysis, polyclonal anti-Gcn4p and anti-GFP antibodies were used. As loading control, expression levels of Aro7p were measured in the same yeast extracts by using a polyclonal anti-Aro7p antibody. Crude protein extracts were prepared from yeast strains RH1376 (GCN4 wild type) and RH1408 (gcn4-103) expressing GFP-Gcn4p from a MET25 promoter on a 2μm plasmid (GFP-Gcn4). The antibodies used are indicated, illustrating the expression of GFP-Gcn4p.
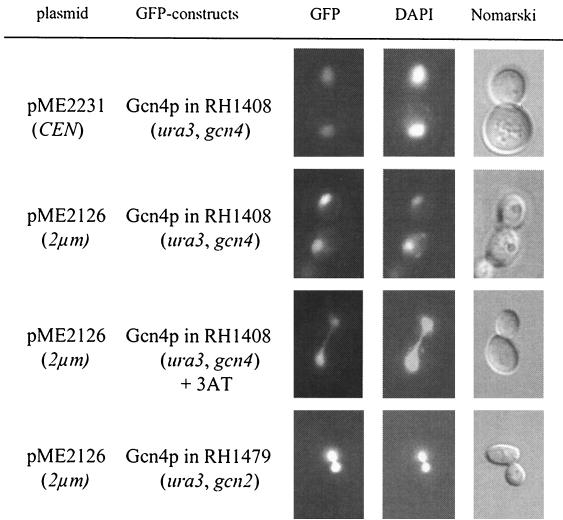
Localization of Gcn4p was examined by fluorescence microscopy in cells grown in the absence of amino acid limitation, which were compared to cells starved for histidine by adding the analogue 3AT. Gcn4p was clearly concentrated in the yeast nucleus, which was visualized by DAPI staining (27). Low amounts of GFP-Gcn4p derived from a centromere plasmid as well as higher amounts derived from a 2μm vector were efficiently transported into the nucleus independently of the absence or presence of 3AT (Fig. 2). The nuclear localization of Gcn4p in 3AT-starved cells was corroborated by testing yeast starved for tryptophan or leucine. Starvation for tryptophan was induced by adding the analogue 5-methyl-tryptophan to the medium (30). Leucine starvation was analyzed without the use of amino acid analogues in a leucine auxotrophic mutant strain (W303) (Table 1) which was transferred from leucine-containing medium to minimal medium lacking leucine. The fluorescence of the GFP-Gcn4p fusion protein of tryptophan- or leucine-starved yeast cells was similar to that of histidine-starved cells and strictly correlated with the DAPI staining of the nucleus. These data imply that most Gcn4p is immediately transported into the nucleus subsequent to translation and that therefore the majority of Gcn4p is localized within the nucleus in both the unstable and stable state.
FIG. 2.
Localization of GFP-Gcn4p in yeast. Yeast cells (RH1408) expressing wild-type GFP-Gcn4p fusion protein from the MET25 promoter on a low-copy plasmid (pME2231) and on a 2μm plasmid (pME2126) were grown to early log phase in selective medium at 30°C and analyzed by DIC microscopy (right column) and fluorescence microscopy (left column, GFP; middle column, DAPI). Nuclear localization of wild-type GFP-Gcn4p (pME2126) was also determined in gcn2Δ (RH1479) mutant cells.
Translation of GCN4 mRNA is regulated by the general control regulatory network. We next asked whether an intact general control system is required for nuclear import of Gcn4p. The kinase Gcn2p senses amino acid starvation and is activated by an increasing concentration of uncharged tRNA molecules in response to limiting amounts of amino acids. Phosphorylation of eIF-2 by Gcn2p leads to an increased translation of the GCN4 mRNA (13). Gcn4p localization was analyzed in a gcn2Δ mutant strain which is unable to induce increased GCN4 mRNA translation. In this strain Gcn4p is also predominantly localized in the nucleus, suggesting that the kinase Gcn2p is not required for its trafficking to the nucleus (Fig. 2).
In summary, these results indicate that Gcn4p transport from the cytoplasm into the nucleus depends on neither the presence or absence of amino acids nor on an intact general control system. These data suggest that the stabilization of Gcn4p in response to amino acid limitation has to be regulated within the nucleus or during a very narrow time window between translation in the cytoplasm and immediate targeting to the nucleus.
Gcn4p nuclear import is triggered by two functional NLS motifs.
We constructed yeast strains mislocalizing the protein to distinguish the Gcn4p population synthesized in the cytoplasm from the bulk of Gcn4p localized in the nucleus. We initially performed deletion and heterologous transfer experiments to identify sequences required for nuclear localization of Gcn4p. Different C-terminal truncations of the GFP-Gcn4p fusion protein were constructed by deleting the corresponding parts of the gene. Mutant proteins were analyzed by fluorescence microscopy. In addition, we verified putative NLS by testing whether they could mislocalize a cytoplasmic protein into the nucleus.
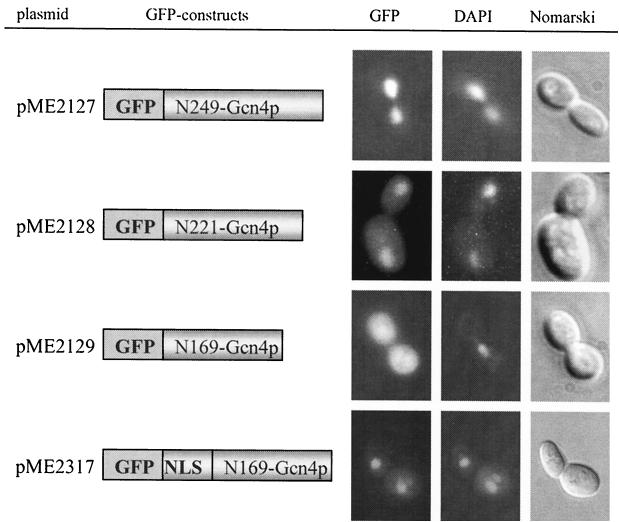
Our results showed that Gcn4p transport into the nucleus does not require the leucine zipper which mediates the dimerization (Fig. 3, pME2127). Even truncated Gcn4p lacking the DNA binding domain and leucine zipper was still predominantly localized in the nucleus (Fig. 3, pME2128), whereas the 169 N-terminal amino acids of Gcn4p were not sufficient to enter the nucleus. The corresponding GFP fusion protein significantly accumulated in the cytoplasm (Fig. 3, pME2129), suggesting the existence of an NLS motif between Gcn4p amino acids 170 and 221. It is known that the activities of the human bZIP proteins c-Jun and c-Fos depend on phosphorylation and dephosphorylation of serine and threonine residues N-terminal of the basic region. While phosphorylation of these residues leads to a deactivation of the protein, their dephosphorylation results in an increased DNA binding activity (3). These serine and threonine residues of the c-Jun protein are conserved in S. cerevisiae Gcn4p.
FIG. 3.
Localization of C-terminal-truncated Gcn4p fused to GFP in yeast. S. cerevisiae cells (RH1408, gcn4-103) expressing different GFP-Gcn4p derivatives from the induced MET25 promoter were grown to early log phase in selective medium at 30°C and analyzed by DIC microscopy (right column) and fluorescence microscopy (left column, GFP; middle column, DAPI). Localization of GFP-Gcn4p derivatives lacking either the leucine-zipper amino acid residues 250 to 281 (pME2127), the leucine zipper and the DNA-binding domain from position 221 to 281 (pME2128), or the 112 C-terminal amino acids of Gcn4p (pME2129) are shown. Fusing back NLS2 from position 215 to 249 to the N terminus of truncated Gcn4p (pME2129) enabled the protein to enter the nucleus again (pME2317). The N-terminal amino acids of Gcn4p fused to GFP are indicated.
The putative phosphorylation sites, serines 214, 218, and 224, are not involved in Gcn4p nuclear import. This was shown by comparing the localization of wild-type Gcn4p and two Gcn4p mutant derivatives where the three serine residues were either mutated to alanine in order to mimic a constitutive dephosphorylation or to aspartic acid in order to mimic a constitutive phosphorylation at these positions. Nuclear import of Gcn4p was never compromised (data not shown).
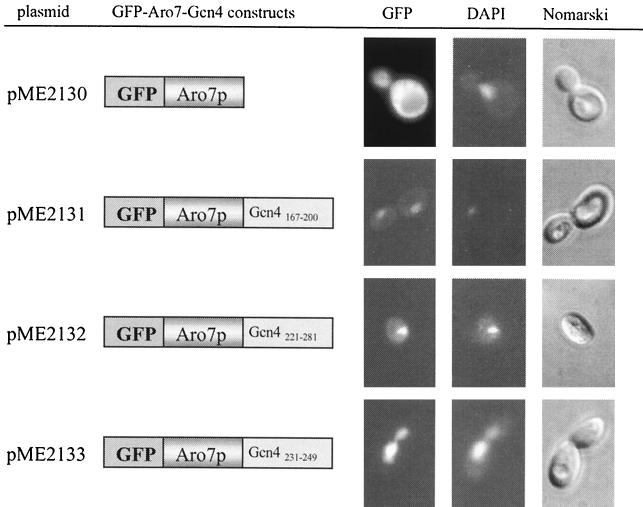
Yeast chorismate mutase (EC 5.4.99.5) was used as a reporter protein to verify putative Gcn4p NLS motifs by fusing different GCN4 fragments to its C terminus. This protein, which is involved in the biosynthesis of the aromatic amino acids tyrosine and phenylalanine, is encoded by the ARO7 gene in S. cerevisiae, and its expression is independent of Gcn4p (29). The GFP ORF was fused to the 5′ end of ARO7. The resulting fusion protein was localized exclusively in the cytoplasm when analyzed by fluorescence microscopy (Fig. 4, pME2130). The protein was also functional, which was shown by its potential to complement an aro7Δ phenotype. Heterologous transfer experiments were carried out to verify the findings of the Gcn4p deletion experiments. A Gcn4p stretch of the 34 amino acid residues from position 167 to 200 fused to the C terminus of Aro7p was able to mislocalize yeast chorismate mutase to the nucleus (Fig. 4, pME2131) and therefore functions as NLS motif 1 (NLS1). The amino acids 221 to 281, consisting of the DNA binding domain and the leucine zipper of Gcn4p, were also able to cause nuclear import of the cytoplasmic chorismate mutase (Fig. 4, pME2132), indicating the existence of a second NLS motif within the 60 C-terminal amino acids of Gcn4p. The second NLS motif (NLS2) was narrowed down to 19 amino acids (amino acids 231 to 249) which are sufficient to mistarget a chimeric chorismate mutase to the nucleus instead of the cytoplasm (Fig. 4, pME2133). NLS2 was further verified by its capability to target the truncated cytoplasmic Gcn4p1-169 back into the nucleus when fused to its N terminus (Fig. 3, pME2317).
FIG. 4.
NLS motifs of Gcn4p target the cytoplasmic Aro7 protein to the nucleus. Yeast cells (RH1347) expressing different GFP-Aro7p constructs from the induced MET25 promoter were grown to early log phase in selective medium at 30°C and analyzed by DIC microscopy (right column) and fluorescence microscopy (left column, GFP; middle column, DAPI). Localization of GFP-Aro7p (pME2130) was compared with that of GFP-Aro7p fused with either Gcn4p amino acids 167 to 200 (pME2131), 221 to 281 (pME2132), or Gcn4p amino acids 231 to 249 (pME2133). Bars illustrate the different GFP constructs, and the fused Gcn4p amino acids are indicated.
NLS2 of Gcn4p resembles a classical bipartite NLS motif, consisting of two basic clusters separated by a 10-amino-acid spacer region. The first cluster is formed from two basic amino acids, whereas the second cluster consists of seven amino acids including four basic residues.
The entire C-terminal amino acid residues 167 to 249 of Gcn4p, containing NLS1 and NLS2 interrupted by a stretch including the three serine residues, were also fused to the C terminus of chorismate mutase. The nuclear localization of this GFP-Aro7p-Gcn4p chimera was indistinguishable from the GFP-Gcn4 protein when analyzed by fluorescence microscopy (data not shown).
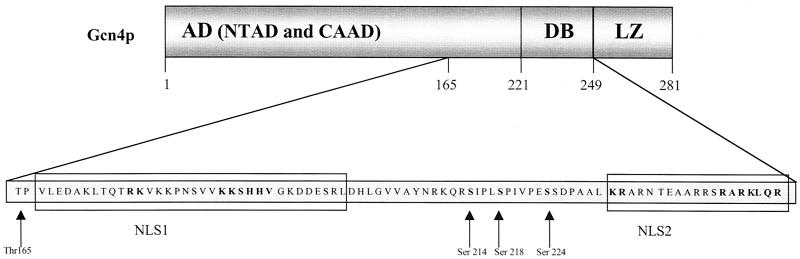
These results suggested two functional NLS motifs located within Gcn4p. There is no hint that phosphorylation of the conserved serine residues 214, 218, and 224, which are located between NLS1 and NLS2, is required for Gcn4p translocation (Fig. 5). The identification of Gcn4p NLS motifs allowed the construction of mislocalized mutant proteins to compare the stability of cytoplasmic and nuclear Gcn4p.
FIG. 5.
Scheme of the two Gcn4p NLS motifs. The positions of the two identified NLS motifs within the amino acid sequence of the transcription factor Gcn4p are shown schematically. The two Gcn4p NLS motifs NLS1 and NLS2 consist of the amino acids 167 to 200 and 231 to 249, respectively. Thr165 is the phosphorylation site of Pho85p (18). The activation domain (AD) consists of an N-terminal activation domain (NTAD) and a central acidic activation domain (CAAD). DB and LZ are the DNA binding domain and the leucine zipper of Gcn4p. The conserved serine residues 214, 218, and 224 regulate the DNA binding activity in the mammalian c-Jun (3).
Mislocalization of Gcn4p in the cytoplasm stabilizes the protein.
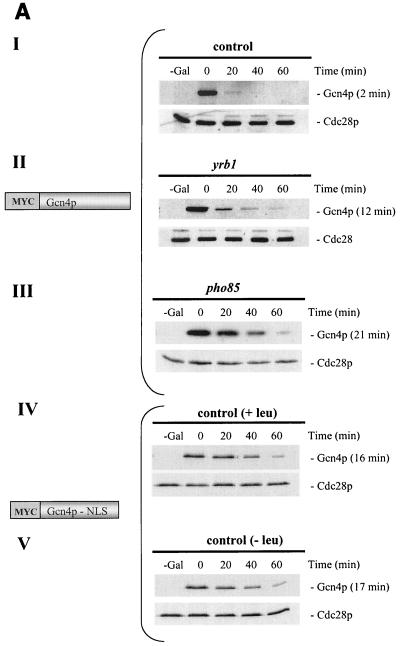
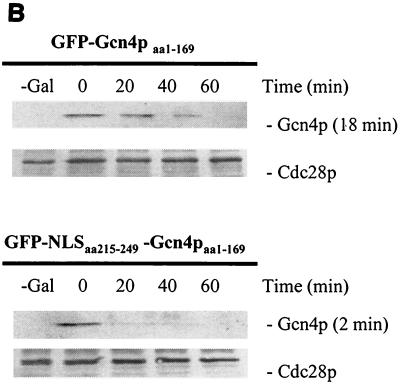
The Gcn4 protein of unstarved yeast cells is an unstable protein (17). We wondered whether the stability of Gcn4p varies depending on the cellular compartment where it is localized. Therefore, we analyzed whether mislocalization of Gcn4p affects protein stability. The myc-tagged Gcn4p lacking the identified NLS motifs was shown to be unable to enter the nucleus (Fig. 3, pME2129). A promoter shut-off experiment of the fusion gene revealed a stabilization of the resulting mutant protein when compared to Gcn4p carrying wild-type NLS1 and NLS2 expressed from the same promoter (Fig. 6A). Therefore, efficient proteolysis of Gcn4p correlates with nuclear localization. No additional protein stabilization of the truncated cytoplasmic Gcn4p could be observed in response to amino acid limitation (Fig. 6A). We also tested protein stability of the truncated Gcn4p (amino acids 1 to 169) where nuclear import was restored by fusing Gcn4p amino acids 215 to 249 to its N terminus (Fig. 4, pME2317). In accordance with the previous data, a more-efficient degradation of the again nuclear protein could be observed (Fig. 6B).
FIG. 6.
Stability analysis of S. cerevisiae Gcn4p. (A) Cytoplasmic Gcn4p and nuclear Gcn4p are stabilized in the absence of Pho85p, compared to wild-type Gcn4p. The isogenic yeast strains W303 (control [I]), RH2663 (yrb1-51 [II]), and EY0140 (pho85Δ [III]) were transformed to express nontruncated GAL-myc-GCN4 on the high-copy-number plasmid (KB294). In addition, cells of the leu2-deficient control strain W303 that expressed GAL- myc-GCN4 lacking the two Gcn4p NLS motifs (amino acids 169 to 281) on the high-copy-number plasmid (pME2140) in the presence (IV) and absence (V) of leucine were pregrown in selective minimal medium containing raffinose. A 2% galactose concentration was added to these cycling cultures to express MYC-GCN4. Cells were collected by filtration and incubated in minimal medium containing glucose. Samples were collected at the indicated time points after the shift to glucose medium (0-min time point). Levels of Myc-tagged Gcn4p were determined by immunoblotting using Myc antibodies. The kinase Cdc28p was used as loading control. The analyzed Myc-Gcn4p fusion proteins with (Myc-Gcn4p) and without (Myc-Gcn4p-NLS) the NLS motifs are illustrated. Protein half-lives were calculated based on the band intensities of at least three independent experiments and are shown in parentheses. (B) Nuclear localization correlates with efficient Gcn4p degradation in nonstarved yeast cells. A control strain (W303) containing GAL-GFP-GCN4aa1-169 lacking the two NLS motifs and GAL-GFP-NLS-GCN4aa1-169 on the high-copy-number plasmids pME2316 and pME2317, respectively, were pregrown in selective minimal medium containing raffinose. A 2% concentration of galactose was added to these cycling cultures to express GFP-GCN4. Cells were collected by filtration and incubated in minimal medium containing glucose. Samples were analyzed at the indicated time points after the shift to glucose medium (0-min time point). Levels of GFP-tagged Gcn4p were determined by immunoblotting, using GFP antibodies. Cdc28p was used as loading control. The analyzed GFP-Gcn4p fusion proteins without (GFP-Gcn4aa1-169) or with (GFP-NLS-Gcn4aa1-169) the NLS motif are illustrated. Protein half-lives were calculated based on the band intensities of at least three independent experiments and are shown in parentheses.
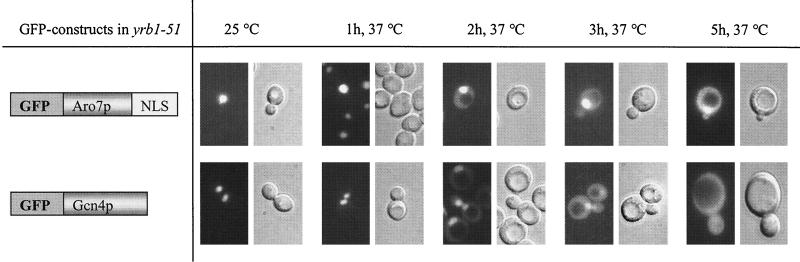
The compartment-specific stability of Gcn4p was verified by mislocalizing an intact protein in a yeast strain impaired in nuclear transport. YRB1 (yeast Ran BP1) is the yeast homologue of the mammalian Ran BP1 (4, 26). Temperature-sensitive mutants of YRB1 show defects in nuclear import at their restrictive temperature (28). The nuclear import of Gcn4p in yrb1-51 mutant cells was inhibited in response to the yrb1 defect after cells had been shifted to their restrictive temperature. Simultaneously, an accumulation of GFP-Gcn4p in the cytoplasm could be observed by fluorescence microscopy (Fig. 7). As an additional control for nuclear import in the cell, the localization of the chimeric GFP-Aro7-Gcn4aa231-249 (pME2133) fusion protein was analyzed in the yrb1-51 mutant strain, confirming the defective nuclear transport machinery (Fig. 7).
FIG. 7.
A yrb1-51 mutation impairs Gcn4p import into the yeast nucleus. Temperature-sensitive yrb1-51 mutant cells (RH2663) expressing either Aro7p-Gcn4p/NLS-GFP (pME2133) or nontruncated Gcn4p-GFP (pME2126) were grown to early log phase in selective medium at the permissive temperature of 23°C and then shifted to the restrictive temperature of 37°C. Samples were collected at the indicated time points after shifting and analyzed by DIC microscopy and fluorescence microscopy.
The stability of intact cytoplasmic Gcn4p in the yrb1-51 mutant was analyzed by a promoter shut-off experiment and Western hybridization. Wild-type and temperature-sensitive yrb1-51 cells were transformed with a plasmid containing a GAL1-promoter-controlled gene construct expressing a myc-tagged Gcn4 protein. The GCN4-containing gene fusion was transiently expressed in exponentially growing wild-type and yrb1-51 mutant cells. This experiment showed that cytoplasmic Gcn4p in yrb1-51 cells is stabilized compared to the nuclear Gcn4p of the control (Fig. 6A).
Taken together, these data suggest that the stability of Gcn4p depends on the compartment of the cell where it is localized. Whereas the cytoplasmic Gcn4p is relatively stable, the nuclear Gcn4p is highly unstable in unstarved yeast cells where sufficient amounts of amino acids are present.
Destabilization of Gcn4p is secured by the nuclear localization of Pho85p protein kinase.
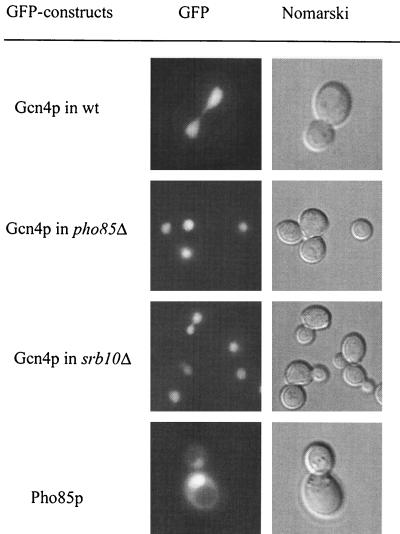
Efficient degradation of the transcription factor Gcn4p requires its phosphorylation by cyclin-dependent kinase Pho85p. A pho85Δ mutation results in Gcn4p stabilization (18). Phosphorylated Gcn4p is a substrate for the SCFCdc4 ubiquitin ligase complex, which labels the protein for degradation by the proteasome. Pho85p phosphorylates Gcn4p at Thr165, which targets it to the SCFCdc4 complex. The F-box protein Cdc4p is exclusively nuclear (2). We asked whether phosphorylation by Pho85p kinase as the initial step leading to Gcn4p instability is a compartment-specific process. Therefore, we examined the localization of Pho85p in S. cerevisiae by using a GFP-Pho85p fusion protein. The kinase Pho85p was detected predominantly inside the nucleus in the presence or absence of amino acid limitation. Therefore, Pho85p colocalized with the large unstable fraction of the Gcn4 protein within the nucleus of unstarved yeast cells, which corresponds to the findings of preceding investigations (15) (Fig. 8). Promoter shut-off experiments confirmed the previously described Gcn4p stabilization in response to the pho85Δ mutation (Fig. 6A). A pho85Δ mutation leads to Gcn4p stabilization without affecting its localization in the nucleus. This was shown by the GFP-Gcn4p fusion in a pho85Δ mutant strain which was not affected in Gcn4p nuclear import (Fig. 8). In addition, we tested Gcn4p localization in a srb10 mutant strain, since the kinase Srb10p was also shown to be involved in Gcn4p degradation. Gcn4p nuclear import is not impaired in an srb10 mutant strain (Fig. 8).
FIG. 8.
Nuclear localization of GFP-Pho85p in wild-type cells (RH1376) and GFP-Gcn4p in pho85 (EY0140) and srb10 (RH2663) mutant strains. GFP-Pho85p localization was identical in the presence or absence of amino acid limitation. Yeast cells were grown to early log phase in selective medium at 30°C and analyzed by DIC microscopy and fluorescence microscopy.
We conclude that the cytoplasmic Gcn4p is relatively stable, whereas destabilization of Gcn4p is spatially separated and restricted to the nucleus. The localization of the Pho85p kinase in the nucleus suggests that the degradation of Gcn4p is initiated within this compartment. The nuclear localization of Cdc4p suggests that also the second step in the degradation pathway is performed in the nucleus. Therefore, the yeast cell strictly separates regulation of Gcn4p synthesis and regulation of Gcn4p stability by restricting them to two different cellular compartments.
DISCUSSION
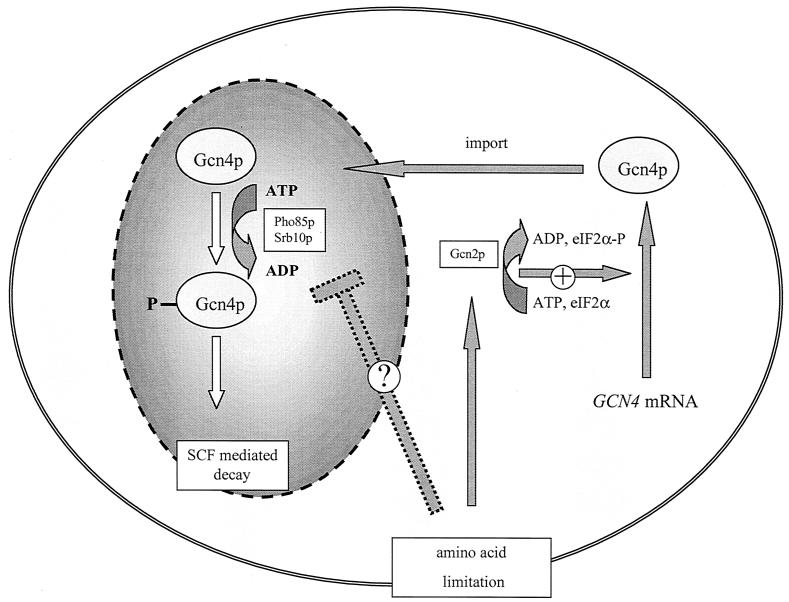
Separation of various processes of gene expression into different subcellular organelles is a typical feature of eukaryotic cells. Whereas transcription occurs inside the nucleus, the built mRNA has to be exported into the cytoplasm in order to be translated. Numerous proteins required for transcription have to be transported into the nucleus after being synthesized in the cytoplasm. We showed here that regulation of synthesis and degradation of the yeast transcriptional activator Gcn4p are spatially separated. Regulation of yeast Gcn4p synthesis is on a translational level in the cytoplasm, whereas Gcn4p degradation is regulated inside the nucleus to further modulate its function as a transcription factor (Fig. 9).
FIG. 9.
Gcn4p translational and stability control are spatially separated processes in yeast. In the presence of amino acids, GCN4 mRNA is weakly translated and efficiently transported into the nucleus. The nuclear Gcn4p in unstarved yeast cells is highly unstable. The labeling for the degradation pathway occurs primarily by phosphorylation by the kinases Pho85p or Srb10p and subsequent SCFCdc4-mediated ubiquitinylation. Amino acid limitation results in increased GCN4 mRNA translation in the cytoplasm and in a more stable Gcn4p within the nucleus. A simple hypothesis for increased stability postulates an inhibition mechanism for the nuclear kinase activities.
Gcn4p in the presence or absence of amino acid limitation.
Expression and stability regulation of the yeast transcriptional activator Gcn4p depend on the amount of available amino acids (17) and on the amount of available nitrogen source (10). In the presence of sufficient amounts of amino acids, Gcn4p is a weakly expressed and unstable protein. GCN4 mRNA translation in the cytoplasm is repressed, and rapid protein degradation is initiated in the nucleus. Gcn4p degradation starts by phosphorylation at Thr165 by the protein kinase Pho85p and is followed by ubiquitinylation by the SCFCdc4 ubiquitin ligase complex (18), which targets it for degradation by the 26S proteasome (12).
When cells are starved for amino acids, increased Gcn4p expression in the cytoplasm is accompanied by protein stabilization in the nucleus. Uncharged tRNA molecules are recognized by the cytoplasmic sensor kinase Gcn2p, which is localized at the ribosome and phosphorylates the general translation initiation factor eIF-2α. Increased eIF-2α-P levels are required to overcome the translational repression of the GCN4 mRNA, resulting in increased amounts of Gcn4 protein (8). The sensor kinase Gcn2p is only involved in regulating Gcn4p synthesis but does not affect its stability (17). We showed here that the cytoplasmic fraction of the Gcn4 protein seems to be relatively small. Therefore, Gcn4p is predominantly a nuclear protein independent of the presence or absence of amino acid limitation. Nuclear import has to occur immediately after the protein has been synthesized in the cytoplasm. Low amounts of Gcn4p are required for the basal level transcription of genes including ADE4, ARO3, and LEU2 under nonstarvation conditions (20, 9). Therefore, even under these conditions the transport of Gcn4p expressed under repressed translational conditions into the nucleus seems to be highly efficient.
We found no indication that nuclear transport of Gcn4p is regulated, as is common for other yeast transcription factors. The metabolic transcription factor Pho4p (25) is required for induction of the PHO5 gene in response to phosphate starvation. Pho4p was shown to be nuclear localized when yeast cells were starved for phosphate and predominantly cytoplasmic in phosphate-rich medium. Amino acid starvation did not affect the predominantly nuclear localization of the metabolic transcription activator Gcn4p. Pho4p as well as Gcn4p activities depend on the kinase Pho85p. The regulation of Pho4p localization is triggered by phosphorylation by the Pho80p-Pho85p cyclin-cyclin-dependent kinase complex, resulting in nuclear export of Pho4p to the cytoplasm (25). In the case of Gcn4p, the same protein kinase, presumably in combination with a yet-undetermined cyclin, initiates a different cellular process by initiating the Gcn4p protein degradation pathway within the nucleus. The nucleus does not necessarily regulate protein stability only for proteins which fulfill their function there. Far1p is required in the cytoplasm to polarize the actin cytoskeleton along a morphogenic gradient (32). Far1p stability is regulated in the nucleus and depends, as does Gcn4p, on SCFCdc4 but has to be exported to the cytoplasm to fulfill its function (2).
Specific protein degradation of Gcn4p within the nucleus.
The amount of Gcn4p is carefully regulated within the cell. The signal transduction pathway resulting in a higher synthesis rate of the protein in the cytoplasm depends on intracellular sensing of the presence or absence of uncharged tRNAs reflecting the availability of protein precursors. It is yet unclear whether uncharged tRNAs are also the signal which is perceived by the nucleus and results in protein stabilization of Gcn4p. In the presence of amino acids, protein destabilization is triggered by at least two different protein kinases. One of these protein kinases is Pho85p, which phosphorylates Gcn4p at residue Thr165 and subsequently targets the protein to the SCFCdc4 ubiquitin ligase complex. Accordingly, a deletion of PHO85 leads to a stabilization of Gcn4p (18). In contrast to the situation with the transcription factor Pho4p, Pho85p phosphorylation of Gcn4p does not change the nuclear localization of the protein. It is unclear how amino acid availability regulates Pho85p activity. We show here that Pho85p is predominantly localized in the nucleus in the presence or absence of amino acid limitation and that a pho85Δ mutation does not affect Gcn4p nuclear import. Furthermore recent studies show evidence that Cdc4p is likewise exclusively nuclear (2). Therefore, a signal monitoring the availability of protein precursors seems to be transduced the nucleus to initiate or inhibit Gcn4p phosphorylation as an initial step in the protein degradation pathway. Regulation of Gcn4p stability is a highly complicated process, because Pho85p is not the only protein kinase which phosphorylates Gcn4p, resulting in protein destabilization. The protein kinase Srb10p also phosphorylates Gcn4p and thereby marks it for recognition by SCFCdc4 ubiquitin ligase (5). Similar to that shown for Pho85p, Srb10p is predicted to be a nuclear and DNA-associated protein (6). It remains to be elucidated whether and how the kinase activities of both proteins Pho85p and Srb10p are coordinated and whether they respond to similar or different signals.
Since Gcn4p stabilization is exclusively regulated inside the nucleus, the next task will be to identify the factors involved in Pho85p and Srb10p regulation. In addition, there has to be a sensor for amino acids. Gcn2p does not seem to be this sensor for the nuclear degradation machinery. Additional factors might be involved in the cross talk between cytoplasm and nucleus.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Volkswagenstiftung, and the Fonds der Chemischen Industrie.
We are grateful to Tim Köhler and Silke Venker for critical reading of the manuscript and all members of the Braus group for helpful discussions. Special thanks to Hans-Ulrich Mösch, Erin K. O'Shea, and Daniel Kornitzer for strains and plasmids.
REFERENCES
- 1.Bäumer, M., M. Künzler, P. Steigemann, G. H. Braus, and S. Irniger. 2000. Yeast Ran-binding protein Yrb1 is required for efficient proteolysis of cell cycle regulatory proteins Pds1p and Sic1p. J. Biol. Chem. 275:38929-38937. [DOI] [PubMed] [Google Scholar]
- 2.Blondel, M., J. M. Galan, Y. Chi, C. Lafourcade, C. Longaretti, R. J. Deshaies, and M. Peter. 2000. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 19:6085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, W. J., T. Smeal, L. H. Defize, P. Angel, J. R. Woodgett, M. Karin, and T. Hunter. 1991. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell 64:573-584. [DOI] [PubMed] [Google Scholar]
- 4.Butler, G., and K. H. Wolfe. 1994. Yeast homologue of mammalian Ran binding protein 1. Biochim. Biophys. Acta 1219:711-712. [DOI] [PubMed] [Google Scholar]
- 5.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, K. F., and R. Strich. 1999. Functional analysis of the Ume3p/Srb11p-RNA polymerase II holoenzyme interaction. Gene Expr. 8:43-57. [PMC free article] [PubMed] [Google Scholar]
- 7.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 8.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translation control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 9.Gedvilaite, A., and K. Sasnauskas. 1994. Control of the expression of the ADE2 gene of the yeast Saccharomyces cerevisiae. Curr. Genet. 25:475-479. [DOI] [PubMed] [Google Scholar]
- 10.Grundmann, O., H. U. Mösch, and G. H. Braus. 2001. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J. Biol. Chem. 276:25661-25671. [DOI] [PubMed] [Google Scholar]
- 11.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-863. [PubMed] [Google Scholar]
- 12.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch, A. G. 1994. Translational control of GCN4: an in vivo barometer of initiation-factor activity. Trends Biochem. Sci. 19:409-414. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272:21661-21664. [DOI] [PubMed] [Google Scholar]
- 15.Kaffman, A., N. M. Rank, E. M. O'Neill, L. S. Huang, and E. K. O'Shea. 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396:482-486. [DOI] [PubMed] [Google Scholar]
- 16.Klopotowski, T., and A. Wiater. 1965. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch. Biochem. Biophys. 112:562-566. [DOI] [PubMed] [Google Scholar]
- 17.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. F. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCdc4 ubiquitin-ligase complex. Mol. Biol. Cell 11:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrik, W. C. 1992. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 56:291-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mösch, H. U., B. Scheier, P. Lathi, P. Mäntsälä, and G. H. Braus. 1991. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J. Biol. Chem. 266:20453-20456. [PubMed] [Google Scholar]
- 21.Mumberg, D., R. Müller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasmyth, K., G. Adolf, D. Lydall, and A. Seddon. 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell 62:631-647. [DOI] [PubMed] [Google Scholar]
- 23.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 13:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niedenthal, R. K., L. Riles, M. Johnston, and J. H. Hegemann. 1996. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12:773-786. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill, E. M., A. Kaffman, E. R. Jolly, and E. K. O'Shea. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209-212. [DOI] [PubMed] [Google Scholar]
- 26.Ouspenski, I. I., U. W. Mueller, A. Matynia, S. Sazer, S. J. Elledge, and B. R. Brinkley. 1995. Ran-binding protein-1 is an essential component of the Ran/RCC1 molecular switch system in budding yeast. J. Biol. Chem. 270:1975-1978. [DOI] [PubMed] [Google Scholar]
- 27.Russel, W. C., C. Newman, and D. H. Williamson. 1975. A simple cytochemical technique for demonstration of DNA in cells infected with mycoplasmas and viruses. Nature 253:461-462. [DOI] [PubMed] [Google Scholar]
- 28.Schlenstedt, G., D. H. Wong, D. M. Koepp, and P. A. Silver. 1995. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 14:5367-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidheini, T., H. U. Mösch, R. Graf, and G. H. Braus. 1990. A GCN4 protein recognition element is not sufficient for GCN4-dependent regulation of the transcription in the ARO7 promoter of Saccharomyces cerevisiae. Mol. Gen. Genet. 224:57-64. [DOI] [PubMed] [Google Scholar]
- 30.Schürch, A., J. Miozzari, and R. Hütter. 1974. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J. Bacteriol. 117:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surana, U., A. Amon, C. Dowzer, J. McGrew, B. Byers, and K. Nasmyth. 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12:1969-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valtz, N., M. Peter, and I. Herskowitz. 1995. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131:863-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voorma, H. O., A. A. M. Thomas, and H. A. A. Van Heugten. 1994. Initiation of protein synthesis in eukaryotes. Mol. Biol. Rep. 19:139-145. [DOI] [PubMed] [Google Scholar]
- 34.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15:4497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]