Abstract
In Saccharomyces cerevisiae, the transcription factors Tec1p and Ste12p are required for haploid invasive and diploid pseudohyphal growth. Tec1p and Ste12p have been postulated to regulate these developmental processes primarily by cooperative binding to filamentous and invasion-responsive elements (FREs), which are combined enhancer elements that consist of a Tec1p-binding site (TCS) and an Ste12p-binding site (PRE). They are present in the promoter regions of target genes, e.g., FLO11. Here, we show that Tec1p efficiently activates target gene expression and cellular development in the absence of Ste12p. We further demonstrate that TCS elements alone are sufficient to mediate Tec1p-driven gene expression by a mechanism termed TCS control that is operative even when Ste12p is absent. Mutational analysis of TEC1 revealed that TCS control, FLO11 expression, and haploid invasive growth require the C terminus of Tec1p. In contrast, the Ste12p-dependent FRE control mechanism is sufficiently executed by the N-terminal portion of Tec1p, which contains the TEA/ATTS DNA-binding domain. Our study suggests that regulation of haploid invasive and diploid pseudohyphal growth by Ste12p and Tec1p is not only executed by combinatorial control but involves additional control mechanisms in which Ste12p activates TEC1 expression via clustered PREs and where Tec1p regulates expression of target genes, e.g., FLO11, by TCS control.
Saccharomyces cerevisiae is a dimorphic fungus that interconverts between unicellular and multicellular filamentous growth modes (15). Upon nitrogen starvation, diploid cells switch from growth as single yeast form cells to a filamentous form consisting of chains of elongated cells called pseudohyphae. A related phenomenon called invasive growth is observed in haploid cells (44). In contrast to pseudohyphal growth, haploid invasive growth is not triggered by nitrogen starvation but occurs on rich medium in response to glucose depletion (7). In addition, low concentrations of α-factor mating pheromone induce haploid invasive growth (9, 43).
Both haploid invasive growth and diploid pseudohyphal development require transcription factors Tec1p and Ste12p (12, 27, 31, 34, 44). Tec1p was originally identified as a regulator of expression of Ty1 transposon insertions (24). Tec1p contains an evolutionarily conserved DNA-binding domain that has been named the TEA (TEF-1, Tec1p, and AbaAp) or ATTS (AbaAp, TEF-1, Tec1p, and Scalloped) motif (5). The TEA/ATTS domain is shared by a group of eukaryotic transcription factors, including Tec1p; human TEF-1, which regulates simian virus 40 and papillomavirus type 16 gene expression; and Aspergillus nidulans AbaAp, a development-specific transcription factor required for asexual spore formation (2, 5, 52). These transcription factors recognize and bind the conserved sequences CATTCC and CATTCT, which have been termed TEA/ATTS consensus sequences or Tec1p-binding site (TCS) elements (1, 3, 20, 31). Ste12p was initially identified as a regulator of mating factor-responsive genes that contain pheromone response elements (PREs) matching the consensus sequence TGAAACA in their promoter regions (11, 23, 51). Ste12p binds poorly to a single PRE but binds cooperatively to multiple PREs, leading to efficient transcriptional activation (8, 53). Clustering of PRE elements is common to mating factor-responsive genes (e.g., FUS1 and SST2) and confers not only mating factor-regulated but also cell cycle-regulated transcription (38, 39, 41). Single PREs confer transcriptional activation only if juxtaposed to recognition sites for other transcription factors, e.g., Mcm1p or Tec1p, that bind cooperatively with Ste12p to combined enhancer elements (3, 10, 31).
Combination of one PRE and one TCS element creates an enhancer element that has been termed a filamentation and invasion response element or FRE (31). Ste12p and Tec1p have been demonstrated to bind cooperatively to FREs and to activate gene expression in a synergistic manner (31). FREs are present in the promoter regions of genes involved in haploid invasive growth and diploid pseudohyphal development and include FLO11, encoding a cell surface flocculin (29, 46), and TEC1 itself (31). These findings have led to the view that FRE-mediated gene expression is the major control mechanism by which Tec1p regulates haploid invasion and diploid pseudohyphal growth and that it confers specificity on the distinct Ste12p-regulated developmental programs (31). However, several lines of evidence indicate that FRE control might not be the sole mechanism by which Ste12p and Tec1p control expression of genes required for haploid invasive and pseudohyphal growth. First, the TEC1 promoter contains several clustered PREs that mediate control of TEC1 transcription by mating pheromone, by Ste12p and other elements of the mating factor signal transduction pathway, and by the cell cycle (38). Second, microarray analysis has identified several genes that are under the control of Ste12p and Tec1p but that do not contain FRE enhancer elements in their promoter regions, e.g., PGU1 encoding a secreted endopolygalacturonase that degrades the plant-specific polysaccharide pectin (32). Instead, the promoter regions of these genes often contain several TCS elements that are not neighbored by PRE sites. Third, the TEA/ATTS family transcription factor AbaAp from A. nidulans activates gene expression by binding on its own to either single or multiple TCS elements present in the promoter regions of target genes (1). Finally, ectopic expression of the Tec1p-related transcription factors AbaAp from A. nidulans and CaTec1p from the human pathogen Candida albicans induce S. cerevisiae haploid invasive and pseudohyphal growth in strains that lack STE12 (12, 47).
These findings prompted us to examine the possibility that Ste12p and Tec1p control gene expression and cellular development by mechanisms distinct from FRE-mediated combinatorial control. We provide evidence that TEC1 gene expression requires Ste12p but is largely independent of Tec1p autoregulation. As a consequence, Tec1p levels drop 20-fold in strains lacking Ste12p. When restored to high levels, Tec1p activates haploid invasive growth and expression of the FLO11 and PGU1 genes even in the absence of STE12. Tec1p efficiently activates gene expression mediated by synthetic single or combined TCS elements inserted upstream of an upstream activating sequence (UAS)-less reporter gene. This Tec1p-mediated transcriptional control mechanism, termed TCS control, is operative even when Ste12p is absent. Mutational analysis of TEC1 reveals that the C terminus of Tec1p is required for haploid invasive growth and TCS-mediated gene expression but is dispensable for the FRE control mechanism. On the basis of our results, we propose that Ste12p and Tec1p control gene expression and cellular development by several distinct mechanisms.
MATERIALS AND METHODS
Yeast strains and growth conditions.
All of the yeast strains used in this study are congenic to the Σ1278b genetic background (Table 1). The tec1Δ::HIS3 and ste12Δ::TRP1 deletion mutations were introduced by using deletion plasmids ptec1Δ::HIS3 (36) and pste12Δ::TRP1. RH2757 was derived from RH2756 by transformation with a linear fragment containing the TRP1 gene. RH2758 was obtained by mating of RH2500 with RH2757, and RH2759 resulted from mating of RH2501 with RH2778. Standard methods for genetic crosses and transformation were used, and standard yeast culture medium was prepared essentially as previously described (18). When required, synthetic complete (SC) medium lacking appropriate supplements was used. Invasive growth tests were performed as described previously (44). Pseudohyphal development was induced by growth on synthetic low-ammonium medium (15).
TABLE 1.
Strains used in this study
| Strain | Relevant genotypeb | Reference |
|---|---|---|
| RH2754 | MATaura3-52 leu2::hisG his3::hisG trp1::hisG | This study |
| RH2500 | MATatec1Δ::HIS3 ura3-52 leu2::hisG his3::hisG trp1::hisG | This study |
| RH2755 | MATaste12Δ::TRP1 ura3-52 leu2::hisG his3::hisG trp1::hisG | This study |
| RH2501 | MATatec1Δ::HIS3 ste12Δ::TRP1 ura3-52 leu2::hisG his3::hisG trp1::hisG | This study |
| RH2756 | MATα tec1Δ::HIS3 ura3-52 leu2::hisG his3::hisG trp1::hisG | This study |
| RH2757 | MATα tec1Δ::HIS3 ura3-52 leu2::hisG his3::hisG | This study |
| RH2778 | MATα tec1Δ::HIS3 ste12Δ::TRP1 ura3-52 leu2::hisG his3::hisG trp1::hisG | This study |
| RH2758 | MATa/MATα tec1Δ::HIS3/tec1Δ::HIS3 ura3-52/ura3-52 leu2::hisG/leu2::hisG his3::hisG/his3::hisG trp1Δ::hisG/TRP1 | This study |
| RH2759 | MATa/MATα tec1Δ::HIS3/tec1Δ::HIS3 ste12Δ::TRP1/ste12Δ::TRP1 ura3-52/ura3-52 leu2::hisG/leu2::hisG his3::hisG/ his3::hisG trp1Δ::hisG/trp1Δ::hisG | This study |
| RH2499 | MATα tec1Δ::HIS3 FRE(Ty1)-lacZ::LEU2 ura3-52 leu2::hisG his3::hisG trp1::hisG | 36 |
| RH2767a | Same as RH2500 except CYC1(ΔUAS)-lacZ::URA3 | This study |
| RH2765a | Same as RH2500 except TCSfw-CYC1-lacZ::URA3 | This study |
| RH2766a | Same as RH2500 except TCSbw-CYC1-lacZ::URA3 | This study |
| RH2764a | Same as RH2500 except TCSfw-6-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2763a | Same as RH2500 except TCSfw-11-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2760a | Same as RH2500 except TCSfw-14-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2761a | Same as RH2500 except TCSbw-13-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2762a | Same as RH2500 except TCSbw-13-TCSbw-13-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2768a | Same as RH2500 except TCSfw-14-PREfw-CYC1-lacZ::URA3 | This study |
| RH2776a | Same as RH2501 except CYC1(ΔUAS)-lacZ::URA3 | This study |
| RH2774a | Same as RH2501 except TCSfw-CYC1-lacZ::URA3 | This study |
| RH2775a | Same as RH2501 except TCSbw-CYC1-lacZ::URA3 | This study |
| RH2773a | Same as RH2501 except TCSfw-6-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2772a | Same as RH2501 except TCSfw-11-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2769a | Same as RH2501 except TCSfw-14-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2770a | Same as RH2501 except TCSbw-13-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2771a | Same as RH2501 except TCSbw-13-TCSbw-13-TCSbw-CYC1-lacZ::URA3 | This study |
| RH2777a | Same as RH2501 except TCSfw-14-PREfw-CYC1-lacZ::URA3 | This study |
Plasmid constructions.
All of the plasmids used in this study are listed in Table 2. Plasmids pME2044, pME2045, and pME2047 were constructed by subcloning of a 3.1-kb PstI-HindIII fragment carrying TEC1 from B3366 (34) into YCplac33, YCplac111, and YEplac181, respectively. For regulated expression of TEC1 from the inducible GAL1-10 promoter, a 2.5-kb Bsp120I/SacI fragment containing a GAL1(p)::TEC1 cassette was subcloned from pME2071 (36) into pRS315, yielding plasmid pME2049. A SalI site was inserted after the ATG start codon of TEC1 by a two-step strategy. (i) The TEC1 promoter region was amplified by PCR with primers TEC1-3 (ACGCGTCGACCATGGTTAAACAGGTATCAGAATTGTTG) and TEC1-4 (CTTCAGGCAAGAGTACGTTCTTCGCTGG) and plasmid B3366 (34) as the template, yielding a 1.05-kb PCR product that was digested with PstI and SalI and inserted into plasmid YCplac33 to obtain plasmid YCplac33-TEC1(p). (ii) A 2.1-kb fragment containing the TEC1 open reading frame (ORF) was amplified from B3366 by using primers TEC1-5 (ACGCGTCGACAGTCTTAAAGAAGACGACTTTGGCAAGG) and TEC1-6 (CGCGGATCCGGCCCCGACTTGAATGATTTTCAAGGTAGG), introducing a SalI site in front of the second codon of TEC1 and a BamHI site 650 bp downstream of the TAA stop codon. This fragment was inserted into the SalI and BamHI sites of YCplac33-TEC1p from step i to obtain plasmid pME2068, carrying a functional TEC1(p)::TEC1 gene with a SalI restriction site after the ATG start codon. Plasmids pME2289 and pME2294 were obtained by subcloning of the TEC1(p)::TEC1 cassette from pME2068 into the PstI and BamHI sites of YCplac111 and YCplac181, respectively. Plasmids pME2279 and pME2280, both expressing a triple myc epitope-tagged version of Tec1p under the control of the TEC1 promoter, were obtained by the following cloning strategy. (i) A 3.1-kb PstI-BamHI fragment containing TEC1(p)::TEC1 was subcloned from pME2068 into YEplac195. (ii) A synthetic BglII linker was ligated into the SalI site following the TEC1 translational start codon. (iii) Finally, a 120-bp BamHI fragment carrying the triple myc epitope (myc3) was inserted into the BglII site to yield plasmid pME2280. The complete TEC1(p)::myc3-TEC1 cassette was isolated by PstI-SmaI digestion and inserted into YCplac33 to yield pME2279. In addition, plasmids pME2295 and pME2296 were constructed, which differ from pME2280 and pME2279 only by the copy number of myc epitope tags. Plasmid pME2295 consists of a TEC1(p)::myc6-TEC1 cassette in YCplac33, and plasmid pME2296 carries the same cassette in YEplac195. Plasmids carrying the different tec1 mutant alleles were identified in a mutant allele library screen (see below). A subset of these tec1 alleles were subcloned as 3.1-kb PstI-BamHI fragments into YCplac111 to obtain plasmids pME2290 to pME2293. The different tec1 mutant alleles were myc epitope tagged by isolation of 2.1-kb SalI-BamHI fragments from the respective plasmids and exchange for the corresponding wild-type TEC1 fragment in plasmids pME2280 and pME2279 to yield plasmids pME2281 to pME2288.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| YCplac33 | URA3-marked centromere vector | 13 |
| YEplac195 | URA3-marked 2μm vector | 13 |
| YCplac111 | LEU2-marked centromere vector | 13 |
| pRS315 | LEU2-marked centromere vector | 49 |
| YEplac181 | LEU2-marked 2μm vector | 13 |
| YEp356R | URA3-marker 2μm vector for lacZ fusions | 37 |
| pME2044 | 3.1-kb fragment containing TEC1 in YCplac33 | This work |
| pME2045 | 3.1-kb fragment containing TEC1 in YCplac111 | This work |
| pME2047 | 3.1-kb fragment containing TEC1 in YEplac181 | This work |
| pME2049 | 2.5-kb GAL1(p)::TEC1 fusion in pRS315 | This work |
| pME2071 | 2.5-kb GAL1(p)::TEC1 fusion in pRS316 | 36 |
| pME2077 | tec1-102 in YCplac33 | This work |
| pME2083 | tec1-103 in YCplac33 | This work |
| pME2085 | tec1-104 in YCplac33 | This work |
| pME2086 | tec1-105 in YCplac33 | This work |
| pME2102 | tec1-201 in YCplac33 | This work |
| pME2096 | tec1-202 in YCplac33 | This work |
| pME2101 | tec1-203 in YCplac33 | This work |
| pME2103 | tec1-204 in YCplac33 | This work |
| pME2290 | tec1-102 in YCplac111 | This work |
| pME2291 | tec1-105 in YCplac111 | This work |
| pME2293 | tec1-201 in YCplac111 | This work |
| pME2292 | tec1-202 in YCplac111 | This work |
| pME2068 | TEC1(p)::TEC1 in YCplac33 | This work |
| pME2289 | TEC1(p)::TEC1 in YCplac111 | This work |
| pME2294 | TEC1(p)::TEC1 in YEplac181 | This work |
| pME2279 | TEC1(p)::myc3-TEC1 fusion in YCplac33 | This work |
| pME2295 | TEC1(p)::myc6-TEC1 fusion in YCplac33 | This work |
| pME2280 | TEC1(p)::myc3-TEC1 fusion in YEplac195 | This work |
| pME2296 | TEC1(p)::myc6-TEC1 fusion in YEplac195 | This work |
| pME2281 | TEC1(p)::myc3-tec1-102 fusion in YEplac195 | This work |
| pME2282 | TEC1(p)::myc3-tec1-103 fusion in YEplac195 | This work |
| pME2283 | TEC1(p)::myc3-tec1-104 fusion in YEplac195 | This work |
| pME2284 | TEC1(p)::myc3-tec1-105 fusion in YEplac195 | This work |
| pME2287 | TEC1(p)::myc3-tec1-201 fusion in YEplac195 | This work |
| pME2285 | TEC1(p)::myc3-tec1-202 fusion in YEplac195 | This work |
| pME2286 | TEC1(p)::myc3-tec1-203 fusion in YEplac195 | This work |
| pME2288 | TEC1(p)::myc3-tec1-204 fusion in YEplac195 | This work |
| pME2043 | 3.5-kb TEC1(p)::abaA fusion in YCplac33 | This work |
| pME2046 | 3.5-kb TEC1(p)::abaA fusion in YCplac111 | This work |
| pME2048 | 3.5-kb TEC1(p)::abaA fusion in YEplac181 | This work |
| pME2050 | 3.1-kb GAL1(p)::abaA fusion in YCplac111 | This work |
| pME2070 | 3.1-kb GAL1(p)::abaA fusion in pRS316 | This work |
| pME2300 | TEC1-lacZ fusion in YCplac33 | This work |
| pME2065 | TEC1-lacZ fusion in YEp356R | This work |
| B3782 | FLO11-lacZ fusion in YEp356R | 46 |
| pME2064 | PGU1-lacZ fusion in YEp356R | 32 |
| pLI4 | CYC1-lacZ fusion in URA3-marked integrative vector | 48 |
| pME1108 | pLI4 with CYC1(ΔUAS)-lacZ | This work |
| pME2051 | pLI4 with TCSfw-CYC1-lacZ | This work |
| pME2052 | pLI4 with TCSbw-CYC1-lacZ | This work |
| pME2056 | pLI4 with TCSfw-6-TCSbw-CYC1-lacZ | This work |
| pME2055 | pLI4 with TCSfw-11-TCSbw-CYC1-lacZ | This work |
| pME2053 | pLI4 with TCSfw-14-TCSbw-CYC1-lacZ | This work |
| pME2057 | pLI4 with TCSbw-13-TCSbw-CYC1-lacZ | This work |
| pME2058 | pLI4 with TCSbw-13-TCSbw-13-TCSbw- CYC1-lacZ | This work |
| pME2066 | pLI4 with TCSfw-14-PREfw-CYC1-lacZ | This work |
| ptec1Δ::HIS3 | Cassette for full deletion of TEC1 ORF | 36 |
| pste12Δ::TRP1 | Cassette for full deletion of STE12 ORF | This work |
For expression of A. nidulans-derived abaA in S. cerevisiae, the abaA ORF was placed behind the S. cerevisiae TEC1 promoter by using the following cloning strategy. (i) The intronless abaA-ORF was amplified by PCR from plasmid pAA35 (2) by using primers ABAA-1 (ACGCGTCGACGCTACTGACTGGCAACCCGAGTGTATGG) and ABAA-2 (ACGCGTCGACCTAGACAGCCTCAACCGCAGTATGTTC), introducing SalI sites at both ends. The resulting 2.4-kb PCR fragment was placed downstream of the TEC1 promoter by insertion into the SalI site of YCplac33-TEC1(p) to yield pME2043. The whole TEC1(p)::abaA cassette of pME2043 was released by PstI-SmaI digestion and inserted into YCplac111 and YEplac181 to yield pME2046 and pME2048, respectively. Plasmids pME2050 and pME2070 were obtained by subcloning of a 3.1-kb GAL1(p)::abaA expression cassette from plasmid pAA35 (2) into YCplac111 and pRS316, respectively.
Plasmid pME1108 was constructed by deletion of a 430-bp XhoI fragment containing the CYC1 UAS of plasmid pLI4 (48). Plasmids pME2051 to pME2058, carrying various combinations of TCS elements upstream of the CYC1-lacZ reporter gene, were constructed by substituting the XhoI fragment of pLI4 for one or several copies of a synthetic linker containing a single TCS element and XhoI-cohesive ends. The linker was prepared by annealing of primers TCS1 (TCGAGTCACATTCTTCTGC [the TCS element is underlined]) and TCS2 (TCGAGCAGAAGAATGTGAC [the reverse complement of the TCS element is underlined]). The number and orientation of the inserted TCS elements were determined by DNA sequence analysis (see Fig. 5). The integrative FRE(Ty1)-lacZ reporter plasmid pME2066 was obtained by subcloning a SalI-BamHI fragment from plasmid FRE(Ty1)::lacZ (31) into pLI4. The TEC1-lacZ reporter plasmid pME2065 was constructed by subcloning of the TEC1 promoter as a 1.05-kb PstI-SalI fragment from pME2041 into YEp356R (37), and plasmid pME2300 was obtained by subcloning the TEC1-lacZ expression cassette from pME2065 into YCplac33.
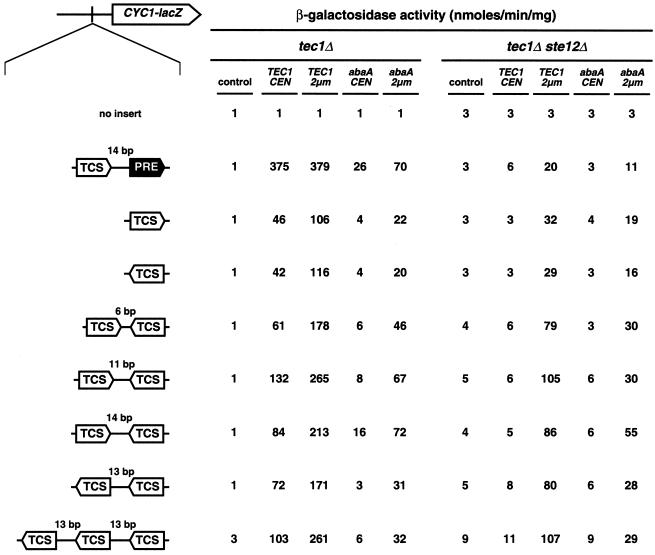
FIG. 5.
Activation of FRE- and TCS-dependent gene expression by TEC1, abaA, and STE12 in yeast. An enhancerless CYC1-lacZ fusion gene carrying no UAS element (no insert), FRE (TCS plus PRE), or TCS elements in different combinations was integrated as a single copy into the genomes of strains RH2500 (tec1Δ) and RH2501 (tec1Δ ste12Δ), and β-galactosidase specific activity was measured with TEC1 and abaA being absent (control) or present at a low (CEN) or high (2μm) copy level. Activities are expressed in nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute times milligrams of protein and are means of four measurements with two independent transformants. The standard deviation was less than 20%.
Library of TEC1 mutants.
TEC1 was mutagenized by PCR amplification of a 2.2-kb fragment of pME2068 containing the TEC1 ORF by using Taq DNA polymerase in the presence of 0.24 mM MnCl2. The resulting DNA was digested with SalI and BamHI and exchanged for the corresponding SalI-BamHI fragment in pME2068, yielding a library of more than 20,000 independent recombinants. Following identification of mutants (see below), TEC1 alleles were sequenced by using the ABI Prism Big Dye terminator sequencing kit and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany).
Screen for TEC1 invasive-growth mutants.
For isolation of mutants with altered invasive growth and FRE-lacZ reporter induction, strain RH2499 carrying a chromosomal deletion of TEC1 and an integrated FRE(Ty1)::lacZ reporter was transformed with the TEC1 mutant library described above. A pool of approximately 30,000 transformants was plated on solid medium lacking uracil (SC-Ura medium) at a density of ∼500 colonies per plate. Invasive-growth mutants were isolated by employing an invasive-growth test (44), and expression of the FRE(Ty1)::lacZ reporter gene was measured by a qualitative filter assay (4). Initial mutant phenotypes were confirmed by isolation of the TEC1-containing plasmids and reintroduction into the parental strain.
Pseudohyphal-development assay.
Qualitative assays for pseudohyphal development were performed as described previously (33). After 3 days of growth on solid synthetic low-ammonium medium, pseudohyphal colonies were viewed with a Zeiss Axiovert microscope and photographed with a Kappa DX30 digital camera and the Kappa ImageBase software (Kappa Opto-Electronics, Gleichen, Germany).
β-Galactosidase assays.
Strains carrying plasmid-borne or integrated lacZ reporters were grown in selective liquid SC medium to exponential growth phase, and extracts were prepared and assayed for β-galactosidase activity as described previously (33). β-Galactosidase specific activity was normalized to the total protein in each extract and equals (optical density at 420 nm × 1.7)/(0.0045 × protein concentration × extract volume × time). Assays were performed on at least three independent transformants, and the mean value is presented. Standard deviations did not exceed 20%.
Northern blot analysis.
Total RNAs were prepared from exponentially growing liquid cultures as described earlier (6). Total RNAs were separated on a 1.4% agarose gel containing 3% formaldehyde and transferred onto nylon membranes as described earlier (35). TEC1, FLO11, and ACT1 transcripts were detected by using gene-specific, 32P-radiolabeled DNA probes. Hybridizing signals were quantified with a BAS-1500 Phospho-Imaging scanner (Fuji, Tokyo, Japan).
Protein analysis.
Yeast strains harboring plasmids encoding myc-Tec1p were grown to exponential phase in liquid SC medium. Preparation of total cell extracts and subsequent Western blot analysis were performed essentially as previously described (45). myc-Tec1p fusion proteins were detected by enhanced-chemiluminescence technology (Amersham, Buckinghamshire, United Kingdom) after incubation of nitrocellulose membranes with mouse anti-myc monoclonal antibodies (9E10) together with a peroxidase-coupled goat anti-mouse immunoglobulin G secondary antibody (Dianova, Hamburg, Germany). For detection of Cdc42p, the membranes were incubated with polyclonal anti-Cdc42p antibodies and a peroxidase-coupled goat anti-rabbit immunoglobulin G secondary antibody (Dianova). Tec1p and Cdc42p signals were quantified with a scanner and Molecular Analyst software (Bio-Rad, Munich, Germany).
Indirect immunofluorescence microscopy.
Yeast strains harboring plasmids encoding myc-Tec1p were cultured to exponential growth phase in liquid yeast nitrogen base medium supplemented with appropriate amino acids. Cells were harvested from 1 ml of the cultures by centrifugation and fixed in 3.7% formaldehyde. Spheroblasts were prepared as described previously (42). 4′,6′-Diamidino-2-phenylindole (DAPI) staining and mouse anti-myc monoclonal antibodies (9E10), together with an Alexa 488-conjugated goat anti-mouse antibody (Molecular Probes, Eugene, Oreg.), were used for visualization of nuclei and myc epitope-tagged proteins, respectively. Cells were viewed on a Zeiss Axiovert microscope by either differential interference contrast microscopy or fluorescence microscopy using standard DAPI and fluorescein isothiocyanate filter sets. Cells were photographed with a Xillix Microimager digital camera and Improvision Openlab software (Improvision, Coventry, United Kingdom).
RESULTS
Tec1p and AbaAp induce haploid invasive growth and pseudohyphal development in the absence of Ste12p.
We tested whether Tec1p is able to induce haploid invasive growth and diploid pseudohyphal development of S. cerevisiae in the absence of Ste12p. Previous work had shown that the Tec1p homologue AbaAp from A. nidulans induces S. cerevisiae pseudohyphal development in strains lacking STE12 but only when expressed from the highly inducible GAL1 promoter (12). We expressed TEC1 and abaA from the endogenous TEC1 promoter on either a low-copy (CEN) or a high-copy (2μm) plasmid in both haploid and diploid tec1Δ and tec1Δ ste12Δ mutant strains. In addition, both genes were expressed from the GAL1 promoter. Haploid strains were assayed for invasive growth by a wash test (Fig. 1), and diploid pseudohyphal development was measured by growth on nitrogen starvation medium (Fig. 2).
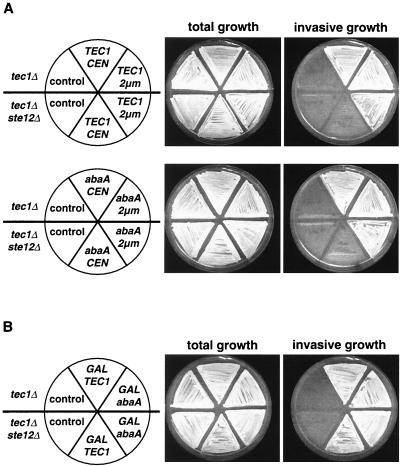
FIG. 1.
Haploid invasive growth of yeast strains expressing TEC1 or A. nidulans abaA driven by the TEC1 promoter. (A) Haploid strains RH2500 (tec1Δ) and RH2501 (tec1Δ ste12Δ) carrying plasmid YCplac111 (control), pME2045 (TEC1 CEN), pME2047 (TEC1 2μm), pME2046 (abaA CEN), or pME2048 (abaA 2μm) were grown on SC-Leu medium containing 2% glucose for 4 days. Plates were photographed before (total growth) and after (invasive growth) cells were washed off the agar surface. (B) Haploid strains RH2500 (tec1Δ) and RH2501 (tec1Δ ste12Δ) carrying plasmid YCplac111 (control), pME2049 (GAL-TEC1), or pME2050 (GAL-abaA) were grown on SC-Leu medium containing 2% galactose to induce expression from the GAL1 promoter. Invasive growth was measured as described for panel A.
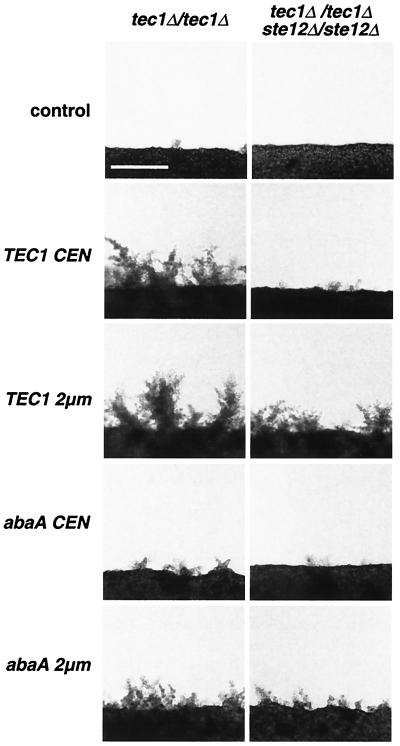
FIG. 2.
Pseudohyphal growth of diploid yeast strains expressing TEC1 or A. nidulans abaA driven by the TEC1 promoter. Diploid strains RH2758 (tec1Δ/tec1Δ) and RH2759 (tec1Δ/tec1Δ ste12Δ/ste12Δ) carrying plasmid YCplac111 (control), pME2045 (TEC1 CEN), pME2047 (TEC1 2 μm), pME2046 (abaA CEN), or pME2048 (abaA 2μm) were transformed with YCplac33 to obtain uracil prototrophy. Transformants were grown on solid nitrogen starvation medium for 4 days before pseudohyphal development was visualized under a microscope and photographed. Bar, 100 μm.
As expected, haploid tec1Δ and tec1Δ ste12Δ mutant strains failed to grow invasively when harboring the empty vectors as control plasmids (Fig. 1). Expression of TEC1 from the low-copy plasmid was sufficient to restore invasive growth in the tec1Δ strain but not in the tec1Δ ste12Δ background. However, defective agar invasion of the tec1Δ ste12Δ strain was fully restored when TEC1 was expressed either from a high-copy plasmid (Fig. 1A) or from the GAL1 promoter (Fig. 1B). Identical results were obtained for strains expressing AbaAp instead of Tec1p. Expression of a single copy of abaA driven by the TEC1 promoter restored agar invasion in the tec1Δ strain but not in the tec1Δ ste12Δ double mutant. As found for TEC1, defective invasive growth of the tec1Δ ste12Δ strain was suppressed by expression of abaA under control of the TEC1 promoter from a high-copy plasmid or when driven by the GAL1 promoter (Fig. 1A and B). These experiments show that both Tec1p and AbaAp can induce haploid invasive growth in the absence of Ste12p.
Pseudohyphal-development assays of diploid strains led to results similar to those obtained for invasive growth in haploids. Pseudohyphal growth was virtually absent in diploid tec1Δ/tec1Δ and tec1Δ/tec1Δ ste12Δ/ste12Δ mutant strains carrying control plasmids. Low-copy expression of TEC1 restored defective pseudohyphal growth of the tec1Δ/tec1Δ mutant but not of the tec1Δ/tec1Δ ste12Δ/ste12Δ double mutant. Again, pseudohyphal-growth defects of strains lacking STE12 could be suppressed by expression of TEC1 from the high-copy plasmid, although suppression was only partial compared to that of the tec1Δ/tec1Δ single mutant (Fig. 2). Low-copy expression of abaA driven by the TEC1 promoter only weakly stimulated pseudohyphal growth in the tec1Δ/tec1Δ mutant. However, expression of abaA driven from a high-copy plasmid was sufficient to induce pseudohyphal growth in the tec1Δ/tec1Δ and tec1Δ/tec1Δ ste12Δ/ste12Δ mutants. In summary, Tec1p and AbaAp are able to at least partially induce pseudohyphal development in the absence of Ste12p.
Expression of TEC1 requires Ste12p but is largely independent of Tec1p itself.
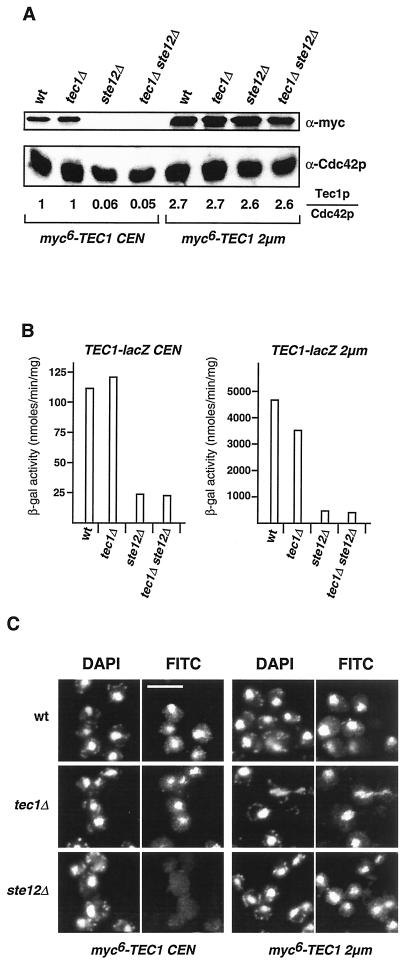
Haploid yeast wild-type, tec1Δ, ste12Δ, and tec1Δ ste12Δ strains were constructed that express a myc epitope-tagged version of Tec1p (myc6-Tec1p) from either a low-copy or a high-copy plasmid to measure intracellular amounts of Tec1p. Strains were tested for invasive-growth behavior and found to be indistinguishable from strains expressing untagged Tec1p (data not shown), demonstrating that the epitope-tagged version is fully functional. When expressed from a low-copy plasmid, myc6-Tec1p levels were identical in wild-type and tec1Δ strains (Fig. 3A). In contrast, myc6-Tec1p levels dropped roughly 20-fold in strains lacking STE12 (ste12Δ and tec1Δ ste12Δ), which consequently were unable to grow invasively. Thus, Ste12p is required for expression of Tec1p to levels required for induction of invasive growth. When expressed from a high-copy plasmid, almost identical amounts of myc6-Tec1p were detectable in all strains, even in the ste12Δ mutants (Fig. 3A). Levels of myc6-Tec1p expressed from high-copy-number plasmids were not more than 2.7-fold higher than levels obtained by low-copy expression of myc6-Tec1p in wild-type or tec1Δ strains. Thus, expression of TEC1 from the high-copy-number plasmid in ste12Δ mutant strains reflects a situation in which, in the absence of Ste12p, the amount of Tec1p is restored and therefore allows invasive growth.
FIG. 3.
Expression and localization of Tec1p in S. cerevisiae. (A) Expression of myc6-Tec1p. Protein extracts were prepared from haploid yeast strains RH2754 (wild type [wt]), RH2500 (tec1Δ), RH2755 (ste12Δ), and RH2501 (tec1Δ ste12Δ) expressing myc6-TEC1 on low-copy plasmid pME2295 (CEN) or high-copy plasmid pME2296 (2μm), and levels of myc6-Tec1p were determined by Western blot analysis with an anti-myc monoclonal antibody. As an internal control, expression levels of Cdc42p were measured in the same extracts by using an anti-Cdc42p polyclonal antibody (bottom). Relative Tec1p (Tec1p/Cdc42p) expression levels are shown in arbitrary units and were obtained by normalizing Tec1p signals to Cdc42p signals and to levels measured in strain RH2500 (tec1Δ) expressing TEC1 from a low-copy plasmid. (B) TEC1-lacZ expression levels. β-Galactosidase (β-gal) specific activity was measured in haploid strains carrying TEC1-lacZ on low-copy plasmid pME2300 (CEN) or high-copy plasmid pME2065 (2μm) and in the presence or absence of TEC1 and STE12 in different combinations. Bars depict means of three independent measurements, with standard deviations not exceeding 15%. (C) Localization of myc6-Tec1p. Haploid strains RH2754 (wt), RH2500 (tec1Δ), and RH2755 (ste12Δ) expressing myc6-TEC1 on low-copy plasmid pME2295 (CEN) or high-copy plasmid pME2296 (2μm) were grown to exponential phase and prepared for an anti-myc immunofluorescence assay. Shown are representative cells that were viewed for nuclear DNA with DAPI imaging or for anti-myc immunofluorescence (FITC). Bar, 5 μm.
Expression of a translational TEC1-lacZ fusion gene from low-copy and high-copy plasmids was measured in haploid wild-type and tec1Δ, ste12Δ, and tec1Δ ste12Δ mutant strains to quantify regulation of TEC1 gene expression by Ste12p and by Tec1p itself. Previous studies had shown that TEC1 transcript levels drop at least sixfold when Ste12p is absent and that activation of TEC1 transcription by Ste12p is predominantly mediated by several PRE sites present in the TEC1 promoter and only to a minor extent by its single FRE site (24, 40). We found that TEC1 is not required for expression of a TEC1-lacZ fusion gene expressed from the low-copy plasmid and only to a minor extent for expression of the high-copy version (Fig. 3B). This indicates that Tec1p does not greatly contribute to its own expression, a conclusion that is supported by the previous finding that high-copy expression of Tec1p induces its own expression not more than twofold (31). In contrast, expression of TEC1-lacZ was reduced between 4.5-fold (low-copy TEC1-lacZ) and 11-fold (high-copy TEC1-lacZ) when Ste12p was absent. High-copy expression of TEC1-lacZ in the ste12Δ background was only 3.8-fold higher than levels obtained by low-copy expression in strains carrying a functional STE12 gene, corroborating the results obtained by measuring Tec1p protein levels (Fig. 3A). However, several differences between TEC1-lacZ expression and Tec1p protein levels were found. In STE12-carrying strains (wild type or tec1Δ), expression of TEC1-lacZ from the high-copy plasmid was 42-fold higher than from the low-copy version (Fig. 3B). In contrast, Tec1p protein levels in STE12 strains are only 2.7-fold higher when TEC1 is expressed from the high-copy plasmid than when it is expressed from the low-copy version (Fig. 3A). Furthermore, TEC1-lacZ expression dropped between 4.5- and 11-fold when STE12 was deleted, whereas Tec1p protein levels decrease by a factor of 20 in the absence of Ste12p. These discrepancies could be explained by a difference in protein stability between Tec1p and β-galactosidase. In summary, Ste12p appears to control the expression of TEC1 to a larger extent than Tec1p itself.
Tec1p enters the yeast nucleus without Ste12p.
Intracellular localization of Tec1p has not been determined directly, although indirect evidence suggests that Tec1p is a nuclear protein (31). Here, subcellular localization of Tec1p was determined to analyze whether Ste12p is required for nuclear transport. Wild-type, tec1Δ, and ste12Δ strains that express myc epitope-tagged Tec1p from a low-copy or high-copy plasmid were used for indirect immunofluorescence microscopy. Specific myc6-Tec1p signals could be detected in the nuclei of wild-type and tec1Δ strains with both low- and high-copy expression (Fig. 3C). In the ste12Δ strain, specific nuclear signals were absent when myc6-Tec1p was expressed from the low-copy plasmid, corroborating the results obtained by Western blot analysis (Fig. 3A). Importantly, nuclear localization of myc6-Tec1p was restored in the ste12Δ strain when expressed from the high-copy plasmid, suggesting that Tec1p is a nuclear protein that enters the nucleus even in the absence of Ste12p.
Tec1p and AbaAp activate expression of FLO11 and PGU1 in the absence of Ste12p.
Our finding that Tec1p can induce invasive growth independently of Ste12p prompted us to measure expression of FLO11 and PGU1 in ste12Δ strains in which Tec1p levels had been restored by using high-copy-number plasmids. Expression of both the FLO11 and PGU1 genes is strongly reduced in the absence of either Tec1p or Ste12p (29, 32, 46). We reasoned that if expression of FLO11 and PGU1 could be activated in strains lacking Ste12p but expressing Tec1p at sufficient levels, activation of FLO11 and PGU1 by Tec1p would have to involve an Ste12p-independent control mechanism.
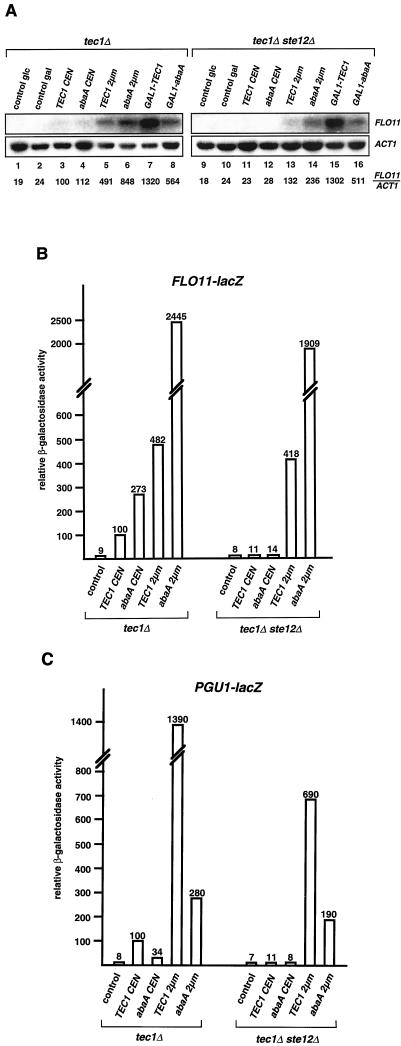
FLO11 transcript levels were measured in tec1Δ and tec1Δ ste12Δ strains expressing TEC1 or A. nidulans derived abaA from either the endogenous yeast TEC1 promoter on low- and high-copy plasmids or from the GAL1 promoter (Fig. 4A). In the tec1Δ genetic background, introduction of single copies of either TEC1 or abaA into yeast induced FLO11 transcription 4.2-fold (TEC1) and 4.7-fold (abaA) compared to that in a control strain expressing no Tec1p. High-copy expression of TEC1 or abaA or expression from the inducible GAL1 promoter led to an increase in FLO11 transcript levels of 4.9-fold (high-copy TEC1), 8.5-fold (high-copy abaA), 13-fold (GAL1-TEC1), or 5.6-fold (GAL1-abaA) compared to those in strains expressing a single copy of TEC1. Thus, expression of FLO11 depends on the dosage of Tec1p and AbaAp. In the tec1Δ ste12Δ strain, introduction of a single copy of TEC1 or abaA was not sufficient to activate normal FLO11 expression. However, high-copy expression of Tec1p or AbaAp restored FLO11 transcription to levels detected in the tec1Δ strain expressing single copies of TEC1 or abaA (corresponding to a wild-type situation). Even stronger induction of FLO11 transcription was measured for GAL1-TEC1 (13-fold) and GAL1-abaA (5.1-fold). These results demonstrate that, in the absence of Ste12p, both Tec1p and AbaAp are able to activate expression of FLO11 to levels sufficient for invasive growth.
FIG. 4.
TEC1 and abaA activate expression of FLO11 and PGU1 in the absence of STE12 in yeast. (A) FLO11 transcript levels. Strains RH2500 (tec1Δ) and RH2501 (tec1Δ ste12Δ) carrying plasmid YCplac111 (control), pME2045 (TEC1 CEN), pME2046 (abaA CEN), pME2047 (TEC1 2μm), pME2048 (abaA 2μm), pME2049 (GAL1-TEC1), or pME2050 (GAL1-abaA) were grown in SC-Leu medium containing 2% glucose (lanes 1, 3, 4, 5, 6, 9, 11, 12, 13, and 14) or 2% galactose (lanes 2, 7, 8, 10, 15, and 16) to exponential phase before total RNA was prepared and used for Northern analysis. ACT1 gene expression served as an internal standard. Relative FLO11 (FLO11/ACT1) expression levels were obtained by using a Phospho-Imaging scanner and normalizing FLO11 transcript levels to ACT1 levels. All values are percentages of the FLO11 expression measured in control strain RH2500 (tec1Δ) expressing TEC1 from a low-copy plasmid (lane 3). (B) FLO11-lacZ expression levels. Strains described in panel A were transformed with plasmid B3782 carrying the FLO11-lacZ reporter (46) and grown to exponential phase before β-galactosidase specific activity was measured. Bars depict relative values normalized to the activity measured in strain RH2500 (tec1Δ) expressing TEC1 from a CEN plasmid, which was set at 100. (C) PGU1-lacZ expression levels. β-Galactosidase activity was measured in strains carrying PGU1-lacZ reporter plasmid pME2064 (32) and expressing TEC1 and A. nidulans abaA driven by the TEC1 promoter at different levels. Activities are given as relative values normalized to that of strain RH2500 (tec1Δ) expressing TEC1 from a CEN plasmid, which was defined as 100. The bars in panels B and C depict means of three independent measurements, with standard deviations not exceeding 20%.
Results obtained by analysis of FLO11 transcript levels were further corroborated by β-galactosidase assays with a FLO11-lacZ reporter gene (46) that was introduced into tec1Δ and tec1Δ ste12Δ yeast strains expressing TEC1 and abaA at different levels (Fig. 4B). In the tec1Δ strain, single copies of TEC1 or abaA increased FLO11-lacZ reporter activity 11.1-fold (TEC1) or 27.3-fold (abaA) in comparison with that of a control strain lacking TEC1 (Fig. 4B). High-copy expression of TEC1 induced the expression of FLO11-lacZ 4.8-fold compared with that of low-copy TEC1, whereas high-copy abaA led to 24.4-fold higher reporter activity. Again, a single copy of TEC1 or abaA was not sufficient to activate FLO11-lacZ expression in the absence of STE12 (tec1Δ ste12Δ background), whereas high-copy expression of both TEC1 and abaA led to high levels of FLO11-lacZ reporter activity that were comparable to those measured in the presence of STE12 (tec1Δ background).
Regulation of PGU1 was measured by use of a PGU1-lacZ reporter gene (32) that was expressed in tec1Δ and tec1Δ ste12Δ strains containing no, single, or high numbers of copies of TEC1 or abaA (Fig. 4C). Expression of PGU1-lacZ was stimulated 12.5-fold by the introduction of a single TEC1 copy into the tec1Δ background and a further 13.9-fold by high-copy expression of TEC1. A 3.4-fold induction was obtained by a single copy, and a further 8.2-fold induction was obtained by high-copy abaA. In the absence of Ste12p (tec1Δ ste12Δ strain), a single copy of TEC1 or abaA was not able to significantly stimulate PGU1-lacZ reporter activity beyond basal levels, but high-copy TEC1 or abaA activated reporter expression 99-fold (TEC1) and 27-fold (abaA), respectively. In summary, Tec1p and AbaAp significantly activate expression of FLO11 and PGU1 in the absence of Ste12p, a fact that might explain why these transcription factors are able to induce invasive growth in ste12Δ strains.
Tec1p activates gene expression via TCS elements in the absence of Ste12p.
The sole mechanism by which Tec1p has been proposed to activate gene expression is cooperative binding together with Ste12p to FRE sites present in the promoter region of target genes (3, 31, 32). In contrast, AbaAp activates target gene expression by binding to repeated TCS elements (1). By a computer search, we found that the promoter regions of both FLO11 and PGU1 contain several TCS sites without neighboring PRE sites, as found in a typical FRE site. The FLO11 promoter region contains four single TCS elements and a sequence composed of two TCS elements in close proximity, whereas the PGU1 promoter region contains three single TCS sites in a different orientation but does not contain a typical FRE. This suggested that Tec1p might activate gene expression not only via FRE sites but also via TCS elements. To test this hypothesis, we constructed a series of TCS-driven reporter genes by insertion of TCS elements in different numbers and orientations upstream of an enhancerless CYC1-lacZ gene (16).
Expression of these reporters was measured in tec1Δ and tec1Δ ste12Δ yeast strains expressing TEC1 at different levels and compared to that of CYC1-lacZ reporter genes containing either no UAS element or a single FRE site (Fig. 5). Without a functional UAS element, the CYC1-lacZ reporter could not be activated beyond basal levels under any condition. A single FRE in front of CYC1-lacZ led to a 375-fold induction of reporter activity that was dependent on TEC1 and STE12. In the presence of STE12 alone (control plasmid in the tec1Δ strain), no activation of the FRE-driven reporter beyond basal levels was found, corroborating the finding that Tec1p and Ste12p cooperatively activate FRE-driven gene expression (31). However, TEC1 alone was sufficient to activate the FRE-driven reporter up to sevenfold even without STE12 (TEC1 on a 2μm plasmid in the tec1Δ ste12Δ strain), suggesting that the TCS element present in the FRE site can be used by Tec1p, mediating significant activation of gene expression. This finding is in agreement with the fact that single TCS elements in a forward or backward orientation were sufficient to mediate at least 10-fold activation of the CYC1-lacZ reporter by Tec1p, even in the absence of Ste12p. In contrast to the FRE-reporter, activation mediated by TCS elements was Tec1p dosage dependent, because induction was 2- to 3-fold (up to 116-fold) higher in strains containing a 2- to 3-fold greater amount of Tec1p (TEC1 expressed from a 2μm plasmid in a tec1Δ background) compared to activation (up to 46-fold) measured in strains expressing lower levels of Tec1p (TEC1 expressed from a CEN plasmid; compare Fig. 3A). Dosage dependence was also observed when STE12 was absent. TCS reporters were not induced in ste12Δ strains expressing TEC1 from low-copy plasmids (TEC1 CEN), a finding that is explained by the fact that Ste12p is required for TEC1 expression and, therefore, in these strains, only low levels of Tec1p can be detected by Western analysis (Fig. 3A). However, restoration of intracellular amounts of Tec1p (TEC1 expressed from a 2μm plasmid in tec1Δ ste12Δ strains) also restored activation of the TCS-driven reporters (up to 10-fold) comparable to a wild-type situation. Double TCS elements led to activation of CYC1-lacZ reporter expression between 61- and 265-fold, depending on the spacing between the TCS elements and the Tec1p levels. Optimal activation was observed when spacing between TCS elements was 11 bp. Stimulation of gene expression was almost threefold better than that obtained by single TCS elements, pointing to a synergistic effect of the double element. In contrast, double TCS elements separated by 6 bp led to only 1.3-fold better activation (61 U) than single TCS elements (46 U). As found for single-TCS-driven reporters, activation mediated by double TCS elements was Tec1p dosage dependent. Again, restoration of Tec1p protein levels in ste12Δ strains by expression of TEC1 from the 2μm plasmid led to activation of the double-TCS reporters comparable to that of the wild-type situation. We further tested a reporter construct composed of three TCS elements in a backward orientation, each separated by 13 bp. In comparison to the reporter with two backward-oriented TCS elements, this triple-TCS reporter could be further activated (261-fold versus 171-fold) when Tec1p was present at high levels.
In summary, these results demonstrate that Tec1p is able to activate gene expression not only via FREs (FRE control) but also via single or multiple TCS elements (TCS control). Interestingly, TCS control depends on Ste12p to some degree because we found a consistent two- to threefold drop in the expression of all TCS reporters in ste12Δ strains compared to STE12 strains (which contain comparable levels of Tec1p). However, Tec1p alone is sufficient to significantly activate TCS-dependent gene expression up to 20-fold even in the absence of Ste12p.
FRE- and TCS-driven reporters were further tested for activation by abaA. abaA activated the FRE reporter 26-fold when expressed from the endogenous TEC1 promoter on a low-copy plasmid. Interestingly, AbaAp appears to activate the FRE reporter cooperatively with Ste12p, because activation was up to 70-fold (2μm plasmid), whereas activation mediated by single TCS elements was not more than 22-fold. This suggests even greater similarity between the mechanisms of Tec1p and AbaAp function than previously assumed. Double-TCS-driven reporters were activated by AbaAp between 31- and 72-fold, depending on the spacing between the TCS elements. The best activation (72-fold) was found for double-TCS elements oriented as inverted repeats and separated by 14 bp. This finding is in agreement with an earlier study showing that inverted TCS repeats with a 13-bp spacing are optimal for activation (1). As found for Tec1p, TCS-mediated activation by AbaAp was functional even in strains lacking Ste12p.
The C-terminal region of Tec1p is required for haploid invasive growth and TCS-mediated reporter gene expression but is dispensable for FRE-driven gene expression.
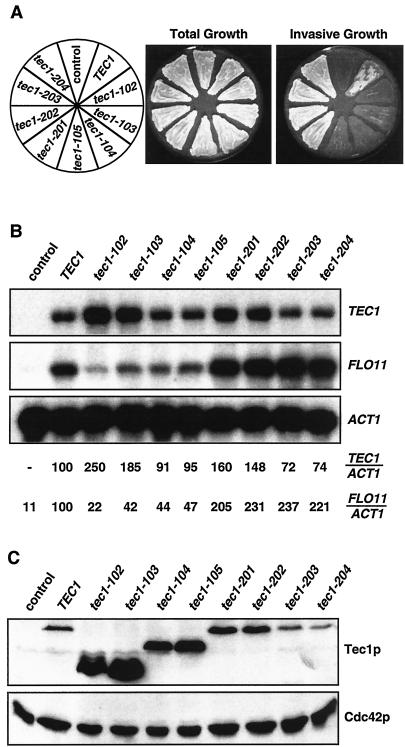
We isolated novel variants of the TEC1 gene to characterize functional domains of the Tec1p protein. A library of PCR-mutagenized TEC1 alleles was created and introduced into a haploid tec1Δ yeast strain that carried a chromosomally integrated FRE-lacZ reporter gene. Resulting transformants were screened for invasive growth and FRE-lacZ reporter activity, yielding two classes of mutants (Table 3 and Fig. 6A). Class I m utants were defective for invasive growth but still promoted induction of the FRE-lacZ reporter. Class II mutants were enhanced for both invasive growth and FRE-lacZ expression. No class of mutants could be isolated that grew invasively but failed to activate FRE-lacZ. Mutants being suppressed for both phenotypes were excluded. Sequence analysis revealed that each of the class I mutant alleles encoded a C-terminally truncated protein and that all of the class II alleles coded for proteins with single or double amino acid substitutions (Table 3).
TABLE 3.
Invasion-defective and hyperinvasive TEC1 mutants
| Class and allele | Mutation(s) | Invasive growtha | Fold FRE-lacZ expressionb |
|---|---|---|---|
| Control | |||
| tec1Δ | Full deletion | − | <1 |
| TEC1 | None | +++ | 100 |
| Class I | |||
| tec1-102 | Δ(K281-Y486) | + | 67 |
| tec1-103 | E222G, Δ(Y257-Y486) | + | 89 |
| tec1-104 | F93L, Δ(P335-Y486) | + | 104 |
| tec1-105 | Δ(I353-Y486) | + | 98 |
| Class II | |||
| tec1-201 | T273M | ++++ | 166 |
| tec1-202 | P274S | ++++ | 157 |
| tec1-203 | K154R, S453P | ++++ | 127 |
| tec1-204 | K154R, S262G | ++++ | 105 |
Invasive growth was measured by a plate-washing assay and quantified as described in reference 34.
Induction of an FRE-lacZ reporter by the different TEC1 alleles was measured in strain RH2499. The values shown are relative β-galactosidase activities normalized to the activity obtained with wild-type TEC1, which was set at 100.
FIG. 6.
Characterization of novel yeast TEC1 alleles. (A) Regulation of haploid invasive growth. Strain RH2499 (tec1Δ) carrying a control plasmid, wild-type TEC1, or different TEC1 mutant alleles on low-copy plasmids was patched on SC-Leu-Ura medium and grown for 4 days before the plate was photographed before (total growth) and after (invasive growth) cells were washed off the agar surface. (B) Expression of FLO11 depending on different TEC1 alleles. Shown are transcript levels of TEC1, FLO11, and ACT1 measured in strain RH2499 (tec1Δ) expressing different TEC1 alleles. ACT1 transcripts served as an internal standard. Relative expression levels of TEC1 alleles (TEC1/ACT1) and FLO11 (FLO11/TEC1) are indicated below and were obtained with a Phospho-Imaging scanner, followed by normalization of TEC1 and FLO11 transcript levels to ACT1 levels and to a strain expressing wild-type TEC1. (C) Western blot analysis. Shown are steady-state levels of the various myc-tagged Tec1p proteins obtained by expression of the indicated TEC1 alleles in strain RH2499. Proteins were detected by using an anti-myc monoclonal antibody, and expression of Cdc42p was measured in the same extracts as an internal control.
A triple-myc epitope tag was inserted just after the start codons of all TEC1 mutant genes to further characterize mutant proteins. No phenotypic differences were detected between the epitope-tagged and nontagged versions (data not shown). TEC1 mRNA and Tec1p protein levels were measured in all mutants to exclude the possibility that reduced expression or stability of Tec1p mutant proteins might account for the phenotypes observed (Fig. 6B and C). No significant decrease in mRNA levels was found for any of the TEC1 mutant alleles. However, we detected increases in the transcript levels of tec1-102 (2.5-fold), tec1-103 (1.9-fold), tec1-201 (1.6-fold), and tec1-202 (1.5-fold). Levels of the Tec1-102p, Tec1-103p, Tec1-104p, and Tec1-105p proteins were markedly higher than that of the wild-type protein, excluding the possibility that low expression of these variants was responsible for the reduced invasiveness of the corresponding mutant strains. In contrast, reduced levels of the Tec1-203p and Tec1-204p proteins were detected, ruling out the possibility that increased expression of these mutant forms was the cause of the enhanced invasive growth of these class II mutants.
FLO11 transcripts levels were measured in the different TEC1 mutant strains (Fig. 6B). All strains expressing class I mutant alleles showed a significant decrease in FLO11 expression, with levels varying between 22 and 47% with respect to the wild-type control, whereas values between 205 and 237% were obtained with strains expressing the class II alleles. Thus, the degree of invasive growth correlates well with expression of FLO11 in both classes of TEC1 mutants.
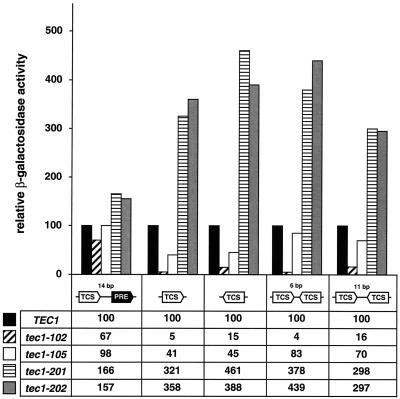
Two TEC1-encoded mutant proteins each from classes I and II were further tested for activation of four different TCS-lacZ reporters and compared to FRE-lacZ reporter activation (Fig. 7). From class I, the severely truncated Tec1-102p mutant protein (ΔK281-Y486) was almost completely deficient in the activation of any of the TCS-lacZ reporters, with values reaching not more than 16% of those of the wild-type control. In contrast, Tec1-102p was still able to efficiently activate expression of FRE-lacZ to levels corresponding to 67% of the level achieved by wild-type Tec1p. The less severely truncated Tec1-105p variant (ΔI353-Y486) activated FRE-lacZ indistinguishably from wild-type Tec1p, whereas expression of the single TCS-lacZ reporters dropped to 41% (forward orientation of TCS) or 45% (backward orientation). Expression of double-TCS-lacZ reporters by Tec1-105p was found to be reduced less significantly, with values of up to 83% of the wild-type control. From class II, Tec1-201p and Tec1-202p strongly activated all TCS-lacZ reporters between 3.0- and 4.6-fold in comparison to wild-type Tec1p. In contrast, these variants stimulated FRE-lacZ no more than 1.6-fold.
FIG. 7.
Activation of FRE- and TCS-dependent CYC1-lacZ reporter genes by TEC1 mutant alleles in yeast. Wild-type TEC1 and tec1-102, tec1-105, tec1-201, and tec1-202 mutant alleles were expressed in tec1Δ mutant strains carrying a CYC1-lacZ fusion gene driven by an FRE (TCS plus PRE) or TCS elements in different combinations, and strains were assayed for β-galactosidase specific activity. Bars and values represent relative activities normalized to the value obtained by expression of wild-type TEC1 (black bars), which was set at 100. All values are means of three independent measurements. The standard deviation was less than 20%.
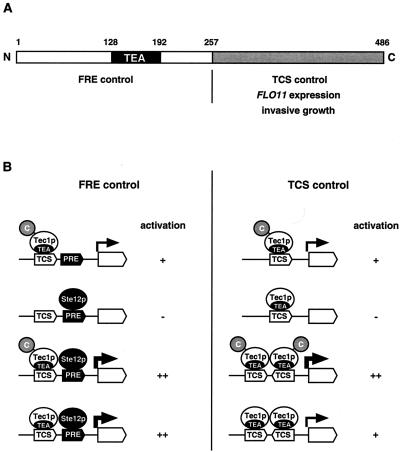
In summary, these data suggest that distinct domains of Tec1p are required for activation of gene expression mediated by either TCS or FRE sites (Fig. 8). The N-terminal region of Tec1p appears to be sufficient for transcriptional activation via FRE but not via TCS sites. TCS-mediated activation by Tec1p additionally requires the C-terminal part of the protein.
FIG. 8.
Domains and functions of yeast Tec1p. (A) Domain structure of Tec1p. The N-terminal part of Tec1p encompassing amino acids 1 to 257 includes the conserved TEA DNA-binding domain and is sufficient for activation of FRE-dependent gene expression together with Ste12p (FRE control). The C-terminal domain spanning amino acids 258 to 486 is required for the activation of gene expression via TCS elements (TCS control). (B) FRE and TCS control mechanisms. High-level activation of gene expression (++) via combined TCS and PRE elements (FRE control) depends on cooperative binding of Tec1p and Ste12p and does not require the C-terminal part of Tec1p. The presence of Ste12p is not sufficient for activation (−). In contrast, the presence of Tec1p alone is sufficient for significant activation (+) of FRE-driven expression via the single TCS site. TCS control depends on the number of TCS elements and the C-terminal part of Tec1p. Single TCS elements mediate significant activation (+) of gene expression by Tec1p depending on its C-terminal part. High-level activation (++) comparable to FRE-mediated expression is achieved by combination of multiple TCS elements but also depends on the C terminus of Tec1p.
DISCUSSION
Ste12p and Tec1p regulate gene expression and cellular development by several distinct mechanisms.
The role of the TEA/ATTS family transcription factor Tec1p from S. cerevisiae in the regulation of gene expression and cellular development was investigated. Tec1p was previously thought to regulate these processes primarily by cooperative binding, together with Ste12p, to FRE sites present in the promoter region of target genes and of TEC1 itself. This combinatorial model of Tec1p regulation is based on the observation that strains lacking either TEC1 or STE12 or both are equally suppressed for haploid invasive and diploid pseudohyphal growth (12, 27, 31, 34, 36, 44). The finding that FRE enhancer elements are bound in vitro by Tec1p and Ste12p in a cooperative manner further supports this model (31). The assumption that combinatorial control is the sole mechanism of Tec1p-regulated processes implies that the presence of either Tec1p or Ste12p alone is not sufficient to activate target gene expression and development. However, this prediction has not been tested experimentally prior to our study. Here, we have shown that strains containing sufficient amounts of Tec1p but lacking Ste12p are able to activate natural target genes and reporter genes that are driven by TCS elements. As a consequence, these strains undergo significant cellular development, although they do not contain any Ste12p. We further present evidence that Ste12p controls expression of TEC1 to a larger extent than Tec1p itself, suggesting the involvement of mechanisms other than combinatorial control. Our data suggest that Ste12p and Tec1p control gene expression and cellular development by several distinct mechanisms (Fig. 9). We propose a model in which Ste12p regulates expression of TEC1 by both combinatorial control together with Tec1p via FRE sites and without Tec1p by acting via clustered PRE sites present in the TEC1 promoter. In turn, Tec1p regulates gene expression by two distinct mechanisms, FRE control in combination with Ste12p and TCS control that is operative even without Ste12p.
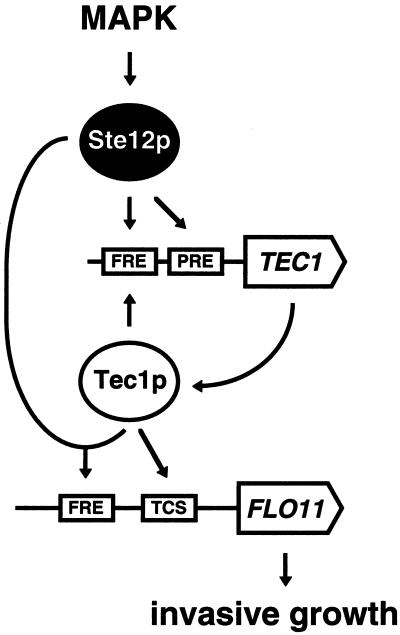
FIG. 9.
Model for regulation of FLO11 expression and invasive growth by Tec1p. The filamentation/invasion mitogen-activated protein kinase (MAPK) cascade controls Ste12p, which activates TEC1 expression either without Tec1p by binding to PRE sites or in combination with Tec1p by binding to a single FRE site present in the TEC1 promoter. Tec1p can activate expression of FLO11 and invasive growth either in combination with Ste12p via the single FRE site or without Ste12p via the TCS sites present in the FLO11 promoter.
TCS control is an efficient mechanism for activation of developmental target genes by Tec1p.
A central finding of our study is that single TCS elements inserted upstream of a reporter gene are sufficient to mediate activation of gene expression by Tec1p. Activation is up to 100-fold and independent of the orientation of the TCS. This finding correlates well with the distribution and arrangement of TCS elements found in natural S. cerevisiae promoter regions. In silico analysis of the complete S. cerevisiae genome reveals a group of more than 100 genes that contain at least four TCS elements in their promoter regions (our unpublished results). Most of these TCS elements are arranged individually; that is, they are separated from each other by at least 20 bp and are not neighbored by an Ste12p-binding site. Remarkably, this group includes FLO11, FLO8, PHD1, DFG10, CLN1, CDC25, and PGU1, which are all genes that have previously been implicated in invasive and pseudohyphal growth (14, 15, 28-30, 32, 34, 46). Here, we have shown that two representatives of these genes, FLO11 and PGU1, are activated by Tec1p even when Ste12p is absent. This suggests that Tec1p, much like other TEA/ATTS family transcription factors, can activate target genes and cellular development also without Ste12p by TCS control. The view that TCS control is evolutionarily conserved is supported by several observations. (i) We demonstrated that in S. cerevisiae, the TEA/ATTS family transcription factor AbaAp from A. nidulans activates not only TCS-driven reporter genes but also FLO11 and PGU1 even when Ste12p is absent. In A. nidulans, AbaAp activates target genes by binding to AbaAp-responsive elements that are identical to TCS elements (1). As found for TCS elements in S. cerevisiae genes, multiple AbaAp-responsive elements are distributed predominantly in an individual arrangement in the promoter region of known target genes of AbaAp, such as brlA, wetA, yA, rodA, and abaA itself (1). Moreover, AbaAp activates target genes and conidiophore development independently of SteAp, a protein with similarity to Ste12p, which regulates sexual but not AbaAp-dependent asexual development (50). In this context, it should be noted that A. nidulans asexual spore formation involves a process that is reminiscent of yeast pseudohyphal development (14). (ii) In the human pathogen C. albicans, the TEA/ATTS transcription factor CaTec1p regulates target genes, serum-induced hyphal growth, and virulence primarily by a mechanism that does not require the Ste12p-like protein CaCph1p (25, 26, 47). (iii) In mammals, members of the transcriptional enhancer factor (TEF-1) family, which have the TEA/ATTS DNA-binding domain in common, activate target genes by binding to the consensus sequence GGAATG, which matches the consensus of TCS elements in a reverse orientation (20-22).
Functions and functional domains of Tec1p.
The hallmark of TEA/ATTS family transcription factors is their DNA-binding domain, which consists of three putative α helices that, in the case of human TEF-1, have been demonstrated to be required for DNA binding (5, 20, 21). DNA binding and transcriptional activation have been proposed to involve either homodimer formation, as found in A. nidulans AbaAp (1), or heterodimer formation together with a second transcription factor, e.g., Ste12p, as shown for S. cerevisiae Tec1p (31), or serum response factor (SRF), as found in the case of human TEF-1 (17). Here, we have performed a functional analysis of S. cerevisiae Tec1p. Our results suggest that TEA/ATTS family transcription factors are able to regulate gene expression by at least two separate modes of action that involve distinct functional domains of the proteins. In the case of Tec1p, we found that gene expression is activated either by FRE control or by TCS control and that these two mechanisms are separable with respect to the structure of the Tec1p protein. We have shown that the N-terminal 257 amino acids of Tec1p, containing the conserved TEA/ATTS DNA-binding domain, are sufficient to confer cooperative FRE control together with Ste12p. This finding suggests that the N-terminal part of Tec1p might contain an Ste12p-interacting domain (Fig. 8). Although this assumption remains to be tested biochemically, it is supported by the observation that A. nidulans AbaAp activates FRE sites in S. cerevisiae in a cooperative manner that depends on Ste12p (Fig. 5). Because AbaAp and Tec1p do not share any regions showing significant similarity, other than the TEA/ATTS DNA-binding domain, cooperativity might be conferred by this conserved part of AbaAp. A previous study has shown that the N-terminal TEA/ATTS DNA-binding domain of mammalian TEF-1 is sufficient for physical interaction with serum response factor, a MADS box family transcription factor that confers combinatorial control together with TEF-1 in gene activation (17). Therefore, one might speculate that the evolutionarily conserved TEA/ATTS domain governs not only the DNA-binding activity but also the domain for heterodimer formation with binding partners that confer combinatorial control and cooperative activation of gene expression.
Tec1p possesses an additional domain in the C-terminal part that is required for gene activation by a second mechanism, TCS control. What is the mechanism by which Tec1p executes this function? An important observation is that TCS control does not appear to involve cooperativity. Our study shows that multiple TCS elements mediate activation in a predominantly additive manner because no significant cooperative effects can be observed in the case of double or triple TCSs (Fig. 5). A slight cooperative effect is observed when two inverted TCS elements are separated by 11 bp. However, cooperativity is not very pronounced (1.4-fold) and not comparable to that observed for an FRE site when Tec1p acts together with Ste12p (19-fold). Remarkably, Tec1p can also activate an FRE-driven reporter gene in the absence of Ste12p. Obviously, Tec1p can bind to the TCS element within the FRE and activate transcription without Ste12p. In this case, however, cooperativity is lost and TCS control depends on the dosage of the Tec1p protein, much like that observed for TCS-driven reporters. Whether Tec1p binds to single TCS elements as a monomer or as a homodimer cannot be concluded from our study. However, biochemical studies with AbaAp suggest that it binds to single TCS elements predominantly as a monomer and not as a homodimer (1). Taken together, these findings support a mechanism by which Tec1p binds as a monomer to single TCS elements and activates transcription by involvement of its C-terminal part (Fig. 8).
What is the function of the C-terminal part of Tec1p, and why is it required for TCS control but not for FRE control? A simple answer to this questions is that the C-terminal portion of Tec1p provides the same function(s) for the TCS control mechanism as does Ste12p for the FRE control mechanism. These functions might include nuclear transport, regulation of DNA-binding activity, or interaction with the transcriptional apparatus. Whether the C-terminal part of Tec1p fulfills these functions on its own or by interaction with further partners remains to be elucidated. Interestingly, Tec1p has been found to be physically associated not only with Ste12p but also with the mitogen-activated protein kinase Kss1p (19). This opens the possibility that FRE control and TCS control involve association of Tec1p with Kss1p. In the case of FRE control, this interaction might be established by the TEA/ATTS domain and involve Ste12p, whereas in the case of TCS control, the C-terminal part of Tec1p could associate with Kss1p independently of Ste12p. However, Ste12p might also be involved in TCS control mediated by the C-terminal part because TCS-driven expression was found to be two- to threefold less efficient in the absence of Ste12p. In this scenario, non-DNA-bound Ste12p would contribute to TCS control by interaction with the C terminus of Tec1p in a manner similar to interaction of Ste12p with the alpha-1 protein at alpha-specific genes (54).
In conclusion, our study shows that the TEA/ATTS transcription factor Tec1p fulfills more than one function in the regulation of gene expression and cellular development. These functions are executed by distinct domains of Tec1p and require combination with additional factors, e.g., Ste12p or maybe Kss1p. It will be interesting to resolve the exact temporal and spatial interactions between the different domains of Tec1p and these factors in the future.
Acknowledgments
We are grateful to Maria Meyer for excellent technical assistance during the course of this work. We thank Alex Andrianopoulos, Gerry Fink, Hiten Madhani, Steffen Rupp, and Bill Timberlake for generously providing plasmids. We thank Sven Krappmann and Ralph Pries for helpful comments on the manuscript and Olav Grundmann for help with some of the experiments.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Volkswagenstiftung.
REFERENCES
- 1.Andrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A., and W. E. Timberlake. 1991. ATTS, a new and conserved DNA binding domain. Plant Cell 3:747-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50:643-650. [DOI] [PubMed] [Google Scholar]
- 5.Bürglin, T. R. 1991. The TEA domain: a novel, highly conserved DNA-binding motif. Cell 66:11-12. [DOI] [PubMed] [Google Scholar]
- 6.Cross, F. R., and A. H. Tinkelenberg. 1991. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65:875-883. [DOI] [PubMed] [Google Scholar]
- 7.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolan, J. W., C. Kirkman, and S. Fields. 1989. The yeast Ste12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86:5703-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdman, S., and M. Snyder. 2001. A filamentous growth response mediated by the yeast mating pathway. Genetics 159:919-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errede, B., and G. Ammerer. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349-1361. [DOI] [PubMed] [Google Scholar]
- 11.Fields, S., and I. Herskowitz. 1985. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell 42:923-930. [DOI] [PubMed] [Google Scholar]
- 12.Gavrias, V., A. Andrianopoulos, C. J. Gimeno, and W. E. Timberlake. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255-1263. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 14.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 16.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, M., P. Kogut, F. J. Davis, N. S. Belaguli, R. J. Schwartz, and M. P. Gupta. 2001. Physical interaction between the MADS box of serum response factor and the TEA/ATTS DNA-binding domain of transcription enhancer factor-1. J. Biol. Chem. 276:10413-10422. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194. [PubMed]
- 19.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, J. J., P. Chambon, and I. Davidson. 1993. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 12:2337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacquemin, P., J. J. Hwang, J. A. Martial, P. Dolle, and I. Davidson. 1996. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J. Biol. Chem. 271:21775-21785. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, S. W., D. Desai, S. Khan, and N. L. Eberhardt. 2000. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 19:507-514. [DOI] [PubMed] [Google Scholar]
- 23.Kronstad, J. W., J. A. Holly, and V. L. MacKay. 1987. A yeast operator overlaps an upstream activation site. Cell 50:369-377. [DOI] [PubMed] [Google Scholar]
- 24.Laloux, I., E. Dubois, M. Dewerchin, and E. Jacobs. 1990. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol. Cell. Biol. 10:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, S., S. Zhou, T. Pan, Q. Dai, and H. Liu. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21:6418-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 28.Liu, H., C. A. Styles, and G. R. Fink. 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb, J. D., T. A. Kerentseva, T. Pan, M. Sepulveda-Becerra, and H. Liu. 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153:1535-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 32.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mösch, H.-U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mösch, H. U., and G. R. Fink. 1997. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145:671-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mösch, H. U., R. Graf, and G. H. Braus. 1992. Sequence-specific initiator elements focus initiation of transcription to distinct sites in the yeast TRP4 promoter. EMBO J. 11:4583-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mösch, H. U., E. Kübler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 38.Oehlen, L., and F. R. Cross. 1998. The mating factor response pathway regulates transcription of TEC1, a gene involved in pseudohyphal differentiation of Saccharomyces cerevisiae. FEBS Lett. 429:83-88. [DOI] [PubMed] [Google Scholar]
- 39.Oehlen, L. J., and F. R. Cross. 1994. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 8:1058-1070. [DOI] [PubMed] [Google Scholar]
- 40.Oehlen, L. J., and F. R. Cross. 1998. The role of Cdc42 in signal transduction and mating of the budding yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:8556-8559. [DOI] [PubMed] [Google Scholar]
- 41.Oehlen, L. J., J. D. McKinney, and F. R. Cross. 1996. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol. Cell. Biol. 16:2830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, R. L., H. U. Mösch, and G. R. Fink. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89:1055-1065. [DOI] [PubMed] [Google Scholar]
- 46.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schroppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 48.Sengstag, C., and A. Hinnen. 1988. A 28-bp segment of the Saccharomyces cerevisiae PHO5 upstream activator sequence confers phosphate control to the CYC1-lacZ gene fusion. Gene 67:223-228. [DOI] [PubMed] [Google Scholar]
- 49.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vallim, M. A., K. Y. Miller, and B. L. Miller. 2000. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36:290-301. [DOI] [PubMed] [Google Scholar]
- 51.Van Arsdell, S. W., G. L. Stetler, and J. Thorner. 1987. The yeast repeated element sigma contains a hormone-inducible promoter. Mol. Cell. Biol. 7:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao, J. H., I. Davidson, H. Matthes, J. M. Garnier, and P. Chambon. 1991. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65:551-568. [DOI] [PubMed] [Google Scholar]
- 53.Yuan, Y. L., and S. Fields. 1991. Properties of the DNA-binding domain of the Saccharomyces cerevisiae Ste12 protein. Mol. Cell. Biol. 11:5910-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, Y. O., Stroke, I. L., and S. Fields. 1993. Coupling of cell identity to signal response in yeast: interaction between the alpha-1 and Ste12 proteins. Genes Dev. 7:1584-1597. [DOI] [PubMed] [Google Scholar]