Abstract
The distribution and phylogenetic affiliations of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV)-degrading denitrifying bacteria in activated sludge were studied by a polyphasic approach including culture-independent biomarker and molecular analyses as well as cultivation methods. A total of 23 strains of PHBV-degrading denitrifiers were isolated from activated sludges from different sewage treatment plants. 16S ribosomal DNA (rDNA) sequence comparisons showed that 20 of the isolates were identified as members of the family Comamonadaceae, a major group of β-Proteobacteria. When the sludges from different plants were acclimated with PHBV under denitrifying conditions in laboratory scale reactors, the nitrate removal rate increased linearly during the first 4 weeks and reached 20 mg NO3−-N h−1 g of dry sludge−1 at the steady state. The bacterial-community change in the laboratory scale sludges during the acclimation was monitored by rRNA-targeted fluorescence in situ hybridization and quinone profiling. Both approaches showed that the population of β-Proteobacteria in the laboratory sludges increased sharply during acclimation regardless of their origins. 16S rDNA clone libraries were constructed from two different acclimated sludges, and a total of 37 clones from the libraries were phylogenetically analyzed. Most of the 16S rDNA clones were grouped with members of the family Comamonadaceae. The results of our polyphasic approach indicate that β-Proteobacteria, especially members of the family Comamonadaceae, are primary PHBV-degrading denitrifiers in activated sludge. Our data provide useful information for the development of a new nitrogen removal system with solid biopolymer as an electron donor.
Biological denitrification is an important process for nitrogen removal in wastewater treatment. Published reports suggest that 10 to 90% of bacteria in the activated-sludge system are capable of denitrification (16, 33, 44). However, the system is often confronted with the problem that the efficiency of nitrogen removal decreases due to low availability of organic matter as the reducing power for denitrification. To overcome this problem, a simple organic compound, such as methanol or acetate, is added intentionally as an electron donor to the denitrification process (16). In recent years, a new biotechnology of nitrogen removal using solid biopolymer as the electron donor has been developed (7). This type of nitrogen removal process, called here the solid-phase denitrification process, may have some advantages, e.g., a constant supply of reducing power, no secondary organic pollution, and ease of operation. A promising solid substrate for denitrification is the bacterial polyester poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) (6, 30, 40, 52), which serves as the source of biodegradable plastic (4, 17, 34, 50). The biodegradability of PHBV in natural environments has been extensively studied, and a number of PHBV-degrading bacteria have been isolated and characterized (1, 11, 38, 39, 41-43, 45, 55). The association of denitrification with intracellular poly(3-hydroxybutyrate) (PHB) metabolism in activated sludge has also been documented (5).
The questions regarding the solid-phase denitrification process using PHBV are what types of bacteria are actually responsible for nitrogen removal in this process and what is the level of phylogenetic variation among the bacteria present. Some denitrifying bacterial strains that are capable of anaerobic degradation of PHBV have been isolated from aquatic environments. These isolates were identified as Acidovorax facilis (40), Acidovorax sp. (52), Brevundimonas sp. (40), and Pseudomonas sp. (6). Our previous study reported the isolation of a new denitrifying β-proteobacterium that exhibits a denitrification rate as high as 19 mg of NO3−-N removed h−1 g−1 with PHBV as the electron donor (30). However, these collective data are not yet enough to provide the microbiological basis of the solid-phase denitrification process using bioplastic.
In order to get more information about the potential of this new biotechnology, we studied the distributions and phylogenetic identities of PHBV-degrading denitrifiers in activated sludge by using a polyphasic approach as reported in this study. Our strategy was a combined use of rRNA-targeted fluorescence in situ hybridization (FISH) (2, 3, 37, 58), quinone profiling (18, 24, 25, 27, 28), and PCR-aided 16S ribosomal DNA (rDNA) cloning and sequencing (8, 9, 54), in addition to culture-dependent isolation and characterization of PHBV-degrading denitrifying bacteria. These methods, differing in the principle of detection, are possibly complementary to each other to correct a technical bias specific to each one. Here, we report that β-Proteobacteria belonging to the family Comamonadaceae are primary PHBV-degrading denitrifiers in activated sludge based on the results of this polyphasic approach.
MATERIALS AND METHODS
Sludge samples.
Activated-sludge samples were collected from the main aerobic treatment tanks of sewage treatment plants located in Nagoya, Osaka, and Toyohashi, Japan, and were designated sludges NS, OS, and TS, respectively (Table 1). These sample names were numbered if sampling from the same plant was performed more than two times: e.g., NS1 and NS2. All samples were taken in polyethylene bottles, kept in an insulated cooler during transportation, and examined immediately upon return to the laboratory, not more than 6 h after sampling.
TABLE 1.
Population densities of bacteria and PHBV-degrading denitrifiers in activated sludge
| Sludge sample | Total counta (109 cells ml−1) | Total plate count (108 CFU ml−1)b | No. of PHBV-degrading denitrifiers
|
|
|---|---|---|---|---|
| CFU ml−1b | MPN ml−1 | |||
| NS1 | 6.9 ± 0.1 | ND | 4.9 ± 3.1 × 104 | ND |
| NS2 | 6.1 ± 0.3 | 3.1 ± 0.4 (2.3 ± 0.8) | 2.5 ± 1.4 × 105 (2.3 ± 1.0 × 105) | 1.2 × 108 |
| OS1 | 5.2 ± 0.5 | 1.3 ± 0.4 | 2.2 ± 1.4 × 104 | 1.0 × 107 |
| OS2 | 4.2 ± 0.3 | ND | 1.9 ± 1.3 × 103 | 9.0 × 106 |
| OS3 | 5.6 ± 0.1 | ND | 3.2 ± 2.0 × 103 | 1.1 × 107 |
| TS | 5.3 ± 0.5 | 0.94 ± 0.31 (1.0 ± 0.1) | 1.1 ± 0.2 × 103 (1.4 ± 0.5 × 103) | 7.3 × 106 |
Direct total count by EtBr staining. The average values and standard deviations from two different determinations are shown.
Average values and standard deviations for triplicate plates are shown. The figures in parentheses show the data obtained at 20 to 22°C for incubation. ND, not determined.
Acclimation of sludge to PHBV.
Four sludges taken from the three sewage treatment plants, i.e., sludges NS1, OS1, OS2, and TS, were collected by centrifugation, washed with phosphate-buffered saline (PBS) (10 mM K2HPO4 and 130 mM NaCl adjusted to pH 7.0 with HCl), and used as the seed sludges for the acclimation study. Screw-cap glass reactors (670-ml capacity) containing 500 ml of an acclimation medium were inoculated with one of the four sludges and incubated at 28°C for 10 weeks for acclimation to PHBV (Japan Monsanto Co., Tokyo, Japan) under denitrifying conditions. The acclimation medium used consisted of mineral base RM2 (20), 2 g of PHBV (5% hydroxyvalerate content) powder, and 2 g of KNO3 per liter of distilled water (pH 7.0). During acclimation, the reactors were gently stirred at 70 rpm min−1 with a magnetic stirrer; under these conditions, the dissolved oxygen tension was at <0.l mg liter−1. Every 3 days of operation, half of the supernatant in the reactors was exchanged with fresh medium, and the concentration of sludge was adjusted to ca. 2,000 mg (dry weight) liter−1. Samples were taken at appropriate intervals from the reactors and subjected to the analyses of denitrification activity and community structure described below. Laboratory sludges originating from the seed sludges NS1, OS1, OS2, and TS were designated NS1L, OS1L, OS2L, and TSL, respectively.
Enumeration, isolation, and cultivation.
For enumeration of bacteria, sludge samples were dispersed by sonication on ice for 90 s (20 kHz; output power, 50 W) and decimally diluted with PBS. Aerobic heterotrophic bacteria were enumerated by the plate-counting method as described previously (22). For the enumeration of PHB-degrading denitrifiers, we used two different enumeration methods, the most-probable number (MPN) method with triplicate tubes and the pour plate counting method. In MPN experiments, 1 ml of sample at each dilution step was inoculated into 20-ml screw-cap test tubes containing 10 ml of PHBN medium (30) and a Durham tube. This medium consisted of mineral base RM2, 2 g of PHB powder, 2 g of KNO3, and 0.1 g of yeast extract (Difco Laboratories) per liter of distilled water (pH 7.0). In some cases, this medium was modified by replacing PHB with an equal weight of PHBV (designated PHBVN medium). The test tubes were completely filled with the same medium just after inoculation and then incubated for 2 weeks before the bacteria were counted. The MPN was calculated based on the number of test tubes positive for gas formation in Durham tubes. For plate counting, 1 ml of diluted samples was plated with PHBVN agar (PHBVN medium plus 1.8% agar) modified by increasing the concentration of yeast extract to 0.05% and was incubated anaerobically using the AnaeroPak system (Mitsubishi Gas Chemicals, Niigata, Japan). After 4 to 5 weeks of incubation, colonies showing a zone of clearance were counted as positive for PHBV degradation and denitrification. Positive colonies were picked up from the plates and purified by repeated streaking on PHBN or PHBVN agar under anaerobic conditions. Incubation was at 28 to 30°C in all enumeration and isolation procedures. In addition, these experiments were performed at 20 to 22°C for some samples. Since all isolates thus obtained were aerobic chemoorganotrophic bacteria, they were maintained aerobically on agar slants containing a complex medium designated PBY (21). This medium was also used for preculture and routine cultivation, whereas PHBVN medium was used to cultivate cells under denitrifying conditions.
One of the isolates, strain NA10B, has been deposited with the Japan Collection of Microorganisms, RIKEN, Wako, Japan, as JCM 11421 and with the Collection de l'Institut Pasteur, Paris, France, as CIP 107294. All other isolates (Table 2) will be made available upon request.
TABLE 2.
Phenotypic characterization and phylogenetic affiliations of the PHBV-degrading denitrifying isolates
| Phylogenetic group and strain | Source | Denitrification rate with PHBV (NO3−-N removed h−1 g[dry wt])−1a | Major quinone type | 16S rDNA sequence comparison
|
|||
|---|---|---|---|---|---|---|---|
| Length of sequence determined (bases) | Species as closest relative | Accession no. | Similarity (%) | ||||
| β-Proteobacteria | |||||||
| NOS3 | NS1 | ND | Q-8 | 1,526 | [Aquaspirillum] psychrophilum | AF078755 | 97.6 |
| NOS8 | NS1 | ND | Q-8 | 1,526 | [Aquaspirillum] psychrophilum | AF078755 | 97.7 |
| NA10Bb | NS1 | 19 | Q-8 | 1,521 | Acidovorax avenae subsp. citrulli | AF078761 | 96.9 |
| NSP4 | NS2 | 12 | Q-8 | 1,521 | Comamonas terrigena | AF078766 | 96.1 |
| NSP5 | NS2 | 12 | Q-8 | 1,521 | C. terrigena | AF078772 | 96.7 |
| NSP7 | NS2 | 13 | Q-8 | 1,521 | C. terrigena | AF078772 | 96.1 |
| NSP8 | NS2 | 14 | Q-8 | 1,521 | C. terrigena | AF078772 | 96.7 |
| NS20-1b,c | NS2 | ND | Q-8 | 695 | A. avenae subsp. citrulli | AF078761 | 96.0 |
| NS20-2c | NS2 | ND | Q-8 | 696 | Acidovorax defluvii | Y18616 | 97.1 |
| KSP1 | OS1 | 18 | Q-8 | 1,521 | A. avenae subsp. citrulli | AF078761 | 98.3 |
| KSP2 | OS1 | 16 | Q-8 | 1,521 | A. avenae subsp. citrulli | AF078761 | 98.7 |
| KSP3b | OS1 | 19 | Q-8 | 1,522 | A. avenae subsp. citrulli | AF078761 | 96.9 |
| KSP4b | OS1 | 19 | Q-8 | 1,523 | A. avenae subsp. citrulli | AF078761 | 96.9 |
| OS-3 | OS2 | ND | Q-8 | 1,495 | C. terrigena | AF078772 | 96.1 |
| OS-6 | OS2 | ND | Q-8 | 1,522 | Acidovorax temperans | AF078766 | 97.5 |
| OS-9 | OS2 | ND | Q-8 | 1,523 | A. temperans | AF078766 | 96.6 |
| OS-14 | OS2 | ND | Q-8 | 1,521 | C. terrigena | AF078772 | 96.7 |
| OS-19 | OS2 | ND | Q-8 | 1,523 | A. avenae subsp. citrulli | AF078761 | 97.2 |
| TS-18 | TS | ND | Q-8 | 1,526 | [Aquaspirillum] psychrophilum | AF078755 | 97.6 |
| TS20-3b,c | TS | ND | Q-8 | 697 | A. avenae subsp. citrulli | AF078761 | 95.8 |
| γ-Proteobacteria | |||||||
| P400Y-1 | NS2 | 10 | Q-9 | 1,498 | Pseudomonas citronellolis | Z76659 | 98.0 |
| PG3-3 | OS1 | ND | Q-8 + MK-8 + DMK-8 | 1,505 | Aeromonas hydrophila | X60404 | 99.7 |
| PG4-1 | OS2 | ND | Q-8 + MK-8 + DMK-8 | 1,505 | A. hydrophila | X60404 | 99.7 |
ND, not determined.
These strains are more closely related to an unidentified bacterium, strain LW1 (99.9% similarity) (Fig. 4).
Strains isolated at 20 to 22°C. All other strains were isolated at 28 to 30°C.
Measurement of denitrification activity.
Cells grown in PHBN or PHBVN medium or sludge were collected by centrifugation, washed twice with 50 mM phosphate buffer (pH 7.0), and concentrated in a small volume of the buffer. Portions of the concentrated cell or sludge suspension were introduced into rubber-plugged test tubes (30-ml capacity) containing 25 ml of PHBVN medium from which (NH4)SO4 and yeast extract were eliminated. Anoxic conditions in the tubes were obtained by sparging argon. In some cases, the test tubes were incubated without sparging argon, because the denitrification rate was not affected by this treatment. The tubes were then incubated at 30°C for 24 to 48 h, and the concentrations of nitrate removed and nitrogen gas produced were monitored by ion chromatography and gas chromatography, respectively, as described previously (30, 35). Preliminary experiments showed that the amounts of nitrite and nitrous oxide produced as intermediates during denitrification were negligible in almost all cases. Therefore, the nitrate removal rate was considered the denitrification rate in this study.
PCR amplification and sequencing of 16S rDNA from isolates.
For PCR use, crude cell lysates as the DNA source were prepared as described previously (23). 16S rRNA gene fragments that corresponded to positions 8 to 1510 or 1543 in Escherichia coli 16S rRNA (12) were amplified from the cell lysate by PCR with Taq DNA polymerase (Takara Shuzo) and a pair of universal primers, 27f and 1492r or 1525r (32). The PCR products were treated with the chloroform-isoamyl alcohol mixture, purified by the polyethylene glycol precipitation method, and sequenced with a SequiTherm Long Read cycle-sequencing kit (Epicentre Technologies, Madison, Wis.). The reaction products were analyzed with a Pharmacia ALFexpress DNA sequencer.
Construction of 16S rDNA clone libraries.
Bulk DNA was isolated and purified from sludges OS1L and TSL according to the protocol previously reported (19). 16S rDNA from the sludge DNA was amplified by PCR with a set of universal primers, 27f and 1492r, as described above. The cycle profile consisted of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min for a total of 17 cycles; the final step was followed by postextension for 5 min. The PCR products were purified using a Geneclean Spin kit (Bio 101, Vista, Calif.) and subcloned with a pTBlue Perfectly Blunt cloning kit (Novagen, Madison, Wis.). Transformation of E. coli competent cells was carried out according to a standard manual of molecular cloning (49). Plasmid DNA was isolated and purified by using a Pharmacia Flexiprep kit according to the manufacturer's instructions. Cloned 16S rDNA was sequenced by the linear PCR sequencing method and analyzed with the Pharmacia DNA sequencer as described above.
Phylogenetic analysis.
Sequence data were compiled by the GENETYX-MAC program (Software Development Co., Tokyo, Japan), analyzed for chimera detection with the CHIMERA_CHECK program version 2.7, and compared with those retrieved from the Ribosomal Database Project II (36). Multiple alignment of sequences and calculation of the nucleotide substitution rate (Knuc) by Kimura's two-parameter model (31) were performed using the CLUSTAL W program (57). Distance matrix trees were constructed by the neighbor-joining method (48), and the topology of the trees was evaluated by bootstrapping with 1,000 resamplings (14). Alignment positions with gaps were excluded from the calculations.
Fluorescence microscopy.
Total bacterial counts were measured by epifluorescence microscopy with ethidium bromide (EtBr) staining as described previously (26, 47). In some cases, another nucleic acid-specific fluorochrome, SYBR Green II (Molecular Probes, Inc., Eugene, Oreg.), was used for cell counting, where the dye solution commercially available was 1/104-fold diluted before use. FISH was performed according to the protocol of Amann and Schleifer (3), with some modifications, in which the FITC- or Cy5-labeled oligonucleotide probes EUB338, ALF-1b, BET42a, and GAM42a, specific to the domain Bacteria (2) and to α-, β-, and γ-Proteobacteria (37), respectively, were used. Sludge was fixed with 3 volumes of paraformaldehyde, washed with PBS, and redissolved in this buffer. Three microliters of cell suspension was put on gelatin-coated slides and dehydrated through a series of 50, 80, and 98% ethanol. Ten microliters of hybridization buffer, along with 1 μl of a probe, was spotted on the fixed cells, and hybridization was carried out at 46°C for 90 min. The cells were observed under an Olympus BX-50 epifluorescence microscope equipped with an FD-120 M digital charge-coupled device camera (Flovel Co., Tokyo, Japan). The number of positive cells was determined with the image analysis program WINROOF; 10 to 15 fields per sample and a total of 1,000 to 2,000 cells per sample were taken to count.
Quinone profiling.
Quinones from sludge samples and the isolates were extracted with an organic solvent mixture and partially purified by column chromatography. Quinone components were separated and identified by reverse-phase high-performance liquid chromatography and photodiode array and mass spectrometry detection with external ubiquinone (Q-n) and menaquinone (MK-n) standards. Detailed information on these analytical procedures has been given previously (25, 28). Differences in quinone profiles among sludge samples were expressed by using the dissimilarity index (D), and the microbial divergence index (MDq) was used to show the extent of diversity of quinones detected (28). Clustering of sludge samples based on D matrix data was performed by the neighbor-joining method (48). Calculation of D and MDq values and construction of a neighbor-joining dendrogram were performed with the BioCLUST program (28). The dendrogram was illustrated using the TreeView program (46).
Nucleotide sequence accession numbers.
The 16S rDNA sequences of the isolates and the uncultured clones determined in this study have been deposited under DDBJ accession numbers AB076842 to AB076859 and AB076860 to AB076886, respectively.
RESULTS
Enumeration and isolation of PHB-degrading denitrifiers.
Population densities of PHBV-degrading denitrifying bacteria, as well as direct total counts and total heterotrophic plate counts in the six sludge samples, which were taken from four different sewage treatment plants, were measured by the MPN and plate-counting methods (Table 1). All of the sludge samples yielded PHBV-degrading denitrifiers at an order of magnitude of 103 to 105 CFU and 106 to 108 MPN per ml. The values estimated by the MPN method might be overestimated, because almost all strains of denitrifying bacteria isolated from the MPN tubes proved to be negative for PHBV degradation (data not shown). It was likely that the cometabolism of PHB-degrading nondenitrifying bacteria and denitrifying bacteria contributed to increasing MPN values. No marked differences in the plate counts were noted between the two temperature ranges (28 to 30°C and 20 to 22°C) of incubation.
Several single colonies that exhibited PHBV degradation were picked up from PHBVN agar plates used for enumeration and subjected to the standard purification procedure. As a result, a total of 23 strains were isolated from sludges NS1, NS2, OS1, OS2, and TS. The denitrification activity of these PHBV-degrading isolates was confirmed by gas formation in Durham tubes in anaerobic PHBVN cultures.
Phenotypic and phylogenetic characterization of isolates.
All of the isolates were found to be motile, rod-shaped bacteria by phase-contrast microscopy. The Gram reaction was negative. In all isolates, nitrate and nitrite reduction and nitrogen gas formation from nitrate were further confirmed by ion chromatography and gas chromatography. The isolates showed an optimum temperature of 28 to 37°C for growth and denitrification, independent of the temperature at which they were isolated.
The denitrification rates and phylogenetic affiliations of the isolates are summarized in Table 2. Of the isolates, 20 were assigned to the β-Proteobacteria and found to be closely related to species of Acidovorax, [Aquaspirillum] (the bracketing denotes a misclassified generic name), and Comamonas, all of which are genera of the family Comamonadaceae. However, many of these isolates had <98% similarity with the known species that were their closest relatives, suggesting that the isolates may be taxonomically new at the species level. The remaining strains were assigned to the genera Aeromonas and Pseudomonas, members of the γ-Proteobacteria. More detailed information about the phylogenetic positions of the isolates, together with uncultured 16S rDNA clones, is presented below.
Denitrification activity in acclimated sludge.
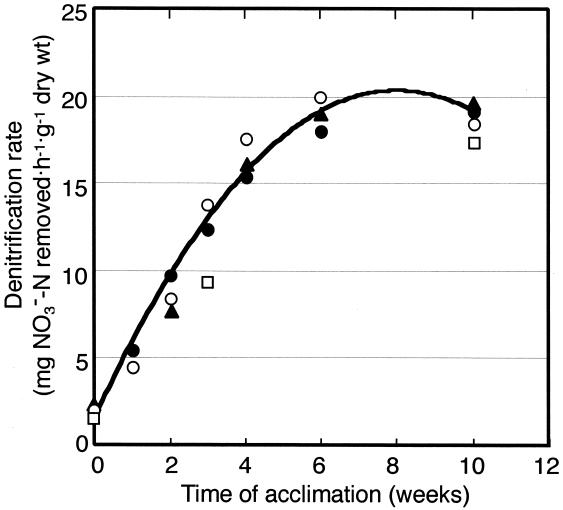
The above-mentioned results provided circumstantial evidence that members of the β-Proteobacteria may constitute the major population of PHBV-degrading denitrifiers in activated sludge. To confirm this, sludges NS1, OS1, OS2, and TS were acclimated with PHBV under denitrifying conditions in laboratory scale reactors and examined for denitrification activity and microbial-community changes. The denitrification rate in the laboratory sludges increased linearly during the first 4 weeks of acclimation and reached around 20 mg NO3−-N removed h−1 g (dry weight) of sludge−1 at the steady state, regardless of their origins (Fig. 1). The sludges at this stage yielded 102- to 103-fold-higher plate counts of PHBV-degrading denitrifiers than before the acclimation (data not shown). Thus, the laboratory sludges after 4 weeks of operation were sampled as acclimated sludges to study microbial-community shifts.
FIG. 1.
Changes in denitrification activities of four laboratory sludges during acclimation with PHBV under denitrifying conditions. Symbols: □, sludge NA1L; ○, sludge OS1L; •, sludge OS2L; ▴, sludge TSL. A polynomial regression curve based on the average values for these sludges is shown.
Microbial-community change during acclimation.
Comparative FISH analyses of the four sludges before and after acclimation showed that the population of β-Proteobacteria increased from 23 to 29 to 60 to 67% of the total population during acclimation (Table 3). In contrast, the population of α-Proteobacteria and γ-Proteobacteria decreased from 8 to 14 to 6 to 10% and from 5 to 7 to 2 to 4%, respectively.
TABLE 3.
Detection by FISH of specific bacterial groups in four different laboratory sludges during acclimation to PHBV under denitrifying conditions
| Target phylogenetic group (FISH probe used) | % of total counta in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| NS1L
|
OS1L
|
OS2L
|
TSL
|
|||||
| 1b | 28 | 0 | 28 | 0 | 28 | 0 | 30 | |
| Domain Bacteria (EUB338) | 72.0 ± 2.6 | 91.7 ± 0.6 | 68.0 ± 2.0 | 84.7 ± 4.0 | 68.7 ± 4.0 | 89.7 ± 1.5 | 64.3 ± 1.5 | 84.3 ± 3.1 |
| α-Proteobacteria (ALF-1b) | 12.4 ± 3.0 | 6.2 ± 1.5 | 10.4 ± 1.4 | 6.8 ± 0.8 | 8.1 ± 0.2 | 7.8 ± 0.6 | 13.7 ± 1.2 | 10.3 ± 1.6 |
| β-Proteobacteria (BET42a) | 28.7 ± 3.5 | 66.7 ± 9.3 | 27.0 ± 3.0 | 61.3 ± 4.5 | 26.3 ± 2.5 | 60.4 ± 4.0 | 23.3 ± 2.1 | 64.6 ± 2.5 |
| γ-Proteobacteria (GAM42a) | 5.3 ± 0.6 | 2.1 ± 1.0 | 7.1 ± 0.5 | 2.1 ± 0.8 | 7.3 ± 0.8 | 4.4 ± 0.6 | 6.2 ± 0.8 | 2.2 ± 0.3 |
Percentage of direct total counts by EtBr or SYBER green II staining. The average values and standard deviations from three different determinations are shown.
Time (days) after which sludge under acclimation was sampled.
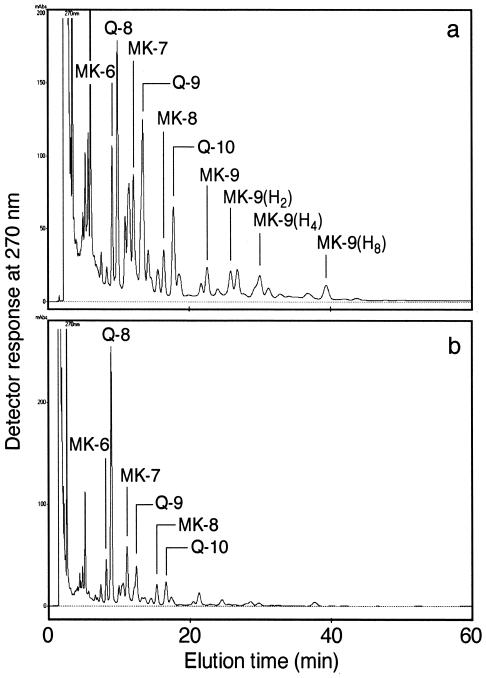
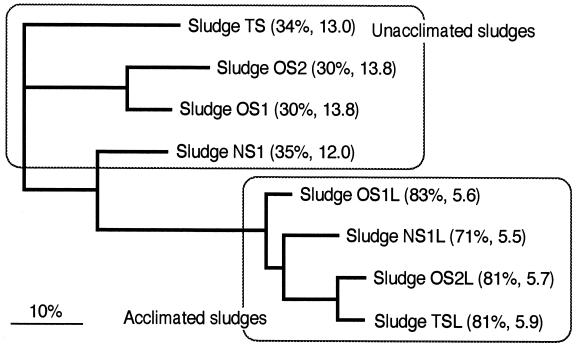
In the quinone analysis, the sludges before acclimation gave a complicated pattern with many quinone components owing to the complexity of the microbial community structure under investigation (Fig. 2a). On the other hand, the acclimated sludges showed a simple quinone profile with Q-8 predominating (Fig. 2b), where Q-8 accounted for 71 to 83% of the total quinone content. A numerical cluster analysis of the sludges based on the quinone profiles showed that the acclimated sludges formed a tight cluster separable from the unacclimated sludges at a D level of more than 40% (Fig. 3). These D values are high enough to justify a statistically significant difference between the acclimated sludges and the unacclimated ones, in light of the numerical standard for D previously reported (27). The MDq values for the sludges before and after acclimation were 12.0 to 13.8 and 5.5 to 5.9, respectively. These decreases in the MDq value shows the simplification of microbial-community structure and the domination of a particular group after acclimation.
FIG. 2.
Examples of high-performance liquid chromatography elution profiles of respiratory quinones from activated sludge before and after acclimation with PHBV under denitrifying conditions. (a) Sludge TSL before acclimation; (b) sludge TSL after 4 weeks of acclimation.
FIG. 3.
Neighbor-joining D matrix dendrogram for clustering of the activated sludges based on quinone profiles. The figures in parentheses following the sludge names indicate the Q-8 contents (mol%) and MDq values. Scale bar = 10% dissimilarity.
16S rDNA clone analysis of acclimated sludges.
16S rDNA clone libraries were constructed from two acclimated sludges, OS1L and TSL, to study whether β-Proteobacteria, especially the Comamonadaceae, actually predominated in these sludges. We obtained and sequenced 25 clones from sludge OS1L and 15 clones from sludge TSL. As a result, two of the OS1L clones and one of the TSL clones were found to be possibly chimerical.
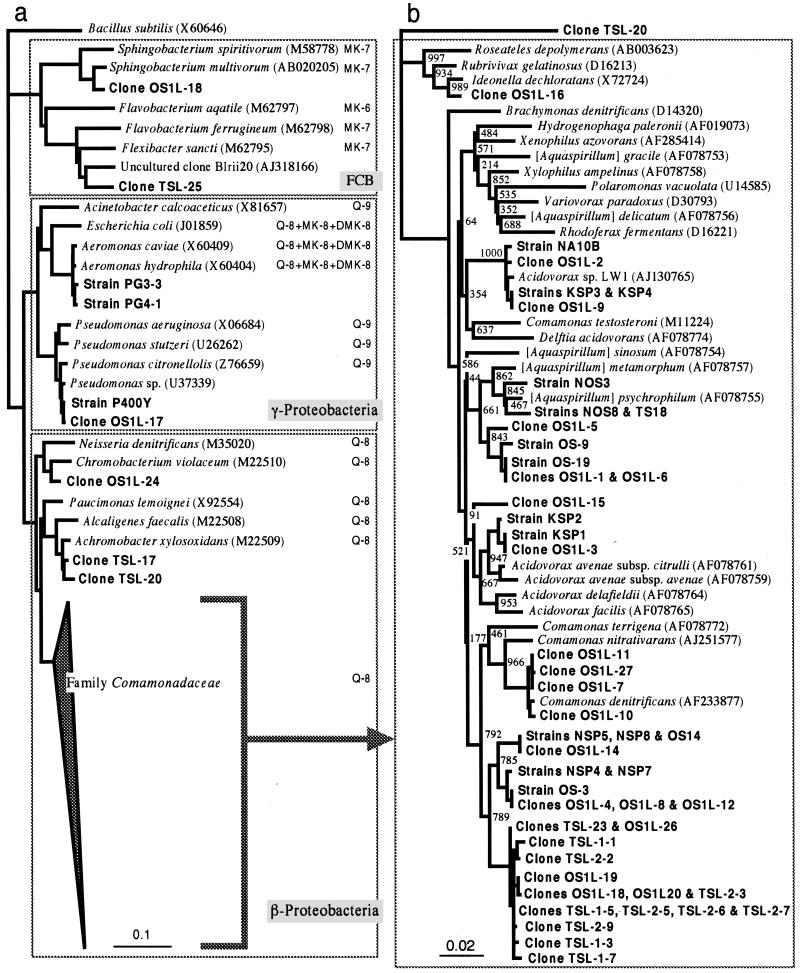
A neighbor-joining phylogenetic tree for a total of 37 uncultured clones, together with the PHBV-degrading denitrifying isolates, is shown in Fig. 4. Most of the clones from the two sludges were positioned within the β-Proteobacteria, especially the family Comamonadaceae, and the remainder belonged to the γ-Proteobacteria and the Bacteroides-Cytophaga-Flexibacter phylum (Fig. 4a). Interestingly, the majority of the clones (8 OS1L clones and 11 TSL clones) and five isolates (NSP4, NSP5, NSP7, NSP8, and OS14) shared a phylogenetic group neighboring with the cluster of the genus Comamonas (Fig. 4b). Some other clones belonging to the Comamonadaceae clustered with Comamonas denitrificans or our isolates (e.g., KSP3 and OS-9) at more than 99% similarity. The majority of our isolates grouped with known species of Comamonadaceae genera, e.g., Acidovarax and [Aquaspirillum], whereas three of the remaining isolates (strains KSP3, KSP4, and NA10B), together with three OS1L clones, represented a distinct line of descent from any previously established genera of this family. This lineage also contained a 1-chloro-4-nitrobenzene-degrading bacterium, strain LW1 (29). One of our isolates, strain NA10B, has been previously reported to represent a distinct lineage within the Comamonadaceae and to exhibit high denitrification activity with PHBV (30).
FIG. 4.
Neighbor-joining distance matrix trees showing phylogenetic positions of uncultured 16S rDNA clones and PHBV-degrading denitrifying isolates from activated sludge. (a) Overall phylogenetic tree with Bacillus subtilis as an outgroup (scale = 0.1 Knuc). The quinone system is shown for each species incorporated and for the family Comamonadaceae. FCB, Flexibacter-Cytophaga-Bacteroides phylum (35). (b) Tree for members of the family Comamonadaceae with clone TSL-20 as an outgroup (scale = 0.02 Knuc).
DISCUSSION
Our culture-dependent experiments for sewage activated sludge indicated that PHBV-degrading denitrifying bacteria occur in relatively small numbers (103 to 105 CFU ml−1) compared to the total bacterial population but are common in sewage treatment plants. The ubiquitous distribution of PHBV-degrading denitrifiers in activated sludge may be expected, in view of the fact that PHBV has been shown to be a readily degradable and usable substrate for microbial communities in the environment, including those in wastewater environments. For example, Briese et al. (11) detected 1.2 × 105 CFU of aerobic PHBV-degrading bacteria ml−1 in sewage sludge. Nishida and Tokiwa (45) reported that PHB-degrading bacteria accounted for 0.2 to 11% of the total colony counts from environmental samples, including sewage sludge. In addition, anaerobic sludge may be a rich source of PHB-degrading bacteria (41).
Most of the PHBV-degrading denitrifiers isolated in this study were identified as members of the family Comamonadaceae, a major group of β-Proteobacteria. Many of these beta-group strains had <98% sequence similarity of 16S rDNA with previously known species of this family, suggesting that they may be taxonomically new. Of special interest is the fact that three of our isolates, strains KSP3, KSP4, and NA10B, together with the 1-chloro-4-nitrobenzene-degrading bacterium LW1 (29), formed a tight cluster distinct from any of the established genera of the family Comamonadaceae. Our phylogenetic data provide circumstantial evidence that this new group of isolates differs taxonomically from the PHBV-degrading denitrifying strains reported so far by other research groups (6, 40, 52)
Culture-independent approaches to the community analysis of activated sludges by FISH and quinone profiling resulted in observations consistent with the culture-dependent data. Namely, FISH analyses with group-specific oligonucleotide probes showed that members of the β-Proteobacteria increased remarkably during acclimation to PHBV under denitrifying conditions in laboratory scale activated-sludge systems, accounting for more than 60% of the total population under fully acclimated conditions. This trend obtained from FISH probing is in good agreement with the results of quinone profiling. The acclimated activated sludges showed a significant increase in Q-8 content. The available information on bacterial quinone systems indicates that Q-8 is the major quinone in all species of β-Proteobacteria and in some members of the γ-Proteobacteria (13, 18, 59). In view of the results of FISH, it is likely that the marked increase in the Q-8 content of the acclimated sludges resulted from domination by members of the β-Proteobacteria. In fact, all of our PHBV-degrading isolates assigned phylogenetically to the β-Proteobacteria contained Q-8 as the major quinone. The numerical cluster analysis of the quinone profiles by using two parameters, D and MDq values, indicate that the acclimation of activated sludge to PHBV under denitrifying conditions brings about a simple community structure with Q-8-containing bacteria, i.e., β-Proteobacteria, predominating, independent of the origin of the sludge.
As the 16S rDNA clone library method is a powerful tool for the characterization of complex bacterial communities without cultivation and isolation, this technique has been used to characterize the whole community structure of activated sludge (8, 9, 54). These previous studies, as well as FISH probing (58), have shown that β-Proteobacteria are the major constituents of the microbial population in sewage activated sludge. In this study, we also applied this technique for the community analysis of the sludges and found that β-Proteobacteria, especially those of the family Comamonadaceae, dominated under PHBV-acclimated denitrifying conditions. The reliability of the clone library method depends upon the efficiency of DNA extraction, PCR biases, and some other factors (56). A previous study showed that 16S rDNA fragments from the Actinobacteria were less amplified by PCR than those from the Proteobacteria in activated sludge (19). However, the results of our clone library approach to the analysis of two different laboratory sludges are not only essentially the same but also very consistent with those of quinone profiling and FISH assays. This demonstrates the reliability of our molecular approach.
A question remains as to whether the uncultured clones obtained from the two acclimated sludges actually represent PHBV-degrading denitrifiers. Although at least the clones showing sequences identical or similar to those of our isolates may be assigned to PHBV-degrading denitrifiers, it is difficult to infer the physiological natures of other clones by using only 16S rDNA sequence information. As the results of MPN experiments suggest that the cometabolism of PHBV-degrading nondenitrifying bacteria and denitrifying bacteria contributes in part to the overall activity of activated sludge, the predominant bacteria in the acclimated sludges may not always be PHBV-degrading denitrifiers. Therefore, further study of other molecular aspects is clearly necessary to characterize the predominant bacteria in this denitrification system. A promising approach is the application of PCR techniques targeting genes involved in PHB degradation (53) and denitrification (10, 15, 51).
In conclusion, the results of this study indicate that members of the β-Proteobacteria, especially those of the family Comamonadaceae, are primary PHBV-degrading denitrifiers in activated sludge or predominate under PHBV-acclimated denitrifying conditions. Such a polyphasic approach as reported in this study is quite useful for describing the community structure of activated sludge. This study provides useful information about microbial ecology for the development of the solid-phase denitrification process with bioplastic as an electron donor and solid matrix.
Acknowledgments
The staffs of the sewage treatment plants from which we were provided with sludge samples are gratefully acknowledged. We also thank Japan Monsanto Co. for generous gifts of PHBV.
REFERENCES
- 1.Abou-Zeid, D., R. Muller, and W. J. Deckwer. 2001. Degradation of natural and synthetic polyesters under anaerobic conditions. J. Biotechnol. 86:113-126. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. L., and K.-H. Schleifer. 1995. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beun, J. J., E. V. Verhoef, M. C. Van Loosdrecht, and J. J. Heijnen. 2000. Stoichiometry and kinetics of poly-β-hydroxybutyrate metabolism under denitrifying conditions in activated sludge cultures. Biotechnol. Bioeng. 68:496-507. [DOI] [PubMed] [Google Scholar]
- 6.Biedermann, J., A. J. Owen, K. T. Schloe, F. Gassner, and R. Süssmuth. 1997. Interaction between poly-3-hydroxybutyrate-co-3-hydroxyvalerate and a denitrifying Pseudomonas strain. Can. J. Microbiol. 43:561-568. [DOI] [PubMed] [Google Scholar]
- 7.Boley, A., W.-R. Müller, and G. Haider. 2000. Biodegradable polymers as solid substrate and biofilm carrier for denitrification in recirculated aquaculture systems. Aquacult. Eng. 22:75-86. [Google Scholar]
- 8.Bond, P. L., R. Erhart, M. Wagner, J. Keller, and L. L. Blackall. 1999. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl. Environ. Microbiol. 65:4077-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bond, P. L., P. Hugenholtz, J. Keller, and L. L. Blackall. 1995. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludge from sequencing batch reactors. Appl. Environ. Microbiol. 61:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briese, B. H., D. Jendrossek, and H. G. Schlegel. 1994. Degradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by aerobic sewage sludge. FEMS Microbiol. Lett. 117:107-111. [DOI] [PubMed] [Google Scholar]
- 12.Brosius, J., J. L. Palmer, J. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S rRNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins, M. D., and D. Jones. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol. Rev. 45:316-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsentein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 15.Hallin, S., and P.-E. Lindgren. 1999. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallin, S., M. Rothman, and M. Pell. 1996. Adaptation of denitrifying bacteria to acetate and methanol in activated sludge. Water Res. 30:1445-1450. [Google Scholar]
- 17.Hankermeyer, C. R., and R. S. Tjeerdema. 1999. Polyhydroxybutyrate: plastic made and degraded by microorganisms. Rev. Environ. Contam. Toxicol. 159:1-24. [DOI] [PubMed] [Google Scholar]
- 18.Hiraishi, A. 1999. Isoprenoid quinones as biomarkers of microbial populations in the environment. J. Biosci. Bioeng. 88:449-460. [DOI] [PubMed] [Google Scholar]
- 19.Hiraishi, A., M. Iwasaki, and H. Shinjo. 2000. Terminal restriction pattern analysis of 16S rRNA genes for the characterization of bacterial communities of activated sludge. J. Biosci. Bioeng. 90:148-156. [DOI] [PubMed] [Google Scholar]
- 20.Hiraishi, A., and H. Kitamura. 1984. Distribution of phototrophic purple nonsulfur bacteria in activated sludge systems and other aquatic environments. Bull. Jpn. Sci. Soc. Fish. 50:1929-1937. [Google Scholar]
- 21.Hiraishi, A., and K. Komagata. 1989. Effects of the growth medium composition on menaquinone homolog formation in Micrococcus luteus. J. Gen. Appl. Microbiol. 35:311-318. [Google Scholar]
- 22.Hiraishi, A., and Y. Morishima. 1990. Capacity for polyphosphate accumulation of predominant bacteria in activated sludge showing enhanced phosphate removal. J. Ferment. Bioeng. 69:368-371. [Google Scholar]
- 23.Hiraishi, A., Y. K. Shin, Y. Ueda, and J. Sugiyama. 1994. Automated sequencing of PCR-amplified 16S rDNA on "hydrolink' gels. J. Microbiol. Methods 145-154.
- 24.Hiraishi, A., Y. Ueda, and J. Ishihara. 1998. Quinone profiling of bacterial communities in natural and synthetic sewage activated sludge for enhanced phosphate removal. Appl. Environ. Microbiol. 64:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiraishi, A., Y. Ueda, J. Ishihara, and T. Mori. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42:457-469. [Google Scholar]
- 26.Hiraishi, A., Y. Yamanaka, and T. Narihiro. 2000. Seasonal microbial community dynamics in a flowerpot-using personal composting system for disposal of household biowaste. J. Gen. Appl. Microbiol. 46:133-146. [DOI] [PubMed] [Google Scholar]
- 27.Hu, H., B. Lim, N. Goto, and K. Fujie. 2001. Analytical precision and repeatability of respiratory quinones for quantitative study of microbial community structure in environmental samples. J. Microbiol. Methods 47:17-24. [DOI] [PubMed] [Google Scholar]
- 28.Iwasaki, M., and A. Hiraishi. 1998. A new approach to numerical analysis of microbial quinone profiles in the environment. Microb. Environ. 13:67-76. [Google Scholar]
- 29.Katsivela, E., V. Wray, D. H. Pieper, and R. M. Wittich. 1999. Initial reactions in the biodegradation of 1-chloro-4-nitrobenzene by a newly isolated bacterium, strain LW1. Appl. Environ. Microbiol. 65:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, S. T., and A. Hiraishi. 2001. Isolation and characterization of a new poly(3-hydroxybutyrate)-degrading, denitrifying bacterium from activated sludge. FEMS Microbiol. Lett. 205:253-257. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques and bacterial systematics. Willey, Chichester, United Kingdom.
- 33.Lemmer, H., D. Roth, and M. Schade. 1994. Population densities and enzymatic activities of heterotrophic bacteria in sewer biofilms and activated sludge. Water Res. 28:1341-1346. [Google Scholar]
- 34.Madison, L., W. Huisman, and W. Gjalt. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Mol. Biol. Rev. 63:21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahne, I., and J. M. Tiedje. 1995. Criteria and methodology for identifying respiratory denitrifiers. Appl. Environ. Microbiol. 61:1110-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manz, W. M., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 38.Mergaert, J., C. Anderson, A. Wouters, and S. Jeans. 1994. Microbial degradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in compost. J. Environ. Polym. Degrad. 2:177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mergaert, J., C. Anderson, A. Wouters, J. Swings, and K. Kersters. 1992. Biodegradation of polyhydroxyalkanoates. FEMS Microbiol. Rev. 9:317-321. [DOI] [PubMed] [Google Scholar]
- 40.Mergaert, J., A. Boley, M. C. Cnockaert, W. R. Müller, and J. Swings. 2001. Identity and potential functions of heterotrophic bacterial isolates from a continuous-upflow fixed-bed reactor for denitrification of drinking water with bacterial polyester as source of carbon and electron donor. Syst. Appl. Microbiol. 24:303-310. [DOI] [PubMed] [Google Scholar]
- 41.Mergaert, J., G. Glorieux, L. Hauben, V. Storms, M. Mau, and S. Jeans. 1996. Biodegradation of poly(3-hydroxyalkanoates) in anaerobic sludge and characterization of a poly(3-hydroxyalkanoates) degrading anaerobic bacterium. Syst. Appl. Microbiol. 19:407-413. [Google Scholar]
- 42.Mergaert, J., A. Webb, C. Anderson, A. Wouters, and S. Jeans. 1993. Microbial degradation of poly(3-hydoxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl. Environ. Microbiol. 59:3233-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mergaert, J., A. Wouters, C. Anderson, and S. Jeans. 1995. In situ biodegradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in natural waters. Can. J. Microbiol. 41:154-159. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen, J. L., and P. H. Nielsen. 2002. Enumeration of acetate-consuming bacteria by microautoradiography under oxygen and nitrate respiring conditions in activated sludge. Water Res. 36:421-428. [DOI] [PubMed] [Google Scholar]
- 45.Nishida, H., and Y. Tokiwa. 1993. Distribution of poly(β-hydroxybutyrate) and poly(ɛ-caprolactone) aerobic degrading microorganisms in different environments. J. Environ. Polym. Degrad. 1:227-233. [Google Scholar]
- 46.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 47.Roser, D. J. 1980. Ethidium bromide: a general purpose fluorescent stain for nucleic acid in bacteria and eucaryotes and its use in microbial ecology studies. Soil Biol. Biochem. 12:329-336. [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Sang, Y. L. 1996. Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49:1-14. [DOI] [PubMed] [Google Scholar]
- 51.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 52.Schloe, K., M. Gillis, B. Hoste, B. Pot, M. Vancanneyt, J. Mergaert, J. Swings, J. Biedermann, and R. Süssmuth. 2000. Polyphasic characterization of poly-3-hydroxybutyrate-co-3-hydroxyvalerate (P(HB-co-HV)) metabolising and denitrifying Acidovorax sp. strains. Syst. Appl. Microbiol. 23:364-372. [DOI] [PubMed] [Google Scholar]
- 53.Sei, K., M. Nakao, K. Mori, M. Ike, T. Kohno, and M. Fujita. 2001. Design of PCR primers and a gene probe for extensive detection of poly(3-hydroxybutyrate) (PHB)-degrading bacteria possessing fibronectin type III linker type-PHB depolymerases. Appl. Microbiol. Biotechnol. 55:801-806. [DOI] [PubMed] [Google Scholar]
- 54.Snaidr, J., R. Amann, L. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suyama, T., Y. Tokiwa, P. Ouichanpagdee, T. Kanagawa, and Y. Kamagata. 1998. Phylogenetic affiliation of soil bacteria that degrade aliphatic polyesters available commercially as biodegradable plastics. Appl. Environ. Microbiol. 64:5008-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer, D. Wedl, and K. H. Schleifer. 1994. Development of an RNA targeted oligonucleotide probe specific for genus Acinetobacter and its application for in-situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokota, A., M. Akagawa-Matsushita, A. Hiraishi, Y. Katayama, T. Urakami, and K. Yamasato. 1992. Distribution of quinone systems in microorganisms: gram-negative eubacteria. Bull. Jpn. Soc. Cult. Collect. 8:136-171. [Google Scholar]