Abstract
We developed a simple and sensitive screening method to investigate the distribution of microbes producing an antimicrobial poly(amino acid), ɛ-poly-l-lysine (ɛ-PL), in microflora. An acidic dye, Poly R-478, incorporated in an agar plate detected ɛ-PL producers by electrostatic interaction with the secreted basic polymers. All ɛ-PL producers, isolated after careful and sufficient screening of soil microflora, belonged exclusively to two groups of bacteria of the family Streptomycetaceae and ergot fungi. They were characterized based on the density and diameter of the concentric zone formed by the secreted polymers. The density depended on each isolate. The increase in the diameter of the concentric zone per unit of time varied among isolates and was negatively correlated with the molecular weight. Although the distribution of ɛ-PL producers was extremely limited, their products were structurally varied. The molecular masses of the secreted polymers among the isolates ranged from 0.8 to 2.0 kDa. There were also isolates producing unknown polymers inconsistent with the correlation or producing a mixture of polymers with original and modified structures. A chemically modified polymer was an ɛ-PL derivative, as determined by mass spectrometry. Since the structural variations had no relation to the phylogenetic position of the isolates, it is possible that enzymes involved in the synthesis diversified after putative horizontal transfers of relevant genes.
ɛ-Poly-l-lysine (ɛ-PL) is a basic homopolymer that consists of 20 to 30 residues of l-lysine with an ɛ amino group-α carboxyl group linkage (10). It was accidentally found as an extracellular material produced by the filamentous bacterium Streptomyces albulus during a screening of Dragendorff's reagent-positive substances (11). This polymer has antimicrobial activity against a wide spectrum of microbes. Moreover, it is harmless to humans and biodegradable; thus, it has been widely used in food manufacturing as a safe preservative. The ɛ-PL-producing strain is resistant to its product, ɛ-PL, but the resistance mechanism is unknown. Recently, ɛ-PL-degrading enzyme has been reported to be a cell membrane-associated protein in the strain (6) as well as in an ɛ-PL-tolerant bacterium, Sphingobacterium (7).
Which microbial species produce ɛ-PL in the natural habitat remains unclear. Aside from S. albulus, a filamentous fungus, Claviceps purpurea, has been described to produce an ɛ-PL-like polymer that is rich in ɛ-amide bonds (12). Although the distribution of ɛ-PL producers beyond the trans-kingdom distance is a subject of interest, there has been no screening method appropriate for an exhaustive exploration. The conventional screening method for detecting ɛ-PL is to assay the extracellular substances secreted into individual culture fluids. However, this is inadequate to examine a large number of microbes. Here, we report a novel, simple, and sensitive screening method that can overcome this problem. The principle of the screening method is to detect the interaction between charged groups of secreted polypeptides and basic or acidic dyes. Because this novel method is applicable to a solid culture medium, it is possible to examine numerous microbes at one time. By using this method, an exhaustive screening was carried out to examine the distribution of ɛ-PL producers.
While the original ɛ-PL-producing strain has been bred to increase its productivity (4), natural microbial resources have been explored to find other strains with more advantageous properties for application: for example, those that produce polymers of lower molecular weight (13). Generally, it is reasonable to seek stronger antimicrobial activity among naturally occurring derivatives. Here, we examined whether this novel screening method can be used to determine strains with potential industrial use or structural derivatives.
MATERIALS AND METHODS
Screening for microbes with ɛ-PL-producing activity.
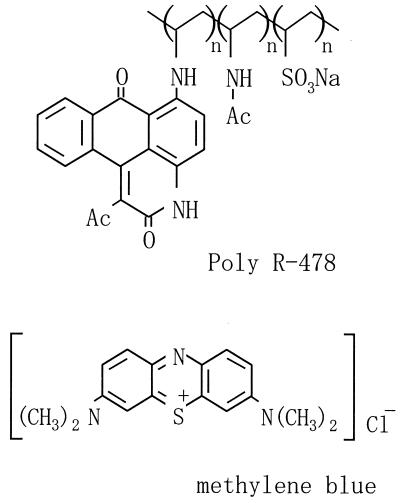
Screenings were carried out by streaking samples on agar plates containing either basic or acidic dyes. Samples were collected mainly from forest soil. The dyes used here, Poly R-478 (Fig. 1) and Remazol Brilliant Blue-R (RBBR), were purchased from Sigma (St. Louis, Mo.). Typically, a synthetic glycerol (SG) medium containing 0.02% Poly R-478 was used for the primary screening. The SG medium was composed of 10 g of glycerol, 0.66 g of ammonium sulfate, 0.68 g of sodium dihydrogen phosphate, 0.25 g of magnesium phosphate heptahydrate, 0.1 g of yeast extract, and 1 ml of Kirk's mineral solution (5) in 1 liter. The pH was adjusted to 7.0, and the medium was solidified by adding 1.5% agar. After incubation at 28°C for about 1 week, microbes forming specific colonies interacting with dyes were picked up and purified after several culture transfers. As a control, an ɛ-PL-nonproducing strain, Streptomyces kifunensis (Kitasatosporia kifunense) IFO15206, which was provided by the Institute for Fermentation, Osaka, Japan, was used.
FIG. 1.
Chemical structures of acidic dye Poly R-478 and basic dye methylene blue.
Production and extraction of ɛ-PL.
Isolates were inoculated on the surface of SG agar plates without dye. To be able to track the diffusion of polymers, isolates were simultaneously cultured on other plates with the same medium, but containing Poly R-478. After 2 weeks of incubation at 28°C, the secreted products were excised from the area surrounding each colony as small agar pieces. To eliminate impurities in the medium ingredients, the agar pieces were immersed in water several times. After freeze-drying, the agar pieces were allowed to swell by immersion in 1 N HCl and heated at 100°C for 1 h to destroy the agar matrix. Alternatively, ɛ-PL was eluted from agar pieces in an electric field in the presence of 0.1% sodium dodecyl sulfate (SDS) buffered with 25 mM Tris HCl (pH 8.0). The eluted polymer was dialyzed against water, and the dialysate was dried in vacuum.
Analytical techniques.
In order to determine the linking pattern of monomers, an extracted polymer was derived by using the 2,4-dinitrophenol (DNP) group and then hydrolyzed completely by heating in 6 N HCl at 100°C for 20 h (10). The reaction products were subjected to thin-layer chromatography (TLC; plate, silica gel 60 [Merck]; developer, n-butanol-acetic acid-pyridine-water at 4:1:1:2; detection, ninhydrin reagent). For the determination of molecular weight, polymers extracted by the moderate acid treatment (1 N HCl, 100°C, 1 h) were further purified with a cation-exchange resin, Amberlite IRC-50 (Organo) (10), and analyzed by polyacrylamide gel electrophoresis (PAGE) in the presence of 0.1% SDS (gel, 16.5% polyacrylamide; stain, CBB-G250). The equipment and all reagents for SDS-PAGE were obtained from Bio-Rad Laboratories. As an authentic standard, commercially supplied ɛ-PL (Wako Pure Chemical Industries, Osaka, Japan) was used. Mass spectrometry (MS) of the polymer was carried out by matrix-assisted laser desorption ionization-time-of-flight MS (MALDI-TOF) MS (model Voyager DE spectrometer; PerSeptive Biosystems). Samples were prepared by mixing electroeluted polymers with α-cyano-4-hydroxycinnamic acid.
Identification and phylogenetic analysis of isolates.
Taxonomic identification of isolates was deputed to the National Collections of Industrial, Food and Marine Bacteria (Shizuoka, Japan). Phylogenetic grouping was carried out by the neighbor-joining method on the basis of 16S and 28S rRNA sequence data for Streptomycetaceae and fungi, respectively.
RESULTS
Screening for ɛ-PL producers on an agar plate containing a charged dye.
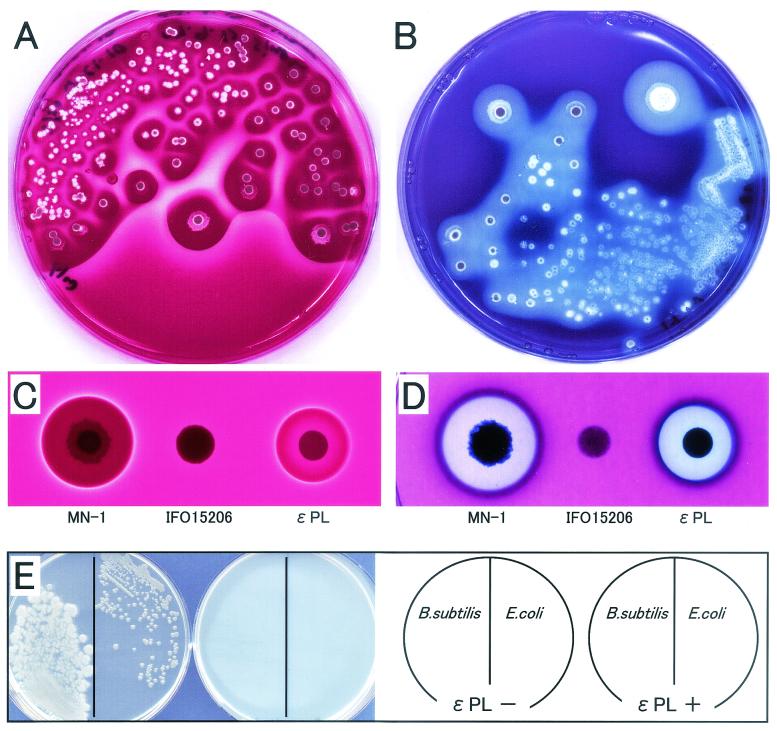
With an acidic polymeric dye, Poly R-478 (0.02%), microbes producing extracellular basic polymers could be visualized as a distinct colony on an agar plate. For example, in the agar plate on which K. kifunense strain MN-1 (Streptomycetaceae), isolated from forest soil, was grown, the acidic dye condensed around the organism's colonies (Fig. 2A). In contrast, a basic dye, methylene blue (0.002%), was excluded from the surrounding zone (Fig. 2B). These results suggest that the substance or substances secreted from MN-1 are basic. Such phenomena were also observed when ɛ-PL, a positively charged substance, was directly applied to the surface of agar plates (Fig. 2C and D). Because K. kifunense strain IFO15206 did not make the characteristic zone around its colonies that MN-1 did, the secretion depended on the strain. These results indicate that charged dyes are very useful for isolation of microbes excreting electrically charged substances. An acidic dye, RBBR (0.02% in the SG agar), completely inhibited the growth of MN-1 originating from a single cell, whereas it did not inhibit the growth originating from a mass of cells. This phenomenon is explainable by assuming that the amount of ɛ-PL secreted from a mass of cells is sufficient to counteract the toxicity of dye. To visualize the interaction between secreted substances and dyes, a zone of at least 5 mm in diameter for each colony was needed. This can permit simultaneous detection of up to 100 to 300 colonies on a single culture dish 9 cm in diameter. Based on this point, this plate-screening method is superior to the conventional method.
FIG. 2.
Detection of secreted basic polymer on agar plate embedded with charged dyes. Areas surrounding colonies of K. kifunense strain MN-1, which extracellularlly produced poly-l-lysine, condensed acidic dye Poly R-478 (A), or excluded basic dye methylene blue (B). For comparison, K. kifunense strain IFO15206, which produces no ɛ-PL, was set as patch inoculation on agar plates containing Poly R-478 (C) or methylene blue (D) with strain MN-1. Authentic ɛ-PL (1 mg, 10 μl) was applied on filter paper disks (φ, 6 mm) set on the surface of the same plates. To test an antimicrobial activity, Bacillus subtilis strain IFO3336 and Escherichia coli strain HB101 were streaked on Luria-Bertani agar plates with and without ɛ-PL produced by strain MN-1, as indicated in panel E. Photographs of the surface of agar plates were taken by front lighting (A, B and E) or by back lighting (C and D).
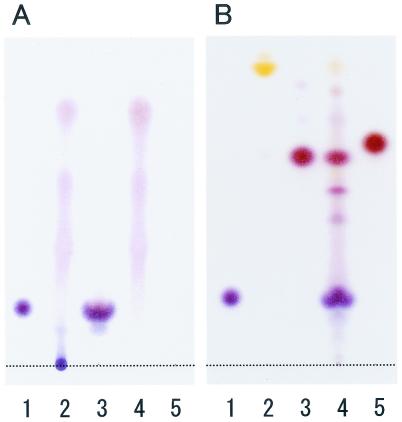
In order to define the structure of the basic substance, the product secreted by MN-1 was analyzed. Hydrolysis showed that the product in the agar matrix was a polymer exclusively composed of lysine (Fig. 3A). The linkage pattern of lysine residues was examined according to the method of Shima and Sakai (10), in which the free amino groups of polylysine were protected by the DNP group and then the obtained polymer was completely hydrolyzed. TLC showed that the main spot corresponded to Nα-DNP-l-lysine, not to Nɛ-DNP-l-lysine (Fig. 3B). Minor spots also appeared because the sample was a mixture of authentic ɛ-PL and its natural derivatives as described below. Thus, the basic substance secreted by MN-1 was identified as polylysine linked by the ɛ-amide bond, namely, ɛ-PL. This polymer had the same antimicrobial activity as that shown in the conventional experiments (Fig. 2E).
FIG. 3.
TLC of ɛ-PL produced by K. kifunense strain MN-1. (A) Lanes: 1, l-lysine (1 μg); 2 and 3, heat-treated polymers in the presence of 1 or 6 N HCl, respectively; 4 and 5, heat-treated blank agar pieces (1 or 6 N HCl). (B) Hydrolysate of DNP-protected polymer. Lanes: 1, l-lysine (1 μg); 2, N,N′-di(2, 4-DNP)-l-lysine (10 μg); 3, Nα-2,4-DNP-l-lysine (10 μg); 4, hydrolysate of DNP-protected polymer produced by MN-1; 5, Nɛ-2,4-DNP-l-lysine (10 μg).
Distribution of ɛ-PL-producing microorganisms.
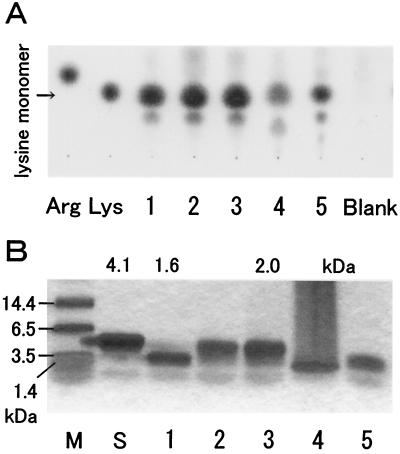
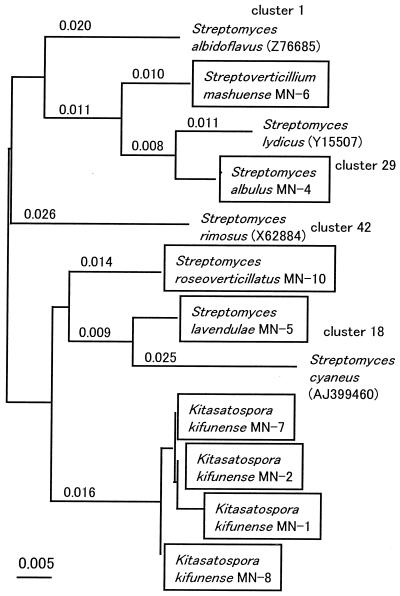
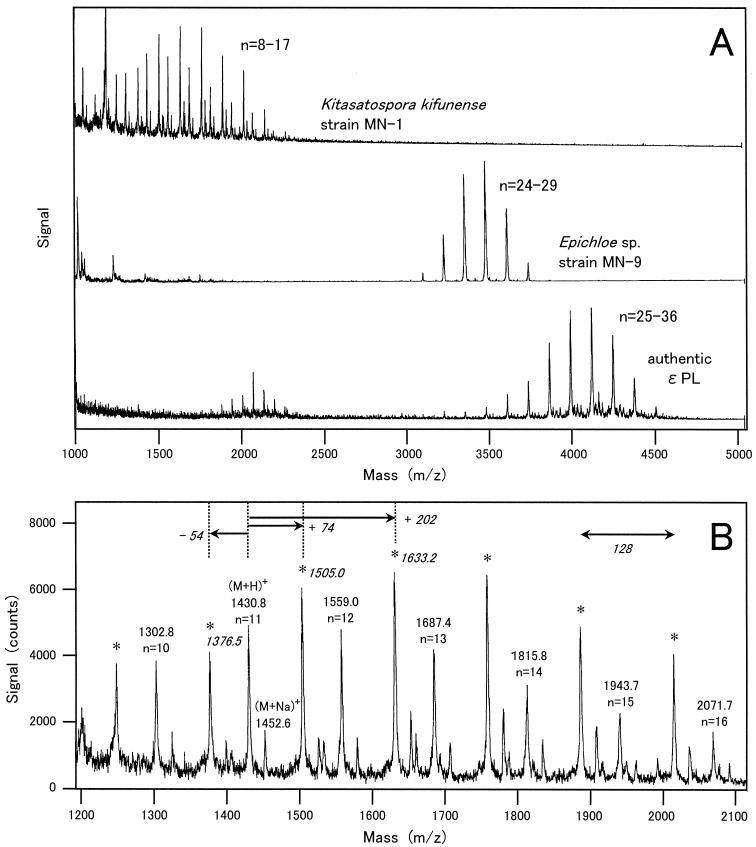
The method presented here enabled screening of a large number of microbes. We carried out careful and extensive screening of soil microflora. After exhaustive screenings of 300 soil samples, more than 10 ɛ-PL producers were isolated. All of the polymers secreted by the representative four isolates had been confirmed to be composed of a lysine monomer (Fig. 4A). Similar data (not shown) were generated for the remaining isolates. The frequent isolation of ɛ-PL producers suggested that production of ɛ-PL is not a unique activity of soil microbes. Based on the 16S ribosomal DNA (rDNA) sequencing data, all bacterial isolates were shown to belong to the genera of Streptomyces and its relative, Kitasatospora (Fig. 5). The distribution of ɛ-PL-producing bacteria was limited within one family, Streptomycetaceae, but they were widely distributed over its genera. By the same method, an ergot fungus producing ɛ-PL, Epichloe sp. strain MN-9, was isolated and found to be evolutionally distant from Streptomycetaceae. MS of the fungal ɛ-PL is shown in Fig. 8A
FIG. 4.
Variation in molecular masses among ɛ-PL-producing isolates. (A) TLC of the complete hydrolysates from ɛ-PL polymers. Arg, l-arginine (1 μg); Lys, l-lysine (1 μg). Lanes: 1 to 5, hydrolysate of agar pieces from strains MN-1, MN-6, MN-4 (from a zone proximal to a colony), MN-4 (from a zone distal to a colony), and MN-10, respectively; Blank, agar pieces from a blank area. (B) SDS-PAGE of uncut ɛ-PL polymers. M, size marker; S, commercially supplied ɛ-PL. Lanes 1 to 5 are the same as in panel A. Molecular masses determined by MALDI-TOF MS are indicated on top. Note that polymers of ɛ-PL do not fit the calibration with standard molecular mass markers.
FIG. 5.
Distribution of the ɛ-PL-producing filamentous bacteria. Strains in boxes are the ɛ-PL producers isolated by the present method. The phylogenetic relationship was drawn in a neighbor-joining tree based on the 16S rRNA sequence. Some representative species in taxonomic clustering (14) are also shown for convenience of explanation.
FIG. 8.
MALDI-TOF MS of ɛ-PL and its derivatives produced by a bacterium, K. kifunense strain MN-1, and an ergot fungus, Epichloe sp. strain MN-9. (A) Comparison of mass spectra among samples of secretions by MN-1 and MN-9 and authentic ɛ-PL. The values of n indicate the numbers of lysine residues. (B) Magnified mass spectrum of MN-1 from the same measurement as in panel A (upper). Peaks representing authentic ɛ-PL polymers are noted by m/z values and the numbers of lysine residues (n). Peaks for derivative polymers are marked by asterisks.
Qualitative variation of ɛ-PL among isolates.
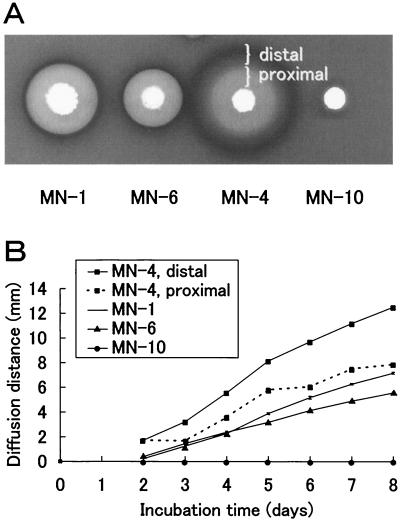
Our method carried out on an agar plate directly indicated both the density and diameter of the concentric zone of secretion. Although the phylogenetic study concluded that all of the isolates were very closely related bacteria, the density and diameter of the concentric zone of secretion depended on the individual isolate. The density reflected the amount of polymer produced, which was determined by the productivity of an isolate (data not shown). On the other hand, the diameter reflected the diffusiveness, which was expected to be partly determined by the molecular mass of the polymer. While each of the ɛ-PL-producing isolates had a particular size in terms of the concentric zone of secretion, S. albulus strain MN-4 produced at least two kinds of secretion zones (Fig. 6A). They were called “distal” for the outside zone and “proximal” for the inside one. K. kifunense strain MN-1 showed similar concentric zones, but they were not as clearly separated as those of MN-4. Streptomyces roseoverticillatus strain MN-10 was unique in that the secretion zone was limited to only around the colony. The time course of the increase in the size was determined by measuring the distance between the margin of diffusion and the margin of a colony (Fig. 6B). The time course was almost linear for the zones of all strains, except for the proximal zone of MN-4. The nonlinearity was likely caused by the proximal zone that spread into the distal zone composed of a rapidly diffusing ɛ-PL polymer. The diffusion velocities of strains MN-4 (distal), MN-1, and MN-6 were about 2.5, 1.3, and 0.9 mm per day, respectively. Secretion by MN-10 was not observed within 8 days (Fig. 6A), but the diffusion velocity was less than 0.1 mm per day after prolonged cultivation.
FIG. 6.
Diffusion of ɛ-PL on an agar plate. (A) Diffusion of secreted PL polymers into agar matrix. Seed cultures were inoculated by patch setting on the SG agar medium containing methylene blue. (B) The diffusion distance is the distance between the margin of the concentric zone of secretion and the margin of a colony. The diffusion distance was measured when ɛ-PL producers grew on GS medium containing Poly R-478. Values are means ± standard deviations (n = 3).
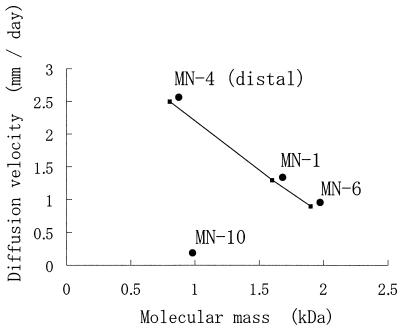
In order to examine whether the diffusion velocity was correlated with the molecular weight distribution, ɛ-PL polymers were recovered from the concentric zones of secretion, and their molecular masses were measured by SDS-PAGE (Fig. 4B). From strain MN-4, in particular, two kinds of secretion products were separately recovered from the proximal and distal zones. The molecular weight distribution varied among isolates. Based on the calibration generated from data obtained by MALDI-TOF MS, the molecular masses of ɛ-PL produced by MN-1, MN-6, MN-4 (distal), MN-4 (proximal), and MN-10 were estimated to be about 1.6, 1.9, 0.8, 2.0, and 0.9 kDa, respectively. As shown in Fig. 7, there was a negative correlation (r2 > 0.99940) between the diffusion velocity and the molecular weight. However, for strain MN-10, such a correlation was not observed. Therefore, this correlation was applicable to the ɛ-PL polymer with a nonderivatized structure in order to estimate the molecular mass from the diffusion velocity.
FIG. 7.
Correlation between the molecular mass of polymer and the diffusion velocity.
Discovering novel ɛ-PL derivatives.
Among our isolates, there were a few particular strains, such as MN-10, which departed from the correlation, and MN-1, which produced a mixture of unknown polymers. Their products were shown to be essentially composed of a lysine monomer by TLC analysis of the hydrolysate (Fig. 4A). However, a question arose as to whether the native structure of the polymers remained after moderate acid hydrolysis (1 N HCl, 100°C, 1 h), because such a treatment might destroy modifications such as acyl substitutions on lysine residues. To answer this question, unknown polymers were extracted by electroelution and then subjected to MALDI-TOF MS. Unfortunately, we could not obtain the MS data for strain MN-10, and only those for strain MN-1 were obtained (Fig. 8). Since ɛ-PL is a chain of lysine residues (molecular weight, 146.19) linked by dehydration of water (molecular weight, 18.02), its molecular weight should be calculated based on the formula 146.19 × n − 18.02 × (n − 1), where n is the number of lysine residues. The polymers produced by MN-1 had a series of (M+H)+ values that fit the formula. For example, the signal at 1430.8 was nearly consistent with the value obtained by substituting 11 for n in the formula. It was a signal from an ɛ-PL undecamer. The molecular mass distribution ranged from 1.2 to 2.1 kDa. In addition, MN-1 also had a series of (M+H)+ values that increased by about 74 or other constants from the original values. These additional peaks were not detected in both MS on authentic ɛ-PL and secretion by a fungus strain, MN-9 (Fig. 8A). Although the actual increment was not determined only from the mass spectra, it was noteworthy that an interval of about 128 was conserved among a series of signals. This means that only one residue, probably either an N- or C-terminal lysine residue, was chemically modified. Thus, MN-1 produces a derivative of ɛ-PL in addition to the original structure.
DISCUSSION
Our improved method has the following three advantages. (i) It can sensitively find ɛ-PL producers. It is based on the strong electrostatic interaction between ɛ-PL and charged dyes. Generally, dyes with an anthraquinone structure are toxic to microbial cells. The acidic dye, Poly R-478, exhibits relatively low toxicity, because the anthrapyridone chromophore is linked to a poly(vinylamine) backbone that is too large to enter the cells (molecular weight, >40,000). Consequently, Poly R-478 is effective in detecting an ɛ-PL producer, even if it secretes only a small amount of polymer. The method is also effective in selecting ɛ-PL producers with high productivity of a charged polymer. A smaller acidic dye, RBBR (molecular weight, 626.6), inhibited cell growth, but its toxicity was counteracted by the secretion of a certain amount of ɛ-PL. Therefore, controlling the concentration of toxic dyes might enable discrimination of ɛ-PL producers with high productivity from other microbes. In principle, the present method is also applicable to the screening of oppositely charged substances, such as acidic polypeptides. (ii) It is applicable to solid culture media in which a large number of microbes can be screened. This simple and sensitive method enabled us to exhaustively screen the soil microflora. (iii) It can provide information about the amount and molecular weight of the secreted ɛ-PL. The molecular weight of a polymer can be estimated from its correlation with the diffusion velocity of the polymer. The correlation covers a molecular mass range of about 0.5 to 2.5 kDa. For polymers longer than 2.5 kDa, it is necessary to change the solidifying agent to allow their diffusion.
By using the newly developed method, we carried out an exhaustive screening of soil microflora. Surprisingly, the distribution of ɛ-PL producers was quite limited among bacteria in the family Streptomycetaceae and ergot fungi, among which the relevant genes might transfer horizontally. It cannot be completely denied that the culture conditions are suited for preferentially isolating members of Streptomycetaceae and ergot fungi. However, the screening method should be applicable to almost all of the neutrophilic and mesophilic microbes, because the culture conditions are generally applicable to the screening of most general microbes. It is very interesting to note that the evolutionally distant groups bacteria and fungi commonly produce ɛ-PL. To prove the possibility of the transkingdom genetic transfer, further investigations are required.
In spite of the limited distribution, there are structural differences, particularly in terms of molecular weight. It is possible that the genes relevant to the control of molecular weight were modified after their putative gene transfer, because the differences are not closely related to the phylogenetic position of the producers. In analogy to the synthesis of poly(glutamic acid), which is coded only by three genes (1, 8), it is possible that ɛ-PL synthesis is controlled by one or a few genes. Our results suggest that a relatively minor genetic alteration occurred in ɛ-PL producers, causing the variation in molecular weight of the polymers.
This is the first report on the chemically modified ɛ-PL derivatives. Very little is known about the mechanism underlying the antimicrobial activity of ɛ-PL. It seems plausible that the positively charged portions initially bind to negatively charged components of the cell membrane, such as phosphatidylglycerol and cardiolipin. Such a model explaining the initial stage of the antimicrobial activity was proposed for natural polycationic antibiotic peptides, such as nisin, magainin, and defensin (2, 3, 9). The increase in hydrophobic interaction besides the electrostatic one may stabilize the binding between the membrane and ɛ-PL, which is expected to intensify the antimicrobial activity. Because hydrophobicity may influence the diffusiveness, the improved method described here should contribute toward finding ɛ-PL derivatives which have hydrophobicity different from that of the original ɛ-PL. The screening, which focused on filamentous bacteria and fungi, will be essential for finding microbes suitable for industrial applications, in view of the limited distribution of ɛ-PL-producing microbes.
Acknowledgments
We are grateful to M. Iwabuchi (RIBS) for helpful discussions.
REFERENCES
- 1.Ashiuchi, M., K. Soda, and H. Misono. 1999. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO 3336: gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 263:6-12. [DOI] [PubMed] [Google Scholar]
- 2.Breukink, E., C. van Kraaij, R. A. Demel, R. J. Siezen, O. P. Kuipers, and B. de Kruijff. 1997. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 36:6968-6976. [DOI] [PubMed] [Google Scholar]
- 3.Fujii, G., M. E. Selsted, and D. Eisenberg. 1993. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. 2:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiraki, J., M. Hatakeyama, H. Morita, and Y. Izumi. 1998. Improved ɛ-poly-l-lysine production of an S-(2-aminoethyl)-l-cysteine resistant mutant of Streptomyces albulus. Seibutsu-Kogaku 76:487-493. [Google Scholar]
- 5.Kirk, T. K., E. Schultz, W. J. Connors, L. F. Lorenz, and J. G. Zeikus. 1978. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch. Microbiol. 117:277-285. [Google Scholar]
- 6.Kito, M., R. Takimoto, T. Yoshida, and T. Nagasawa. 2001. ɛ-Poly-l-lysine-degrading enzyme in Streptomyces albulus, p. 142. In Proceedings of the Annual Meeting of the Society for Bioscience and Bioengineering, Osaka, Japan.
- 7.Kito, M., Y. Onji, T. Yoshida, and T. Nagasawa. 2002. Occurrence of ɛ-poly-l-lysine-degrading enzyme in ɛ-poly-l-lysine-tolerant Sphingobacterium multivorum OJ10: purification and characterization. FEMS Microbiol. Lett. 207:147-151. [DOI] [PubMed] [Google Scholar]
- 8.Makino, S., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzaki, K., M. Harada, S. Funakoshi, N. Fujii, and K. Miyajima. 1991. Physicochemical determinants for the interactions of magainins 1 and 2 with acidic lipid bilayers. Biochim. Biophys. Acta 1063:162-170. [DOI] [PubMed] [Google Scholar]
- 10.Shima, S., and H. Sakai. 1981. Poly-l-lysine produced by Streptomyces. III. Chemical studies. Agric. Biol. Chem. 45:2503-2508. [Google Scholar]
- 11.Shima, S., and H. Sakai. 1981. Poly-l-lysine produced by Streptomyces. II. Taxonomy and fermentation studies. Agric. Biol. Chem. 45:2497-2502. [Google Scholar]
- 12.Szokan, G., M. Almas, K. Krizsan, A. R. Khlafulla, E. Tyihak, and B. Szende. 1997. Structure determination and synthesis of lysine isopeptides influencing on cell proliferation. Biopolymers 42:305-318. [DOI] [PubMed] [Google Scholar]
- 13.Takehara, M., Y. Aihara, S. Kawai, H. Yoshikabu, Y. Inoue, and H. Hirohara. July 1999. Strain producing low molecular weight ɛ-poly-l-lysine and production of low molecular weight ɛ-poly-l-lysine using the strain. Japan patent 2001-017159.
- 14.Williams, S. T., M. Goodfellow, G. Alderson, E. M. Wellington, P. H. Sneath, and M. J. Sackin. 1983. Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 129:1743-1813. [DOI] [PubMed] [Google Scholar]