Abstract
Recently, the feasibility of using Escherichia coli for the heterologous biosynthesis of complex polyketides has been demonstrated. In this report, the development of a robust high-cell-density fed-batch procedure for the efficient production of complex polyketides is described. The effects of various physiological conditions on the productivity and titers of 6-deoxyerythronolide B (6dEB; the macrocyclic core of the antibiotic erythromycin) in recombinant cultures of E. coli were studied in shake flask cultures. The resulting data were used as a foundation to develop a high-cell-density fermentation procedure by building upon procedures reported earlier for recombinant protein production in E. coli. The fermentation strategy employed consistently produced ∼100 mg of 6dEB per liter, whereas shake flask conditions generated between 1 and 10 mg per liter. The utility of an accessory thioesterase (TEII from Saccharopolyspora erythraea) for enhancing the productivity of 6dEB in E. coli was also demonstrated (increasing the final titer of 6dEB to 180 mg per liter). In addition to reinforcing the potential for using E. coli as a heterologous host for wild-type- and engineered-polyketide biosynthesis, the procedures described in this study may be useful for the production of secondary metabolites that are difficult to access by other routes.
Complex polyketide natural products include many medicinally important compounds, such as the antibiotic erythromycin, the cholesterol-lowering agent lovastatin, and the recently discovered anticancer agent epothilone. These therapeutic agents are typically synthesized by bacteria or fungi through the action of multienzyme catalysts known as polyketide synthases (PKSs) (10). Notwithstanding major advances in synthetic chemistry, the structural complexity of pharmacologically important polyketides precludes the development of practical synthetic processes for their manufacture, leaving microbial fermentation as the only economical route to large-scale production. However, the production of complex polyketides via fermentation processes typically suffers from two major problems: the nonideal growth characteristics of the producing microorganisms (typically Streptomyces spp. that possess especially long doubling times and mycelial morphologies) and the need to develop a scalable process for each newly discovered polyketide-producing organism. Over the past decade, methods for identifying polyketide biosynthetic genes have become well-established, and it has become possible to transfer the genetic code for polyketide biosynthesis from a poorly characterized microorganism into a genetically, physiologically, and metabolically well-characterized heterologous host. In addition to providing a facile route for the scale-up of such a process, the use of a heterologous host also provides an effective strategy for the directed manipulation of the PKS to produce “unnatural” natural products (7).
Recently, we have demonstrated the feasibility of using Escherichia coli for the heterologous biosynthesis of complex polyketides (9). Specifically, the large PKS genes responsible for the biosynthesis of 6-deoxyerythronolide B (6dEB), the macrocyclic core of the antibiotic erythromycin, were functionally expressed in E. coli. In parallel, the host was engineered to efficiently convert exogenous propionate into propionyl-coenzyme A (CoA) and (2S)-methylmalonyl-CoA, the building blocks for 6dEB biosynthesis. Together, these features yielded a cellular catalyst capable of producing 6dEB at concentrations of ∼1 to ∼10 mg per liter. For comparison, both the wild-type strain of Saccharopolyspora erythraea and engineered Streptomyces coelicolor hosts produce 50 to 200 mg of 6dEB per liter (6), whereas industrial strains of Saccharopolyspora erythraea, evolved over decades, have the ability to produce 2.5 to 5 g of 6dEB per liter (8). Since future fundamental and applied studies of polyketide biosynthesis in E. coli will depend substantially on the productivity of this heterologous host, we embarked upon a concerted effort, using genetic, physiological, and fermentation strategies, to improve the titers of 6dEB in E. coli. Here we report the results of our efforts that have led to an ∼100-fold enhancement of the 6dEB titer. In the process, we have also established that a recently identified mechanism for enhancing polyketide biosynthesis in natural-polyketide-producing hosts is also effective in a heterologous host such as E. coli (10, 12).
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
All experiments used E. coli BAP1 as the host cell (9) [F−ompT hsdSB(rB− mB−) gal dcm (DE3) ΔprpRBCD (sfp)]. 6dEB was produced by BAP1/pBP130(Carr)/pBP144(Kanr) (9). Briefly, pBP130 contains the genes for DEBS2 and DEBS3 under coordinate control of the T7 promoter; pBP144 contains DEBS1 (under its own T7 promoter) and the individual subunits for a propionyl-CoA carboxylase (PCC) under coordinate control of a second copy of the T7 promoter. DEBS1, -2, and -3 are responsible for 6dEB biosynthesis, whereas the PCC complex acts to convert propionyl-CoA (supplied by exogenously fed propionate) to (2S)-methylmalonyl-CoA; propionyl- and methylmalonyl-CoA are the substrates used by DEBS1, -2, and -3 to produce 6dEB. The components of fermentation media used in these experiments (expressed in quantities per liter) were as follows: for F1 flask studies, 3 g of KH2PO4, 6.62 g of K2HPO4, 4 g of (NH4)2SO4, 150.5 mg of MgSO4, 5 g of glucose, 1.25 ml of trace metal solution, and 1.25 ml of vitamin solution; for F1 fed-batch fermentation, 1.5 g of KH2PO4, 4.34 g of K2HPO4, 0.4 g of (NH4)2SO4, 150.5 mg of MgSO4, 5 g of glucose, 1.25 ml of trace metal solution, and 1.25 ml of vitamin solution; for feed medium, 110 g of (NH4)2SO4, 3.9 g of MgSO4, 430 g of glucose, 10 ml of trace metal solution, and 10 ml of vitamin solution; for trace metals solution, 27 g of FeCl3 · 6H2O, 2 g of ZnCl2 · 4H2O, 2 g of CaCl2 · 6H2O, 2 g of Na2MoO4 · 2H2O, 1.9 g of CuSO4 · 5H2O, 0.5 g of H3BO3, and 100 ml of concentrated HCl; and for vitamin solution, 0.42 g of riboflavin, 5.4 g of pantothenic acid, 6 g of niacin, 1.4 g of pyridoxine, 0.06 g of biotin, and 0.04 g of folic acid.
Cell stocks were prepared by growing a culture of BAP1/pBP130/pBP144 in Luria-Bertani (LB) medium supplemented with carbenicillin (100 mg per liter) and kanamycin (50 mg per liter) at 37°C and shaking it at 250 rpm (Lab-line model 3527 shaker). After the culture reached an optical density at 600 nm (OD600) between 0.5 and 1, the cells were centrifuged and resuspended in half of the initial culture volume in F1 medium with 8% glycerol. The resuspended cells were aliquoted and frozen at −80°C.
For small-scale experiments, 25 ml of F1 medium in 250-ml flasks was inoculated with 0.5 ml of a prefrozen glycerol cell stock, grown at 37°C, and shaken at 250 rpm (with carbenicillin and kanamycin concentrations at 100 and 50 mg per liter, respectively) to a suitable OD600 (between 0.2 and 1, as specified below). At this point the culture was cooled to 22°C and supplemented with sodium propionate (Sigma-Aldrich. St. Louis, Mo.) to a final concentration between 10 mg and 10g per liter, as specified below. Gene expression was then induced with IPTG (isopropyl-β-d-thiogalactopyranoside; GibcoBRL, Grand Island, N.Y.) between 10 μM and 10 mM, as specified below. The cultures were then incubated at 22 or 25°C and shaken at 200 rpm until no further increase in production was observed.
Fed-batch aerated fermentations were conducted with a 3-liter Biobundle system (Applikon Inc., Foster City, Calif.). A starter culture was grown in 1.5 ml of LB medium (with 100 mg of carbenicillin per liter and 50 mg of kanamycin per liter). After reaching late exponential phase at 37°C and being shaken at 250 rpm, the culture was centrifuged and resuspended in 50 ml of LB medium (with 100 mg of carbenicillin per liter and 50 mg of kanamycin per liter). The culture was grown overnight at 30°C and shaken at 200 rpm to stationary phase, centrifuged again, and resuspended in 20 ml of phosphate-buffered saline for inoculation into the 3-liter Biobundle vessel containing 2 liters of F1 medium. Growth was conducted at 37°C with pH maintained at 7.1 throughout the experiment with 1 M H2SO4 and concentrated NH4OH. Aeration was maintained at 2.8 liters per min and agitation was controlled at 600 to 900 rpm to maintain the level of dissolved oxygen above 50% air saturation. The salt solution [KH2PO4, K2HPO4, and (NH4)2SO4] in the fermentation apparatus was autoclaved, whereas the additional feed components (MgSO4, glucose, trace metals, and vitamins) were filter sterilized and added aseptically prior to inoculation along with carbenicillin at 150 mg per liter and kanamycin at 75 mg per liter. The feed was also filter sterilized. Once the glucose was exhausted from the starting medium (as indicated by a sudden decrease in the oxygen requirement of the culture), the temperature was reduced to 22°C and IPTG (100 μM) and sodium propionate (2 g per liter) were added. At that point a peristaltic pump started to deliver 0.1 ml of the feed medium per min, and samples were typically taken twice daily thereafter. Additional sodium propionate was added approximately every 48 h (to 2 g per liter, assuming no propionate remained) to avoid the depletion of this precursor for polyketide biosynthesis.
Plasmid construction.
The ery-ORF5 gene from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea encodes a homolog of thioesterase (also designated thioesterase II, or TEII) (14) and has been shown to enhance polyketide biosynthesis in actinomyces hosts by an as-yet-unknown mechanism (Z. Hu, unpublished result). To test the effect of TEII coexpression on polyketide biosynthesis in E. coli, we constructed an expression plasmid in which the ery-ORF5 gene was cloned under the control of an IPTG-inducible T7 promoter on a chloramphenicol-resistant plasmid, pGZ119EH (5), that is compatible with plasmids pBP130 and pBP144. The resulting plasmid, pBP190, was cotransformed into BAP1 along with pBP130 and pBP144 and studied by using the fermentation protocol outlined above (including chloramphenicol at 34 mg per liter in the starter cultures and 20 mg per liter in the fermenter).
Analytical methods.
OD600 measurements were made with a Beckman DU650 spectrophotometer, with the necessary dilutions being made with phosphate-buffered saline. Glucose concentrations were measured with an enzymatic hexokinase detection kit (Sigma-Aldrich). Acetate and propionate concentrations were analyzed by using an isocratic high-pressure liquid chromatography (HPLC) method with 5 mM H2SO4 (Agilent 1100 HPLC series). The column used was a Bio-Rad HPLC organic acid analysis column (Aminex HPX-87H) maintained at 55°C with the refractive index used to detect acetate and propionate from clarified fermentation broth samples (20-μl injections). A separate HPLC assay was used to detect 6dEB as described previously (6). Protein levels were monitored after cell disruption (sonication of 2-ml samples), clarification, and detection via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and SimplyBlue SafeStain (Invitrogen, Carlsbad, Calif.) staining.
Theoretical conversion of propionate to 6dEB.
The theoretical conversion of propionate to 6dEB was calculated assuming that all of the propionate added to the experimental cultures was converted to propionyl-CoA and subsequently to 6dEB {1 mol of 6dEB = 7 mol of propionyl-CoA [1 mol of proionyl-CoA and 6 mol of (2S)-methylmalonyl-CoA, which derives from propionyl-CoA in this system]}. The percent conversion was then calculated based on the actual amount of 6dEB produced.
RESULTS
Development of a fed-batch fermentation procedure for 6dEB biosynthesis in E. coli.
The effects of various growth and process parameters on the production and titer of 6dEB in cultures of E. coli BAP1/pBP130/pBP144 were studied in shake flask cultures. Initially, three media that are compatible with fed-batch cultivation of E. coli were tested (data not shown). Of these, fermentation medium F1 gave the best yields (∼1 mg per liter over 19 h). The effects of IPTG and propionate concentrations, postinduction temperature, and time of induction were investigated by using this medium. Here, the OD600 values tested at the time of induction ranged from 0.25 to 4.5, with the optimal level between 0.25 and 1.0. IPTG values between 10 μM and 10 mM were tested, and 100 μM was determined to be the optimal concentration. The propionate levels tested were between 10 mg and 10 g per liter, with the optimal level being between 50 mg and 2 g per liter. Finally, 22 and 25°C were tested as postinduction temperatures, with no difference observed between the two temperatures. For the best conditions, 6dEB titers were consistently found to be between 1 and 3 mg per liter over 19 h.
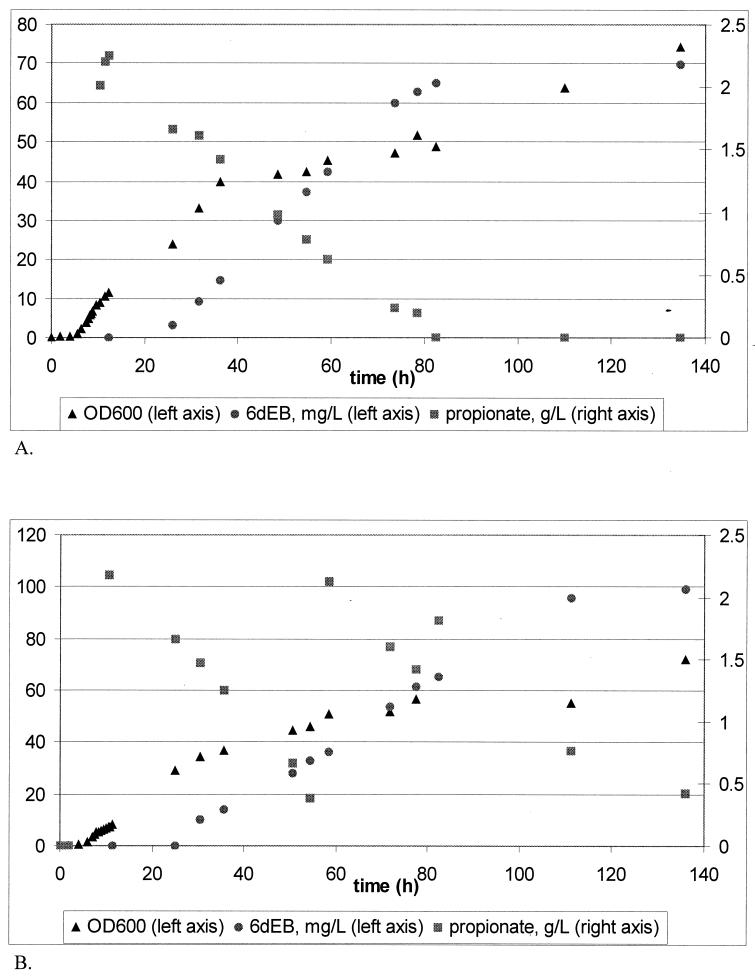
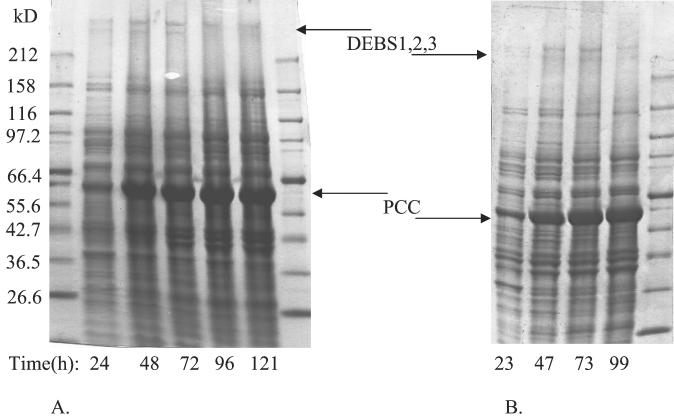
To increase the volumetric productivity and titer, the above-described conditions were used as a foundation to develop a high-cell-density fermentation scheme analogous to earlier procedures developed for recombinant polypeptide production in E. coli (13). As shown in Fig. 1A, cell growth was extended to a final OD600 between 50 and 70 (24 and 33 g [dry weight] of cells per liter) by the controlled addition of a nutrient feed stream that maintained glucose and acetate concentrations below 1 g per liter throughout the course of the experiment. Polyketide biosynthesis was induced at an OD600 between 5 and 10 by the addition of 100μM IPTG and 2 g of propionate per liter at 22°C. Intracellular protein was analyzed periodically thereafter via SDS-PAGE, and the expressed levels of soluble PKS proteins were found to remain relatively constant throughout the length of the experiment (beginning approximately 4 h after induction) (Fig. 2). This fermentation procedure yielded a final 6dEB titer of 70 mg per liter over a 110-h period. Precursor and product analysis showed that the titer reached a plateau after all the exogenous propionate had been consumed. As shown in Fig. 1B, propionate supplementation in the fermentation medium reproducibly resulted in a titer increase to ∼100 mg of 6dEB per liter. The maximum conversion of propionate into 6dEB was 6%. Fermentor- and shake flask-specific productivities were similar; however, the fermentor volumetric productivity was 17-fold higher than the shake flask value.
FIG. 1.
E. coli fed-batch fermentation experiments with no additional propionate added over time (A) and with additional propionate feeding as specified in Materials and Methods (B). The experiment whose results are shown in panel B was repeated two times with an average final 6dEB titer of 95.2 mg per liter, with a standard deviation of 7.7 mg per liter; the average volumetric productivity during the linear production phase was 1.1 mg per liter per h, with a standard deviation of 0.007 mg per liter per h.
FIG. 2.
Protein expression profiles for E. coli fed-batch fermentation experiments without TEII coexpression (A) and with TEII coexpression (B).
Metabolic engineering of E. coli for enhanced 6dEB biosynthesis.
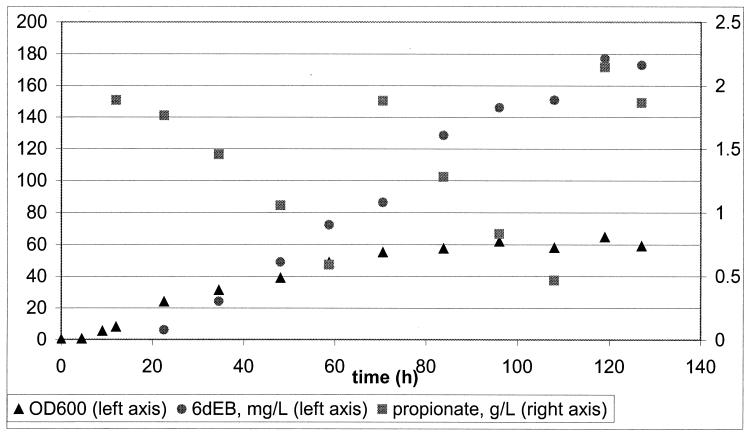
Recent studies of several Streptomyces spp. have demonstrated the beneficial impact of a thioesterase-like enzyme on polyketide productivity (1, 12). Homologs of these enzymes (termed TEII) are encoded within the biosynthetic gene clusters of many macrolides. The mechanism of Saccharopolyspora erythraea TEII is unclear, and it has been proposed that these enzymes play an editing role in polyketide biosynthesis (1, 4). To test the effect of the Saccharopolyspora erythraea TEII, encoded by ery-ORF5, on 6dEB biosynthesis in E. coli, the recombinant strain BAP1/pBP130/pBP144/pBP190 was subjected to a high-cell-density fermentation using the procedure described above. The growth characteristics of this recombinant strain mirrored those of previous experiments, but the titer of 6dEB doubled to ∼180 mg per liter (Fig. 3). These experiments were reproducible with 6dEB levels between 180 and 200 mg per liter and suggest that TEII coexpression enhances the polyketide productivity in E. coli in a manner analogous to its effect inStreptomyces spp. (where 6dEB production also doubled compared to levels in cultures without TEII [Z. Hu, unpublished result]).
FIG. 3.
E. coli 6dEB fed-batch fermentation, including TEII gene expression. For three separate experiments, the average final 6dEB level was 184.6 mg per liter, with a standard deviation of 9.96 mg per liter. The volumetric productivity average was 2.05 mg per liter per h, with a standard deviation of 0.40 mg per liter per h.
In the context of fermentation experiments, TEII coexpression did not significantly improve the previously noted plasmid stability properties of BAP1 (9), making it unlikely that the TEII expression vector beneficially altered cellular plasmid retention. In addition, the fermentation gene expression profile showed no qualitative difference compared to profiles from fermentations without TEII expression (Fig. 2). In this case, unlike the deoxyerythronolide-B synthase and PCC proteins, TEII was not readily visible via SDS-PAGE analysis. However, when expressed in isolation under shake flask conditions meant to mirror fermentor conditions, TEII (∼30 kDa) could be readily purified with an N-terminal six-histidine tag (3). This suggests that, once expressed, TEII is present within the cellular background of E. coli. Small-scale in vivo radioactive experiments (9) helped to support the notion that the titer increase was due to TEII coexpression. Here, a strain containing pBP130, pBP144, and pBP190 was compared to BAP1/pBP130/pBP144/pGZ119EH. Additionally, these two strains were compared to BAP1/pBP130/pBP144. Each comparison showed an approximately twofold increase in 6dEB titer with TEII coexpression.
DISCUSSION
In this communication we have described the development of a robust high-cell-density fed-batch protocol for the efficient production of complex polyketides in the well-studied laboratory host E. coli. To accomplish this, we have built upon established fermentation protocols, typically used for protein production, towards the goal of polyketide production. Our initial results for the production of 6dEB in E. coli are encouraging and suggest that further gains could be made in improving the biosynthetic potential of this heterologous system by the installation of more sophisticated metabolic control strategies. For example, in the existing system, biosynthesis of 6dEB requires the expression of seven heterologous genes, two in the chromosome and five borne by two plasmids. It is likely that at least some of these genes are overexpressed relative to the metabolic demand. By using alternative promoters, regulatory elements, and plasmids (or perhaps even by altogether eliminating the plasmids), it may be possible to minimize the metabolic burden and, in the process, to increase the polyketide productivity of recombinant E. coli.
Another limitation of the existing system is the relatively low yield of 6dEB from exogenous propionate. In earlier work we have confirmed that E. coli BAP1 is unable to utilize propionate as a sole carbon source to support growth, as the only known pathway for propionate catabolism has been deleted from this recombinant strain (9). However, since <10% of exogenous propionate is converted into 6dEB, it appears that propionate is catabolized by E. coli BAP1 into unknown and potentially undesirable side products. Since the ability of E. coli to activate propionate as propionyl-CoA has been retained (and intentionally amplified) in BAP1, it is possible that biosynthesis of odd-chain fatty acid synthesis provides one route for the utilization of exogenous propionate; indeed, when radiolabeled propionate is fed to E. coli BAP1/pBP130/pBP144, radiolabeled products which are even more lipophilic than 6dEB (as judged by radiolabeling-thin-layer chromatography TL analysis; data not shown) are observed. Another possibility is the potential utilization of propionate via nonspecific transacylation mechanisms that can utilize propionyl-CoA in lieu of acetyl-CoA (the most common acyl donor in vivo). The identification of such “dead-end” products may lead to the genetic or metabolic attenuation or elimination of such nonproductive pathways from the E. coli host, thereby further enhancing its capacity for polyketide biosynthesis.
Finally, our results have demonstrated the utility of an accessory thioesterase present in Saccharopolyspora erythraea, TEII, for enhancing the productivity of 6dEB in E. coli. The exact mechanism of TEII is presently not clear, and it is believed that this enzyme plays an editing role by hydrolyzing incorrectly processed intermediates off the multifunctional PKS (1, 4). Alternatively, TEII may increase the intracellular activity of PKS enzymes by purging acyl carrier protein (ACP) domains that have been posttranslationally modified with an inappropriate phosphopantetheine donor. Ordinarily, ACP domains on PKSs are posttranslationally modified by the attachment of the phosphopantetheine arm at an active-site serine (2). This reaction is catalyzed by a phosphopantetheinyl transferase. In BAP1, a heterologous enzyme, Sfp, from Bacillus subtilis catalyzes this reaction. The primary advantage of using Sfp is that it possesses a broad substrate specificity for virtually any ACP domain (11); however, since Sfp can utilize acyl-CoA donors with a specificity comparable to that of CoA-SH, the possibility exists for it to erroneously utilize substrates such as acetyl-CoA or propionyl-CoA. If so, the acylated phophopantetheine would effectively block polyketide intermediates from being processed past this misprimed ACP domain until the thioester bond is hydrolyzed. To the extent that the TEII domain can selectively hydrolyze acetyl-ACP or propionyl-ACP domains in preference to the corresponding CoA thioesters, it may act as an enhancer of newly synthesized PKS activity in vivo. Further biochemical analysis should address this mechanism.
In summary, this study illustrates the potential for complex natural-product biosynthesis in E. coli. Through genetic and physiological manipulations, rapid and dramatic improvements were made to the ability of this heterologous host to produce polyketides. We believe that these studies are only the beginning for future improvements, given some of the remaining limitations of the system that could be readily overcome. Efficient polyketide biosynthesis in E. coli could make this bacterium the host of choice for future efforts aimed at the protein engineering of PKS pathways.
Acknowledgments
This work was supported in part by a grant from the National Science Foundation (BES 9806774) to C.K. B. Pfeifer is a recipient of a predoctoral fellowship from the Stanford-NIH training program in biotechnology.
The assistance of Kara Calhoun during HPLC analysis and Jessica Wuu during fermentation analysis is also gratefully acknowledged.
REFERENCES
- 1.Butler, A. R., N. Bate, and E. Cundliffe. 1999. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem. Biol. 6:287-292. [DOI] [PubMed] [Google Scholar]
- 2.Cane, D. E., C. T. Walsh, and C. Khosla. 1998. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282:63-68. [DOI] [PubMed] [Google Scholar]
- 3.Gokhale, R. S., D. Hunziker, D. E. Cane, and C. Khosla. 1999. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem. Biol. 6:117-125. [DOI] [PubMed] [Google Scholar]
- 4.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 5.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombo, F., B. Pfeifer, T. Leaf, S. Ou, Y. S. Kim, D. Cane, P. Licari, and C. Khosla. 2001. Enhancing the atom economy of polyketide biosynthetic processes through metabolic engineering. Biotechnol. Prog. 17:612-617. [DOI] [PubMed] [Google Scholar]
- 7.McDaniel, R., A. Thamchaipenet, C. Gustafsson, H. Fu, M. Betlach, and G. Ashley. 1999. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc. Natl. Acad. Sci. USA 96:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minas, W., P. Brunker, P. T. Kallio, and J. E. Bailey. 1998. Improved erythromycin production in a genetically engineered industrial strain of Saccharopolyspora erythraea. Biotechnol. Prog. 14:561-566. [DOI] [PubMed] [Google Scholar]
- 9.Pfeifer, B. A., S. Admiraal, H. Gramajo, D. Cane, and C. Khosla. 2001. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790-1792. [DOI] [PubMed] [Google Scholar]
- 10.Pfeifer, B. A., and C. Khosla. 2001. Biosynthesis of polyketides in heterologous hosts. Microbiol. Mol. Biol. Rev. 65:106-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quadri, L. E., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585-1595. [DOI] [PubMed] [Google Scholar]
- 12.Tang, L., H. Fu, M. C. Betlach, and R. McDaniel. 1999. Elucidating the mechanism of chain termination switching in the picromycin/methymycin polyketide synthase. Chem. Biol. 6:553-558. [DOI] [PubMed] [Google Scholar]
- 13.Tsai, L. B., M. Mann, F. Morris, C. Rotgers, and D. Fenton. 1987. The effect of organic nitrogen and glucose on the production of recombinant human insulin-like growth factor in high cell density Escherichia coli fermentations. J. Ind. Microbiol. 2:181-187. [Google Scholar]
- 14.Weber, J. M., J. O. Leung, G. T. Maine, R. H. Potenz, T. J. Paulus, and J. P. DeWitt. 1990. Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J. Bacteriol. 172:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]