Abstract
Rhodococcus sp. strain DK17 was isolated from soil and analyzed for the ability to grow on o-xylene as the sole carbon and energy source. Although DK17 cannot grow on m- and p-xylene, it is capable of growth on benzene, phenol, toluene, ethylbenzene, isopropylbenzene, and other alkylbenzene isomers. One UV-generated mutant strain, DK176, simultaneously lost the ability to grow on o-xylene, ethylbenzene, isopropylbenzene, toluene, and benzene, although it could still grow on phenol. The mutant strain was also unable to oxidize indole to indigo following growth in the presence of o-xylene. This observation suggests the loss of an oxygenase that is involved in the initial oxidation of the (alkyl)benzenes tested. Another mutant strain, DK180, isolated for the inability to grow on o-xylene, retained the ability to grow on benzene but was unable to grow on alkylbenzenes due to loss of a meta-cleavage dioxygenase needed for metabolism of methyl-substituted catechols. Further experiments showed that DK180 as well as the wild-type strain DK17 have an ortho-cleavage pathway which is specifically induced by benzene but not by o-xylene. These results indicate that DK17 possesses two different ring-cleavage pathways for the degradation of aromatic compounds, although the initial oxidation reactions may be catalyzed by a common oxygenase. Gas chromatography-mass spectrometry and 300-MHz proton nuclear magnetic resonance spectrometry clearly show that DK180 accumulates 3,4-dimethylcatechol from o-xylene and both 3- and 4-methylcatechol from toluene. This means that there are two initial routes of oxidation of toluene by the strain. Pulsed-field gel electrophoresis analysis demonstrated the presence of two large megaplasmids in the wild-type strain DK17, one of which (pDK2) was lost in the mutant strain DK176. Since several other independently derived mutant strains unable to grow on alkylbenzenes are also missing pDK2, the genes encoding the initial steps in alkylbenzene metabolism (but not phenol metabolism) appear to be present on this approximately 330-kb plasmid.
Methylbenzenes are used in various industrial processes, including the production of chemicals, drugs, paints, and enamels (19, 28), and they are thus widespread environmental contaminants (5). Several bacterial strains are known that have the ability to grow on different methylbenzenes, including the three xylene isomers, as the sole source of carbon and energy. Bacteria that degrade xylenes commonly fall into two classes: those that can degrade both m- and p-xylene, and those that can degrade o-xylene only. Very rarely are the two abilities found together in the same organism. The positions of the methyl groups on the aromatic ring thus play a major role in the selection of bacteria able to grow on the xylene isomers (5, 14).
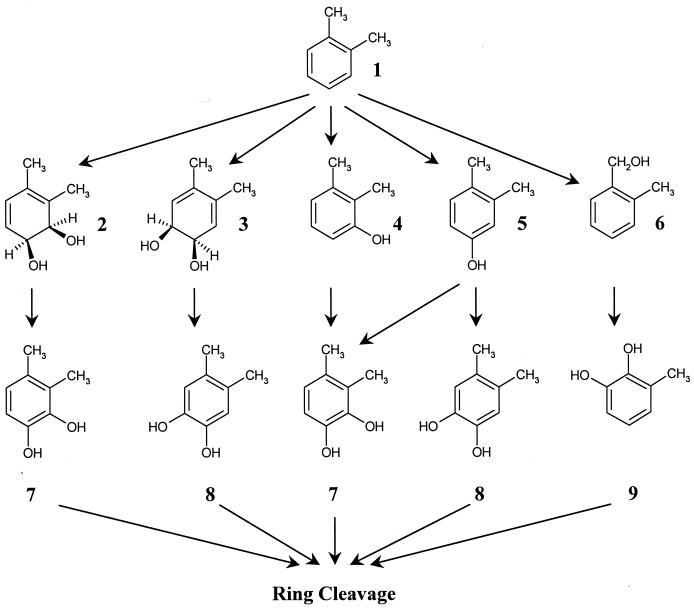
During the past three decades much research has centered on elucidating the metabolism of m- and p-xylene at the biochemical and molecular level, and the details of these metabolic pathways have been well documented (2, 10, 13, 20, 24, 37, 38). In contrast, little in-depth work has been reported for the degradation of o-xylene. Several different pathways have been proposed in a variety of different microorganisms (Fig. 1). It has been proposed that a Nocardia sp. strain (21) and Rhodococcus sp. strain C125 (originally Corynebacterium sp. strain C125, renamed by van der Meer et al. [34]) (30) metabolize o-xylene through an initial aromatic dioxygenase to form a cis-dihydrodiol, but there is no direct evidence for the presence of a dioxygenase in either strain.
FIG. 1.
Catabolic pathways for the aerobic degradation of o-xylene. For simplicity, only selected metabolites are shown. The pathway intermediates are as follows: compound 1, o-xylene; compound 2, 1,2-dihydroxy-3,4-dimethylcyclohexa-3,5-diene; compound 3, 1,2-dihydroxy-4,5-dimethylcyclohexa-3,5-diene; compound 4, 2,3-dimethylphenol; compound 5, 3,4-dimethylphenol; compound 6, 2-methylbenzylalcohol; compound 7, 3,4-dimethylcatechol; compound 8, 4,5-dimethylcatechol; compound 9, 3-methylcatechol.
In Rhodococcus sp. strain B3 two pathways, both initiated by monooxygenases, are found to operate simultaneously. One pathway involves the oxidation of a methyl group to form 2-methylbenzylalcohol, which is oxidized via 2-methylbenzoic acid to 3-methylcatechol. The second catabolic pathway is thought to proceed through oxidation of the aromatic ring to form 2,3-dimethylphenol, but the subsequent oxidation product, dimethylcatechol, has not yet been identified (8). In the well-studied Pseudomonas stutzeri strain OX1, o-xylene is degraded initially through two successive monooxidations of the aromatic ring to form 2,3-dimethylphenol and 3,4-dimethylphenol simultaneously, which are converted into 3,4-dimethylcatechol and 4,5-dimethylcatechol, respectively (4, 7). In this work, we isolated a new o-xylene-degrading Rhodococcus strain and characterized its ability to degrade o-xylene and related compounds.
MATERIALS AND METHODS
Isolation of bacteria and growth conditions.
A standard enrichment method was used to isolate o-xylene degraders from a crude oil-contaminated plant site in Yeochon, Korea. A few grams of soil was added to a 250-ml flask containing 50 ml of mineral salts basal (MSB) medium (32). o-Xylene was provided as sole carbon and energy source in the vapor phase in a glass bulb. The flask was incubated for 48 h at 30°C with shaking (200 rpm). Subsequently, 5 ml of the culture was transferred to 50 ml of fresh MSB medium for another subsequent 2-day incubation. Serial dilutions of the second culture were plated out on MSB agar plates and incubated at 30°C in the presence of o-xylene provided in the vapor phase in a cotton-stoppered glass vial. The fastest-growing colony was selected for further characterization. Volatile compounds used for growth substrate analyses (o-, m-, and p-xylene isomers, ethylbenzene, isopropylbenzene, toluene, benzene, and phenol) were all provided in the vapor phase. Stock solutions (0.5 M) of 2,3- and 3,4-dimethylphenol were prepared in N,N-dimethylformamide and when needed as substrate were added to MSB medium at concentrations of 0.01 to 0.1%.
Determination and analysis of 16S ribosomal DNA.
Total genomic DNA was isolated from Rhodococcus sp. strain DK17 following standard procedures (3). The 16S rRNA gene was PCR amplified using the 27F and 1522R primers (23) with Taq DNA polymerase (Sigma, St. Louis, Mo.) as described previously by others (15). The PCR product was purified using a spin column (Qiagen, Valencia, Calif.) and used directly as the template for sequencing. Automated DNA sequencing reactions were performed and resolved on an ABI 377 automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.). The forward primers 27F, 357F, 704F, 926F, and 1242F and the reverse primers 321R, 685R, 907R, 1220R, and 1522R (23) were utilized in the DNA sequencing reactions to ensure full overlapping coverage in both directions. The resulting DNA sequences were aligned with the DNAStar Lasergene program (DNAStar, Madison, Wis.) and analyzed using the GenBank database (1) and the Ribosomal Database Project (25).
Generation of mutants.
UV mutagenesis was performed according to the method of Carlton and Brown (11) with slight modification. Overnight-grown cells were harvested, washed twice with 0.1 M MgSO4 by centrifugation, and resuspended in the same solution. After serial dilutions with 0.1 M MgSO4, each cell suspension was transferred to sterile glass petri plates and exposed to 254-nm UV light positioned 22 cm above the open petri plates for 90 s (cell survival rate of 0.1 to 1.0%). The treated cell suspensions were plated out on MSB agar plates containing 20 mM glucose (master plates) and incubated at 30°C in the dark. Toothpicks were used to transfer colonies from the master plates onto MSB plates, which were subsequently incubated under a saturated atmosphere of o-xylene and monitored for the loss of ability to grow on o-xylene as sole carbon and energy source.
Ring-cleavage dioxygenase assays.
Bacterial cells reaching the exponential phase on 20 mM glucose were harvested and resuspended in 200 ml of fresh MSB medium. In order to induce the ring-cleavage pathway, o-xylene, toluene, or benzene was directly added to the suspension at a final concentration of 0.1% and further incubated at 30°C for 12 h. The induced cells were harvested; washed in a half volume of 1× PBS (phosphate-buffered saline; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]); suspended in 5 ml of 50 mM 3-morpholinopropanesulfonic acid buffer (pH 7.8) containing 1 mM ascorbic acid, 10% acetone, 10% glycerol, and 100 μM FeSO4; and disrupted by sonication. Unbroken cells and cell debris were removed by centrifugation at 10,000 × g for 30 min. The resulting supernatant was used as the enzyme solution. Catechol 2,3-dioxygenase activity was assayed spectrophotometrically by measuring the increase in the absorbance at the corresponding wavelength of each meta-cleavage product formed from the following substrates: catechol, λmax = 375 nm and ɛ = 33,400 cm−1 M−1 (6); 3-methylcatechol, λmax = 388 nm and ɛ = 13,800 cm−1 M−1 (6); 4-methylcatechol, λmax = 382 nm and ɛ = 28,100 cm−1 M−1 (6). The reaction mixture contained 100 mM phosphate buffer (pH 7.4) and an appropriate substrate at a final concentration of 0.4 mM. The activity of catechol 1,2-dioxygenase was assayed spectrophotometrically by monitoring the increase in the absorbance at 260 nm for cis,cis-muconate (ɛ = 16,800 cm−1 M−1 [16]), 260 nm for 2-methyl-cis,cis-muconate (ɛ = 18,000 cm−1 M−1 [16]), and 255 nm for 3-methyl-cis,cis-muconate (ɛ = 14,300 cm−1 M−1 [16]). The reaction mixture contained 50 mM Tris-HCl (pH 8.0), 1.3 mM EDTA, and an appropriate substrate at a final concentration of 0.4 mM. Protein content was determined by the method of Bradford (9) with bovine serum albumin as the standard. One unit of enzyme activity was defined as the formation of 1 μmol of product per min.
Analysis and identification of o-xylene and toluene metabolites.
The UV-generated mutant strain DK180 was inoculated in MSB containing 20 mM glucose and incubated overnight at 30°C with shaking. Ten milliliters of the overnight culture was transferred to 400 ml of fresh medium containing 5 mM glucose and incubated for 24 h under the same conditions, in the presence of o-xylene or toluene provided in the vapor phase. Cells were removed by centrifugation at 10,000 × g for 30 min, and the supernatant was extracted twice with an equal volume of ethyl acetate. After concentration by a rotary evaporator, the residue was dissolved in a small volume of methanol and analyzed by high-pressure liquid chromatography (HPLC). HPLC analysis was performed with a Hewlett-Packard model 1100 HPLC apparatus equipped with a 5-μm ZORBAX column (4.6 by 250 mm). The mobile phase was a 45-min linear gradient of methanol-water (from 5:95 to 95:5) at a flow rate of 1.0 ml/min. Gas chromatography-mass spectrometry (GC-MS) analysis of metabolites was carried out with a Hewlett-Packard 5973 mass spectrometer (electron impact ionization, 70 eV) connected to a 6890 gas chromatogram fitted with a fused silica capillary column (HP-5; 0.25 by 30 m; film thickness, 0.25 μm). The following conditions were used for the GC: 1 ml of He/min, on-column injection mode; oven temperature, 60°C for 2 min; thermal gradient, 5°C/min to 220°C, and then held at 220°C. Prior to injection, the sample was treated with N-methyl-N-trimethylsilyltrifluoroacetamide, methaneboronic acid, or 9-phenanthreneboronic acid in pyridine at 80°C for 30 min to produce the trimethylsilyl ether (TMSi), methaneboronate, or phenanthreneboronate derivatives, respectively. o-Xylene metabolites were acetylated for stabilization prior to nuclear magnetic resonance (NMR) analysis. The dried residue was dissolved in acetic anhydride (1 ml) and pyridine (2 ml) and incubated at room temperature overnight. Subsequently, the product was adjusted to pH 7.0 with an HCl solution and extracted with 30 ml of n-hexane. After removing n-hexane, the acetate of the metabolite was collected and subjected to a normal-phase HPLC (6 by 150 mm; Senshu Pak Aquasil SS-532N) at a flow rate of 1 ml/min with a mixture of n-hexane-isopropanol as an elution solvent (5 min with 100% n-hexane followed by a 20-min gradient to 5% isopropanol in n-hexane). The collected sample was dissolved in CdCl3 and analyzed by 300-MHz 1H-NMR (Varian Gemini 2000) using tetramethylsilane as an internal standard.
PFGE.
Agarose plugs containing genomic DNA were prepared as described by Saeki et al. (29), with slight modification. Bacterial cultures were harvested and resuspended in EET [0.1 M EDTA, 10 mM ethylene glycol-bis(β-aminoethyl ether)-N, N, N′,N′-tetraacetic acid, 10 mM Tris-HCl, pH 8.0], and mixed with 2.0% (wt/vol) low-melting-point agarose in distilled water. The plugs were incubated in TE buffer (50 mM Tris, 20 mM EDTA; pH 8.0) containing lysozyme (2 mg/ml), proteinase K (2 mg/ml), and sodium N-lauroylsarcosine (0.5 mg/ml) for 48 h at 30°C, and then incubated in EET buffer containing 1% (wt/vol) sodium dodecyl sulfate for 12 h at 55°C. The plugs were washed three times in TE buffer and stored at 4°C in the same buffer. Digestion of the genomic DNA with S1 nuclease was performed essentially as described by Barton et al. (B. Barton, G. Harding, and A. Zuccarelli, Abstr. 94th Gen. Meet. Am. Soc. Microbiol., abstr. H-249, p. 244, 1994). Three 4.5- by 4.5-mm pieces of agarose containing intact genomic DNA were incubated in 1 ml of S1 nuclease buffer (50 mM NaCl, 5 mM ZnCl2, 30 mM sodium acetate; pH 4.5) for 0.5 h. The plugs were then incubated in 0.2 ml of the same buffer containing 1 U of S1 nuclease for 45 min at 37°C. The reaction was stopped by placing the tubes in ice and adding 20 μl of 0.5 M EDTA (pH 8.0) and 20 μl of 1 M Tris-HCl (pH 8.0). Pulsed-field gel electrophoresis (PFGE) was performed using a Bio-Rad Laboratories CHEF DRIII system. Gels (1.0% agarose in 0.5× TBE buffer [1× TBE is 89 mM Tris-borate, 2.5 mM EDTA; pH 8.0]) were run at 6 V/cm at 14°C. The pulse duration increased from 15 to 60 s during a 16-h run.
Chemicals.
Aromatic compounds used in this study were obtained from Sigma-Aldrich Korea (Seoul, Korea). All solvents were purchased from Mallinckrodt Baker, Inc. (Phillipsburg, N.J.). All chemicals were analytical-grade purity or above.
Nucleotide sequence accession number.
The Rhodococcus sp. strain DK17 16S rRNA sequence was deposited into GenBank with the accession number AF468521.
RESULTS
Isolation, identification, and characterization of strain DK17.
A new bacterial strain was isolated from a crude oil-contaminated site in Yeochon, Korea, for the ability to grow on o-xylene as the sole source of carbon and energy by enrichment culture, as described in Materials and Methods. The strain, designated DK17, is a gram-positive, oxidase-negative, catalase-positive bacterium with a growth temperature optimum of 30°C. DK17 shows a pleomorphic cell morphology depending on the growth substrate. For example, DK17 is rod-shaped when growing on glucose, while it exhibits branching but not mycelial morphology during growth on o-xylene. In order to taxonomically identify strain DK17, its 16S rRNA gene was amplified and the PCR product was directly sequenced with internal primers. Analysis of the nucleotide sequence using either the GenBank (1) or Ribosomal Database Project (25) databases revealed that the DK17 16S rRNA sequence has high levels of identity (ca. 97%) with those from several different Rhodococcus species. DK17 was thus positively identified as a Rhodococcus sp.
The ability of Rhodococcus sp. strain DK17 to grow on different monocyclic aromatic compounds was determined by the rate of colony formation on MSB plates in the presence of each substrate as the sole carbon source (Table 1). DK17 grew well on o-xylene, excreting a yellow compound into the medium around the colonies and forming 1-mm colonies in less than 3 days. The doubling time of DK17 in MSB liquid medium with o-xylene provided as vapor phase was 2.4 h. The other xylene isomers (m- and p-xylene) were not used as growth substrates by DK17. However, DK17 was able to grow on ethylbenzene, isopropylbenzene, toluene, benzene, and phenol. DK17 grows significantly faster on o-xylene, ethylbenzene, and isopropylbenzene than on benzene and toluene.
TABLE 1.
Growth characteristics of Rhodococcus sp. strains DK17, DK176, and DK180 on various monocyclic aromatic hydrocarbons
| Compound | DK17 | DK176 | DK180 |
|---|---|---|---|
| o-Xylene | ++++a | − | − |
| m-Xylene | −b | − | − |
| p-Xylene | − | − | − |
| Ethylbenzene | ++++ | − | − |
| Isopropylbenzene | ++++ | − | − |
| Toluene | ++ | − | − |
| Benzene | + | − | + |
| Phenol | +++ | +++ | +++ |
Four, three, two, and one plus signs indicate the formation of a 1.0-mm-diameter colony within 3, 5, 7, and 9 days, respectively.
A minus sign denotes the compound could not serve as sole carbon and energy source.
In order to begin characterization of the catabolic pathway for o-xylene degradation, the ability of DK17 to grow on potential metabolic intermediates of o-xylene was analyzed. DK17 is unable to grow on the hydroxylated derivatives of o-xylene, 2,3-dimethylphenol, and 3,4-dimethylphenol, as well as the methyl group oxidized derivatives 2-methylbenzylalcohol, 2-methylbenzylaldehyde, and 2-methylbenzoic acid. Bickerdike and coworkers (8) previously reported that Rhodococcus sp. strain B3 failed to grow on 2-methylbenzylalcohol, 2-methylbenzylaldehyde, and 3,4-dimethylphenol mainly due to toxicity to cells, because the strain's growth on glucose was completely inhibited by these compounds. A similar inhibitory effect was observed when the growth of DK17 on glucose was examined in the presence of 2-methylbenzylaldehyde, 2,3-dimethylphenol, or 3,4-dimethylphenol above concentrations of 0.1%, while its growth was not affected at all by 2-methylbenzylalcohol or 2-methylbenzoic acid.
Mutant construction and characterization.
In order to analyze the DK17 catabolic pathway for o-xylene in more depth, UV mutagenesis was performed. After screening approximately 5,500 colonies, a total of 54 mutant strains were selected for the inability to grow on o-xylene, and 49 lost the ability to convert indole to indigo when grown on glucose in the presence of o-xylene vapors, which is indicative of the loss of an initial oxygenase (17, 18). One mutant strain in this class, designated DK176, was selected for further characterization. DK176 is unable to grow on ethylbenzene, isopropylbenzene, toluene, or benzene, although it is still able to grow on phenol (Table 1). These observations suggest that the same oxygenase is involved in the initial steps in the metabolism of ethylbenzene, isopropylbenzene, benzene, toluene, and o-xylene by DK17. One mutant strain in the second class of mutants, DK180, was also chosen for further analysis. This strain is unable to grow on all of the alkylbenzenes tested but retains the ability to grow on benzene and phenol (Table 1). Aerobic degradation of benzene is processed via catechol, while methylbenzenes are converted to methylcatechols. Accordingly, the inability of DK180 to grow on o-xylene and toluene raises a possibility that the enzymatic steps handling methylcatechols are disrupted in this strain.
Induction of meta- and ortho-cleavage dioxygenases in DK17 and DK180.
The above results indicate that DK17 possesses at least two different pathways for the degradation of (methyl)catechols, although the initial oxidation reactions may be catalyzed by a common oxygenase. It is generally accepted that methylcatechols are metabolized more efficiently by the meta-cleavage pathway than by the ortho-cleavage pathway, due to the alkyl substituent (22). Thus, the wild-type DK17 and the mutant strain DK180 were analyzed for the type of ring-cleavage dioxygenase physiologically induced during growth in the presence of o-xylene, toluene, or benzene. As summarized in Table 2, a large amount of meta-cleavage dioxygenase (catechol 2,3-dioxygenase) activity was detected in cells of the wild-type DK17 grown in the presence of o-xylene, toluene, or benzene, while no meta-cleavage dioxygenase activity was detected under similar conditions in the mutant strain DK180. The detected meta-cleavage dioxygenase had maximal activity against 3-methylcatechol and significant activity against 4-methylcatechol (30.5% of that for 3-methylcatechol). However, the enzyme showed very low activity against catechol (1.9% of that detected for 3-methylcatechol). It is apparent that the same meta-cleavage enzyme was induced in the presence of benzene, toluene, and o-xylene, since the ratios of activities on the three substrates were the same under the three conditions. The ortho-cleavage dioxygenase (catechol 1,2-dioxygenase), however, was induced only when DK17 was grown in the presence of benzene (Table 3). The same level of induction is seen in both the wild-type and the meta-cleavage dioxygenase mutant strain DK180. This result, coupled with the low level of activity of the meta-cleavage dioxygenase against catechol and the decreased level of induction of the meta-cleavage dioxygenase in the presence of benzene, suggests that the same amount of metabolic flux occurs through catechol in both DK17 and DK180 when grown on benzene. It is not simply a rerouting of metabolism through a different downstream pathway in the mutant strain.
TABLE 2.
Comparison of meta-cleavage dioxygenase activities in cell extracts of wild-type strain DK17 and mutant strain DK180
| Strain | Inducer | Enzyme activity (U/mg of protein)a against: |
||
|---|---|---|---|---|
| Catechol | 3-Methylcatechol | 4-Methylcatechol | ||
| DK17 | Glucose | NDb | ND | ND |
| o-Xylene | 32.0 ± 1.8 | 1,666.3 ± 71.1 | 507.9 ± 11.1 | |
| Toluene | 15.8 ± 2.3 | 818.6 ± 78.9 | 253.9 ± 29.9 | |
| Benzene | 6.8 ± 2.3 | 226.3 ± 27.3 | 60.1 ± 10.7 | |
| DK180 | o-Xylene | ND | ND | ND |
| Toluene | ND | ND | ND | |
| Benzene | ND | ND | ND | |
Enzyme activities are the averages from at least three independent experiments.
ND, not detected.
TABLE 3.
Comparison of ortho-cleavage dioxygenase activities in cell extracts of wild-type strain DK17 and mutant strain DK180
| Strain | Inducer | Enzyme activity (U/mg of protein)a against: |
||
|---|---|---|---|---|
| Catechol | 3-Methylcatechol | 4-Methylcatechol | ||
| DK17 | Glucose | NDb | ND | ND |
| o-Xylene | ND | ND | ND | |
| Toluene | ND | ND | ND | |
| Benzene | 577.0 ± 5.6 | 55.1 ± 7.2 | 156.7 ± 13.5 | |
| DK180 | Benzene | 480.3 ± 18.9 | 63.6 ± 14.7 | 185.5 ± 24.8 |
Enzyme activities are the averages from at least three independent experiments.
ND, not detected.
Identification of metabolites.
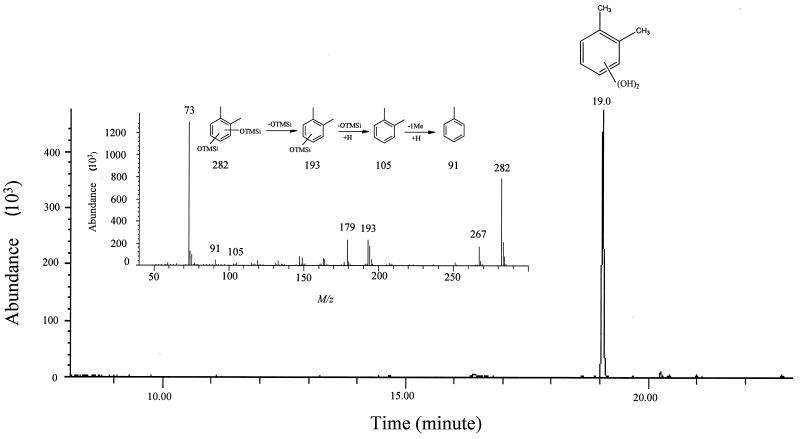
Since the above enzyme assay data demonstrated that the inability of DK180 to grow on o-xylene resulted from a mutation in the gene encoding the meta-cleavage dioxygenase, attempts were made to identify the corresponding metabolite in the o-xylene catabolic pathway. DK180 was grown on glucose in the presence of o-xylene, the culture supernatant was extracted with ethyl acetate, and potential metabolites were derivatized with N-methyl-N-trimethylsilyltrifluoroacetamide. Analysis by capillary GC-MS revealed a major peak for an o-xylene metabolite compound at 19.0 min on the total ion chromatogram (Fig. 2). The metabolite has a molecular ion at m/z 282 (Fig. 2, inset) and a prominent ion due to fission of TMSi at m/z 193 (M+-OTMSi), suggesting that the original metabolite is a dihydroxylated o-xylene, as expected. To examine whether the dihydroxyls are vicinal, this metabolite was also derivatized with methaneboronic acid and 9-phenanthreneboronic acid, which form the respective boronate derivative only by reacting with vicinal dihydroxyls. Subsequently, the expected molecular ions for methane boronate and phenanthreneboronate derivatives of the metabolite were successfully detected at m/z 162 and 324, respectively. Thus, the metabolite at 19.0 min must either be 1,2-dihydroxy-3,4-dimethyl-benzene (3,4-dimethylcatechol) or 1,2-dihydroxy-4,5-dimethyl-benzene (4,5-dimethylcathechol). It was practically impossible to purify the major metabolite for further NMR analysis because the extracted metabolite was extremely unstable at room temperature. In order to overcome this problem, the metabolite was stabilized by acetylation, purified by normal-phase HPLC, and analyzed by 300-MHz 1H-NMR. As summarized in Table 4, the acetate derivative of the major metabolite gave a singlet at δ 2.27 for six protons (6H), which are assignable to the two equivalent methyls in the diacetyls. The two methyls derived from o-xylene were detected at δ 2.08 (s, 3H) and 2.32 (s, 3H). Two aromatic protons that are reciprocally coupled were absorbed at δ 7.05 (d, J = 8.3 Hz) and δ 6.92 (d, J = 8.3 Hz), proving that the aromatic protons are attached at C-5 and C-6. Based on the GC-MS and NMR data, the major metabolite was rigorously identified as 3,4-dimethylcatechol.
FIG. 2.
Total ion chromatogram of metabolites formed during a 24-h incubation of the mutant strain DK180 with o-xylene. The inset shows the electron ionization mass spectrum and the fragmentation pattern of the metabolite at Tr = 19.0 min.
TABLE 4.
300 MHz 1H-NMR data (TMS internal standard) for the acetylated o-xylene metabolite

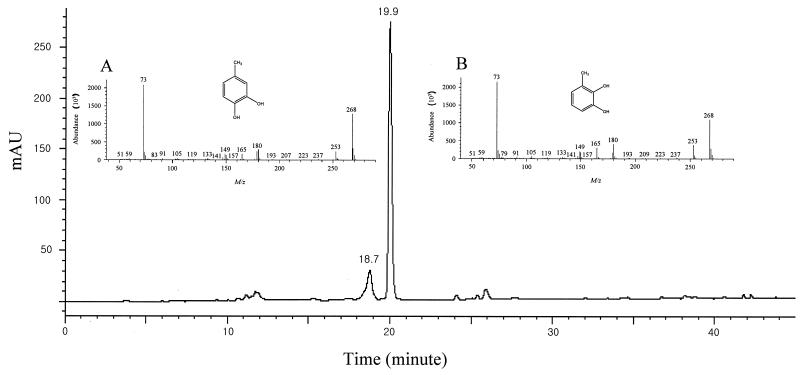
As indicated by the enzyme assay data (Table 2), toluene is also degraded via the same meta-cleavage pathway. An HPLC elution profile of the ethyl acetate-extractable metabolites of toluene formed by DK180 is shown in Fig. 3. Two major peaks eluted at 18.7 and 19.9 min. GC-MS analysis of each peak readily identified the metabolites at 18.7 and 19.9 min as 4-methylcatechol and 3-methylcatechol, respectively, since they showed the same mass spectra and GC retention times as those of the authentic compounds (Fig. 3, insets A and B).
FIG. 3.
HPLC elution profile of metabolites formed from a 24-h incubation of Rhodococcus sp. strain DK180 with toluene. Electron ionization mass spectra of the metabolites at 18.7 and 19.9 min are shown in insets A and B, respectively.
PFGE separation of genomic DNA from Rhodococcus sp. strains DK17, DK176, and DK180.
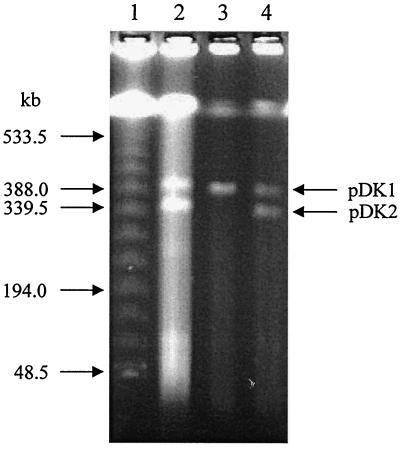
As described above, 49 mutant strains including DK176 showed the same phenotype (loss of an initial oxygenase) from a total of 54 mutant strains selected for the inability to grow on o-xylene. Furthermore, no revertant was obtained from the mutant strains in this class. These observations strongly suggest that the genes for an initial oxygenase are present on a plasmid, which could be lost due to exposure to UV light. PFGE analysis of genomic DNA was performed to separate large plasmids from each other and from the chromosome. Indeed, as shown in Fig. 4, Rhodococcus sp. strain DK17 contains at least two megaplasmids, pDK1 (approximately 380 kb) and pDK2 (approximately 330 kb). The latter plasmid is missing in the mutant strain DK176 but not from the mutant strain DK180. This result indicates that at least some genes for the degradation of o-xylene, benzene, and alkylbenzenes are present on pDK2.
FIG. 4.
PFGE separation of genomic DNAs prepared from Rhodococcus sp. strains DK17, DK176, and DK180. Lane 1, λ ladder standard; lane 2, Rhodococcus sp. strain DK17; lane 3, Rhodococcus sp. strain DK176; lane 4, Rhodococcus sp. strain DK180.
DISCUSSION
The genus Rhodococcus is a group of bacteria that exhibit a diverse range of metabolic activities, including the degradation of various aromatic hydrocarbons (see the review by Warhurst and Fewson [36]). Although much is known about the ability of Rhodococcus strains to grow on toluene (33, 36) and biphenyl (3, 26, 35), relatively little is known about the metabolism of o-xylene by members of this genus. Rhodococcus sp. strain DK17 is able to grow on o-xylene, ethylbenzene, isopropylbenzene, toluene, benzene, and phenol, but not on m- and p-xylene. This inability of DK17 to grow on the latter two compounds reinforces the theory that the xylenes are metabolized by two mutually exclusive pathways (5, 14). As described in Results, since the toxic properties of potential intermediates of the ring oxidation raised an obstacle for determining the catabolic pathway for o-xylene degradation by DK17, attempts were made to generate blocked mutants which would accumulate metabolic intermediates.
A total of 54 mutant strains unable to grow on o-xylene were generated by treatment with UV light and, interestingly, 49 mutant strains neither consumed o-xylene nor oxidized indole to indigo. These data suggest that these strains are either regulatory mutants or are blocked in an initial enzyme in the catabolic pathway. The mutant strain DK176 from this group is unable to grow on o-xylene, ethylbenzene, isopropylbenzene, toluene, and benzene but retains the ability to grow on phenol. Therefore, DK17 apparently employs the same oxygenase to catalyze the initial oxidation of all of the alkylbenzenes tested. In contrast, the mutant strain DK180 oxidizes o-xylene and accumulates metabolites from o-xylene when grown on a second carbon source. One interesting aspect of DK180 is that it retains the ability to grow on benzene although it lost the ability to grow on all of the alkylbenzenes tested. Enzyme assays under different growth conditions showed that DK180 no longer had meta-cleavage dioxygenase activity but retained the separate ortho-cleavage dioxygenase activity. Interestingly, the enzyme assays also demonstrated that the wild-type DK17 normally metabolizes benzene through an ortho- rather than a meta-cleavage pathway. This conclusion is based on the very low specific activity of the meta-cleavage dioxygenase for catechol (less than 2% of that for 3-methylcatechol) and the fact that the ortho-cleavage dioxygenase is induced to high levels in both the DK17 wild-type and DK180 mutant strains only in the presence of benzene. This is unlike other organisms, where metabolism of the aromatic hydrocarbon degradation metabolites catechol and alkyl-substituted catechol proceed through the same meta-cleavage pathway. For instance, in Pseudomonas putida F1, metabolism of benzene, toluene, and ethylbenzene normally proceeds through the intermediates catechol, 3-methylcatechol, and 3-ethylcatechol, respectively, and the meta-cleavage pathway (38). However, in mutant strains lacking the meta-cleavage dioxygenase, F1 metabolizes only benzene, using the now-induced ortho-cleavage pathway (39). Similar observations were made with the TOL plasmid catabolic pathway, which metabolizes toluene and m- and p-xylene through catechol, 3-methylcatechol, and 4-methylcatechol, respectively, and the meta-cleavage pathway. TOL plasmid mutants lacking the meta-cleavage enzyme now channel metabolism of toluene through the ortho-cleavage pathway (27).
The inability of DK180 to grow on o-xylene and the fact that it is blocked in the meta-cleavage step allowed for the unambiguous determination of the identity of the ring-cleavage substrate for o-xylene metabolism in DK17. GC-MS analysis of both trimethylsilyl and boronate derivatives identified a major metabolite with a mass spectrum consistent with a dihydroxyxylene compound where the hydroxyl groups are adjacent to one another. Purification of this major metabolite and determination of the NMR spectrum of the acetylated derivative unambiguously identified the intermediate as 1,2-dimethyl-3,4-dihydroxybenzene (3,4-dihydroxy-o-xylene). Metabolism of o-xylene (and by analogy toluene and benzene) must therefore proceed through either a ring-oxidizing dioxygenase enzyme or two successive monooxygenase enzyme additions of molecular oxygen to the aromatic ring, as expected from the data obtained from growth substrate experiments coupled with inhibition tests. This is in contrast to the metabolism of o-xylene in Rhodococcus sp. strain B3, for which Bickerdike et al. (8) proposed two separate catabolic pathways. The first pathway involves the oxidation of a methyl group to form 2-methylbenzylalcohol and subsequent metabolism to form 3-methylcatechol via 2-methylbenzoic acid. The second proposed pathway for Rhodococcus sp. strain B3 involved initial oxidation of the aromatic ring first to 2,3-dimethylphenol and then to dimethylcatechol. However, the authors failed to identify dimethylcatechol in culture supernatants, probably due to the inherent instability of the compound and the fact that it is readily oxidized. We surmounted this problem by stabilizing the dimethylcatechol by acetylation and successfully identified it as 3,4-dimethylcatechol by NMR. Since Bickerdike et al. (8) detected 2,3- and 3,4-dimethylphenol as oxidation products of o-xylene by using a monooxygenase, we thoroughly examined all of the significant peaks of the total ion chromatogram of metabolites formed during the incubation of DK180 with o-xylene, although no o-xylene metabolites other than 3,4-dimethylcatechol were detected. Unlike Rhodococcus sp. strain B3, Rhodococcus sp. strain C125 is known to initiate o-xylene degradation by directly oxidizing the aromatic ring (31). The authors showed that whole cells of C125 produced the corresponding cis-dihydrodiol from tetralin when incubated in the presence of cis-toluenedihydrodiol as a competitive inhibitor of the cis-dihydrodiol dehydrogenase, and they proposed that o-xylene was degraded via the same type of pathway. However, efforts to detect the formation of cis-dihydrodiol from toluene by DK17 using cis-benzenedihydrodiol as a competitive inhibitor were unsuccessful (data not shown)
As indicated by the PFGE analysis, the genes for the initial oxygenase and the meta-ring cleavage dioxygenase are apparently present on a large plasmid designated as pDK2 (of approximately 330 kb). The fact that the mutant strain DK176, which could not grow on the alkylbenzenes due to the loss of pDK2, was still able to grow on phenol clearly indicates that there is an additional catabolic pathway exclusively for this monohydroxylated aromatic compound. Indeed, Dabrock et al. (12) similarly reported that the degradation of phenol by Rhodococcus erythropolis BD2 was not affected by the loss of a 208-kb linear plasmid, pBD2, containing the catabolic genes for isopropylbenzene degradation.
Acknowledgments
This work was supported by a grant from the Korea Science and Engineering Foundation through the Research Center for Proteineous Materials, 2000.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myera, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Assinder, S. J., and P. A. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv. Microbiol. Physiol. 31:1-69. [DOI] [PubMed] [Google Scholar]
- 3.Asturias, J. A., and K. N. Timmis. 1993. Three different 2,3-dihydroxybiphenyl-1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J. Bacteriol. 175:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baggi, G., P. Barbieri, E. Galli, and S. Tollari. 1987. Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl. Environ. Microbiol. 64:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri, P., L. Palladino, P. Di Gennaro, and E. Galli. 1993. Alternative pathways for o-xylene or m-xylene and p-xylene degradation in a Pseudomonas stutzeri strain. Biodegradation 4:71-80. [Google Scholar]
- 6.Bayly, R. C., S. Dagley, and D. T. Gibson. 1996. The metabolism of cresols by species of Pseudomonas. Biochem. J. 101:293-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertoni, G., F. Bolognese, E. Galli, and P. Barbieri. 1996. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 62:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bickerdike, S. R., R. A. Holt, and G. M. Stephens. 1997. Evidence for metabolism of o-xylene by simultaneous ring and methyl group oxidation in a new soil isolate. Microbiology 143:2321-2329. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Burlage, R. S., S. W. Hooper, and G. S. Sayler. 1989. The TOL (pWW0) catabolic plasmid. Appl. Environ. Microbiol. 55:1323-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlton, B. C., and B. J. Brown. 1981. Gene mutation, p. 224-225. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. Briggs Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 12.Dabrock, B., M. Kesseler, B. Averhoff, and G. Gottschalk. 1994. Identification and characterization of a transmissible linear plasmid from Rhodococcus erythropolis BD2 that encodes isopropylbenzene and trichloroethene catabolism. Appl. Environ. Microbiol. 60:853-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey, J. F., and D. T. Gibson. 1974. Bacterial metabolism of para- and meta-xylene: oxidation of a methyl substituent. J. Bacteriol. 119:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, R. S., F. S. Hossler, and R. W. Stone. 1968. Metabolism of p- and m-xylene by species of Pseudomonas. Can. J. Microbiol. 14:1005-1009. [DOI] [PubMed] [Google Scholar]
- 15.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorn, E., and H. J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds: substituted effects on 1,2-dioxygenation of catechol. Biochem. J. 174:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton, R. W., and K. N. Timmis. 1986. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J. Bacteriol. 168:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensley, B. D., B. J. Ratzkin, T. D. Osslund, M. J. Simon, L. P. Wackett, and D. T. Gibson. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222:167-169. [DOI] [PubMed] [Google Scholar]
- 19.Fishbein, L. 1985. An overview of environmental and toxicological aspects of aromatic hydrocarbons. III. Xylene. Sci. Total Environ. 43:165-183. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, D. T., V. Mahadevan, and J. F. Davey. 1974. Bacterial metabolism of para- and meta-xylene: oxidation of the aromatic ring. J. Bacteriol. 119:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson, D. T., and V. Subramanian. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-252. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, New York, N.Y.
- 22.Harayama, S., N. Mermod, M. Rekik, P. R. Lehrbach, and K. N. Timmis. 1987. Roles of the divergent branches of the meta-cleavage pathway in the degradation of benzoate and substituted benzoates. J. Bacteriol. 169:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, J. L. 1994. Similarity analyses of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 24.Kunz, D. A., and P. J. Chapman. 1981. Catabolism of pseudocumene and 3-ethyltoluene by Pseudomonas putida (arvilla) mt-2: evidence for new functions of the TOL (pWW0) plasmid. J. Bacteriol. 146:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, K., C. J. Duggleby, J. M. Sala-Trepat, and P. A. Williams. 1972. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur. J. Biochem. 28:301-310. [DOI] [PubMed] [Google Scholar]
- 28.Patty, F. A. 1963. Industrial hygiene and toxicology, vol. 2. Toxicology. Interscience Publishers, New York, N.Y.
- 29.Saeki, H., M. Akira, K. Furuhashi, B. Averhoff, and G. Gottschalk. 1999. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology 145:1721-1730. [DOI] [PubMed] [Google Scholar]
- 30.Schraa, G., B. M. Bethe, A. R. W. Vanneerven, W. J. J. Vandentwel, E. Vanderwende, and A. J. B. Zehnder. 1987. Degradation of 1,2-dimethylbenzene by Corynebacterium strain C125. Antonie van Leeuwenhoek 53:159-170. [DOI] [PubMed] [Google Scholar]
- 31.Sikkema, J., and J. A. de Bont. 1993. Metabolism of tetralin (1,2,3,4-tetrahydronaphthalene) in Corynebacterium sp. strain C125. Appl. Environ. Microbiol. 59:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stainer, R. Y., N. J. Palleroni, and M. Duodoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 33.Vanderberg, L. A., R. Krieger-Grumbine, and M. N. Taylor. 2000. Evidence for diverse oxidations in the catabolism of toluene by Rhodococcus rhodochrous strain OFS. Appl. Microbiol. Biotechnol. 53:447-452. [DOI] [PubMed] [Google Scholar]
- 34.van der Meer, J. R., T. N. P. Bosma, W. P. de Bruin, H. Harms, C. Holliger, H. H. M. Rijnaarts, M. E. Tros, G. Schraa, and A. J. B. Zehnder. 1992. Versatility of soil column experiments to study biodegradation of halogenated compounds under environmental conditions. Biodegradation 3:265-284. [Google Scholar]
- 35.Wang, Y., J. Garnon, D. Labbe, H. Bergeron, and P. C. Lau. 1995. Sequence and expression of the bpdC1C2BADE genes involved in the initial steps of biphenyl/chlorobiphenyl degradation by Rhodococcus sp. M5. Gene 164:117-122. [DOI] [PubMed] [Google Scholar]
- 36.Warhurst, A. M., and C. A. Fewson. 1994. Biotransformations catalyzed by the genus Rhodococcus. Crit. Rev. Biotechnol. 14:29-73. [DOI] [PubMed] [Google Scholar]
- 37.Worsey, M. J., and P. A. Williams. 1975. Metabolism of toluene and xylene by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zylstra, G. J. 1994. Molecular analysis of aromatic hydrocarbon degradation, p. 83-115. In S. J. Garte (ed.), Molecular environmental biology. Lewis Publishers, Inc., Boca Raton, Fla.
- 39.Zylstra, G. J., W. R. McCombie, D. T. Gibson, and B. A. Finette. 1988. Toluene degradation by Pseudomonas putida: genetic organization of the tod operon. Appl. Environ. Microbiol. 54:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]