Abstract
A mesophilic toluene-degrading consortium (TDC) and an ethylbenzene-degrading consortium (EDC) were established under sulfate-reducing conditions. These consortia were first characterized by denaturing gradient gel electrophoresis (DGGE) fingerprinting of PCR-amplified 16S rRNA gene fragments, followed by sequencing. The sequences of the major bands (T-1 and E-2) belonging to TDC and EDC, respectively, were affiliated with the family Desulfobacteriaceae. Another major band from EDC (E-1) was related to an uncultured non-sulfate-reducing soil bacterium. Oligonucleotide probes specific for the 16S rRNAs of target organisms corresponding to T-1, E-1, and E-2 were designed, and hybridization conditions were optimized for two analytical formats, membrane and DNA microarray hybridization. Both formats were used to characterize the TDC and EDC, and the results of both were consistent with DGGE analysis. In order to assess the utility of the microarray format for analysis of environmental samples, oil-contaminated sediments from the coast of Kuwait were analyzed. The DNA microarray successfully detected bacterial nucleic acids from these samples, but probes targeting specific groups of sulfate-reducing bacteria did not give positive signals. The results of this study demonstrate the limitations and the potential utility of DNA microarrays for microbial community analysis.
Although hydrocarbons such as benzene, toluene, ethylbenzene, and the xylene isomers are among the most tractable of environmental contaminants to eliminate by natural or stimulated activities of native microbiota, their degradation is constrained by the natural system. For example, aerobic degradation of hydrocarbons is generally much faster than anoxic processes (13, 25). However, most polluted environments (e.g., soils, sediments, and groundwater) are oxygen limited or anoxic. Although many pathways of aerobic degradation of hydrocarbons are well known, their degradation under anaerobic conditions has been little studied. Thus, the fate of hydrocarbons in anoxic settings is much less predictable. Since pollutant fate is largely controlled by the native microbiota, a more complete understanding of community structure and activity should provide for better prediction and process control. Since most environmental bacteria cannot be cultured by conventional culture methods (6, 29), molecular methods (often based on 16S rRNA) have served to provide a more explicit accounting of the genetic diversity (6).
A variety of genetic fingerprinting and hybridization formats have been developed to resolve sequence diversity and abundance. Fingerprinting methods are often a prelude to more quantitative methods. For example, denaturing gradient gel electrophoresis (DGGE) of PCR-amplified fragments provides for the evaluation of genetic diversity and the monitoring of succession in microbial communities (21, 39). However, it is well recognized that PCR biases compromise quantitative interpretation of amplified products (8, 50) and that variable rRNA gene copy number further complicates this assessment (22). Thus, more direct methods such as quantitative membrane hybridization of total 16S rRNA extracted from environmental samples provide a better estimate of relative abundance with good sensitivity (45, 48, 49, 55). However, available formats cannot provide for intensive monitoring.
Recently, DNA microarrays have been developed for medical and diagnostic applications (e.g., detection of single-nucleotide polymorphism, sequencing by hybridization with oligonucleotide in a matrix, and evaluation of gene expression) (14, 53, 59, 63). With this format, thousands of genes can be simultaneously assessed by using a large set of probes miniaturized on one glass slide. This is also an ideal format to assess the sequence diversity of 16S rRNA in natural environmental samples (27).
Sulfate-reducing bacteria are thought to play an important role in degrading hydrocarbons in marine environments because the concentration of sulfate in seawater is very high (28 mM) (31). This is supported by the enrichment and isolation of hydrocarbon-degrading organisms under sulfate-reducing conditions (9, 10, 11, 24, 28, 42, 46, 47, 51). However, their isolation from anaerobic consortia is usually difficult, presumably because they often depend on syntrophic associations (42). Therefore, studies of their natural diversity and abundance in relationship to hydrocarbon degradation should be greatly facilitated by application of microarray technology. The aim of this study is the parallel detection of specific 16S rRNAs in microbial consortia by using the novel microarray technique.
MATERIALS AND METHODS
Microorganisms.
Desulfobacter latus (DSM 3381), Desulfosarcina variabilis (DSM 2060), Desulfobacterium anilini (ATCC 49792), and Desulfovibrio africanus (ATCC 19996) were used as reference strains in this study. They were cultured with the sulfide-reduced bicarbonate-buffered defined medium described by Widdel and Bak (61). The medium for seawater and freshwater contained 20 and 1 g of NaCl liter−1, respectively. The substrate for each microorganism was added according to the recipe on the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH web site.
Bacterial enrichment and sample collection.
The mesophilic toluene- and ethylbenzene-degrading consortia were established from sediment obtained from the Tokyo Bay estuary, Japan, in 1993. The enrichment of these consortia was established by Nakagawa et al. (41). An aliquot (5 to 10 ml) of sediment was inoculated into 115 ml of medium in 150-ml serum bottles, which were then sealed with butyl rubber stoppers under a headspace of N2-CO2 (80:20, vol/vol). Sulfate-reducing enrichment cultures were established with crude oil as the sole source of carbon and energy. Incubations were carried out at 28°C in the dark. An aliquot (3 ml) of enrichment culture was transferred to freshly prepared medium every 4 months until the sediment was removed. From this enrichment, toluene (TDC)- and ethylbenzene (EDC)-degrading consortia were established. To avoid toxicity, toluene and ethylbenzene were diluted with 2,2,4,4,6,8,8-heptamethylnonane as the carrier phase containing 2% (vol/vol) toluene or ethylbenzene (28, 47). An aliquot (5 ml) of carrier phase was added to the medium.
To confirm sulfate-reducing bacterial growth, sulfide production was occasionally monitored by mixing 0.2 ml of the culture and 1 ml of sulfide detection reagent (5 mM CuCl2, 50 mM HCl) (17), producing a dark brown color in the presence of sulfide. The dissolved sulfide concentration in the aqueous phase of the enrichment cultures was also colorimetrically measured by using the methylene blue reaction (16), as modified by Aeckersberg et al. (1). The sulfide concentration data were obtained from a single experiment and measurement.
A crude oil-contaminated sediment sample was obtained at Shuaiba in the coastal area of Kuwait in June 1995 by using a van Veen-type grab sampler. The composition of the oil recovered from the sediment was 7% asphaltene, 43% polar moltene, 29% saturate neutral, 6% monoaromatics, 11% diaromatics, and 4% triaromatics (52). The sample was transferred to the laboratory in an icebox for analysis.
DNA extraction.
DNA extraction from cell pellets was carried out as described previously (62). Cells were disrupted by sodium dodecyl sulfate (SDS) and proteinase K treatment. Polysaccharides and other contaminating macromolecules derived from bacterial cells were removed by using hexadecyltrimethyl ammonium bromide (CTAB). After chloroform-isoamyl alcohol (24:1) and phenol-chloroform-isoamyl alcohol (PCI; 25:24:1, pH 8.0) extractions, total nucleic acid was recovered by ethanol precipitation.
Extraction and purification of DNA from 2 g of washed sediment were performed by the hydroxylapatite (HTP) (Bio-Gel HTP Gel, Bio-Rad Laboratories, United Kingdom) spin-column method (44). The samples were freeze-thawed in 0.9 ml of lysing buffer (50 mM Tris-HCl [pH 8.0], 0.1 mM EDTA [pH 8.0], 25% sucrose, and 10 mM sodium pyruvate). After centrifugation, the supernatants were removed, and the following solutions added to each tube: 0.7 ml of 120 mM sodium phosphate (pH 8.0), 0.5 ml of PCI, and 50 μl of 20% SDS. The cells were then mechanically disrupted by bead beating (FastPrep FP120; Bio 101, Inc., Vista, Calif.) for 2 min at the max speed (37).
For removal of humic substances, we used an HTP spin-column constructed from a 2.5-ml plastic syringe (Terumo Corp., Tokyo, Japan). After loading extracts, the column was washed four times by spinning at 100 × g for 4 min with 0.8 ml of 50 mM sodium phosphate (pH 6.8) for each wash. Following successive passages of 0.4 ml of 140 mM potassium phosphate (pH 6.8) and 0.8 ml of 300 mM potassium phosphate (pH 6.8), total nucleic acid was recovered in two 1.5-ml microtubes by eluting twice with 0.8 ml of 300 mM potassium phosphate, pH 6.8. Salts and humic contaminants were removed from the elution with a 2.5-ml Sephadex G-75 spin column (38), and the nucleic acid was collected by isopropyl alcohol precipitation.
DGGE analysis.
PCR for DGGE analysis was performed as described by Muyzer et al. (40) but with the complement of probe EUB338 (4) as a forward primer with GC-clamp (338f, 5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′). As a reverse primer, 907r (5) was used.
PCR-amplified 16S rDNA fragments were analyzed by DGGE, followed by sequencing. DGGE was performed with a D-gene or D-code system (Bio-Rad Laboratories, Hercules, Calif.), as described previously (39, 40). The denaturing gradient ranged from 20 to 60%. Approximately 500 ng of PCR products and 100 ng of reamplified DNA fragments were electrophoretically fractionated for 4 h at a constant voltage of 200 V and a temperature of 60°C. After electrophoresis, the gels were incubated for 10 min in Milli-Q water containing ethidium bromide (1.0 mg liter−1), rinsed for 10 min in Milli-Q water, and photographed with a UV (302 nm) transillumination system equipped with an Atto Printgraph (Atto, Tokyo, Japan).
DGGE bands excised from the gel were reamplified, and their purity was verified by DGGE. After purification of PCR products with the QIAquick PCR purification kit (Qiagen, Hiden, Germany), sequences were determined by cycle sequencing with a dye-labeled primer.
RNA sample preparation.
For native RNA from microorganisms, total RNA was extracted from cell pellets by the low-pH bead-beating method (37, 55).
For recovery of native RNA from sediments, extraction and purification of nucleic acids were performed by using the HTP spin-column method described above. DNA was degraded by using DNase I as previously described (38). RNA was recovered by PCI extraction followed by ethanol precipitation.
In vitro transcription was used to produce 16S rRNA from the major DGGE bands and from genomic DNA of individual isolates. PCR amplification was carried out with the primer pair of T7 promoter-conjugated 338f (5′-TGAATTGTAATACGACTCACTATAGGGCGAATTC-3′) and 907r or T7 promoter-conjugated BACT11F (32) and S-D-Bact-1512-a-A-16 (23) in a thermal cycler (Thermo Hybaid US, Franklin, Mass.) in 100-μl aliquots under the following conditions. Each tube contained 1× PCR buffer, deoxynucleoside triphosphate mixture (2.5 mM each), 25 μM each primer, 5 U of Taq DNA polymerase (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) μl−1, and 100 ng of template DNA. An initial denaturation step of 3 min at 95°C was followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C, then 5 min at 72°C. RNA was transcribed from these PCR-generated templates by using a commercial RNA transcription kit (New England Bio Labs Inc., Beverly, Mass.) in 150-μl aliquots according to the manufacturer's protocol.
Oligonucleotide probe design and determination of washing temperature.
Four oligonucleotide probes were designed for the major DGGE bands by using the Probe_Design tool of the ARB software package (O. Strunk, O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckman, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig, 1998, http://www.mikro.biologie.tu-muenchen.de/pub/ARB). Since probe S-∗-Tsrda-0641-a-A-20 had self-complementarity near the termini, an alternative probe (probe S-∗-Tsrda-0644-a-A-19) was designed (Table 1). The Probe_Check program provided by the Ribosomal Database Project II (35) and BLAST search (3) were both used to evaluate probe specificity. The Td (temperature of dissociation) values and the specificities of the oligonucleotide probes were assessed by the general method described by Zheng et al. (64). All oligonucleotide probes were synthesized with an amino linker at the 3′ end by Operon Inc. (Alameda, Calif.). The amino linker is used for immobilization of the oligonucleotide probes within the DNA microarrays.
TABLE 1.
Probe used in this study
| Probea and microorganisms | Target sequencee | GC% | Membrane
|
DNA microarray
|
Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Td (°C) | Tw (°C) | % Washed off at Tw | Td (°C) | Tw (°C) | % Washed off at TW | ||||
| S-*-Tsrda-0641-a-A-20 | 5′CATACTCAAGCCTGGCAGTA3′ | 50.0 | 46 | ||||||
| T1 | 3′GUAUGAGUUCGGUCCGUCAU5′ | 54 | 58 | 70 | 42 | 46 | 70 | This study | |
| Desulfosarcina variabilis | --------------U----- | 52 | 96 | 36 | 88 | ||||
| S-*-Tsrda-0644-a-A-19 | 5′CCCATACTCAAGCCTGGCA3′ | 57.9 | 46 | ||||||
| T1 | 3′GGGUAUGAGUUCGGACCGU5′ | 51.5 | 60 | 77 | 44 | 46 | 60 | This study | |
| Desulfosarcina variabilis | ----------------U-- | 51.5 | 100 | 38.5 | 79 | ||||
| S-*-Edn-0656-a-A-18 | 5′CGTCTTCCCCCACCTTAC3′ | 61.1 | 46 | ||||||
| E1 | 3′GCAGAAGGGGGUGGAAUG5′ | 66 | 66 | 50 | 40.5 | 46 | 68 | This study | |
| Lactobacillus catenaforme | --G-----A--G------ | ||||||||
| S-*-Edsrb-0656-a-A-19 | 5′CGAATCTCCTCTCCCATAC3′ | 52.6 | 46 | ||||||
| E2 | 3′GGUUAGAGGAGAGGGUAUG5′ | 62.5 | 53 | 40.5 | 46 | 68 | |||
| T1 | ---G-AG------------ | 63 | 39.5 | 46 | 98 | This study | |||
| Desulfobacterium anilini | --G---------------- | 54.5 | 100 | 33 | 77 | ||||
| S-*-Univ-1390-a-A-18b | 5′GACGGGCGGTGTGTACAA3′ | 61.1 | 44 | 46 | 64 | ||||
| S-*-Univ-0907-a-A-22 | 5′CCCCGTCAATTCCTTTGAGTTT3′ | 45.5 | 46 | ||||||
| T1 | 3′GGGGCAGUUAAGGAAACUCAAA5′ | 53 | 6 | 47 | 46 | 36 | |||
| E1 | ---------------------- | 45.6 | 46 | 54 | |||||
| E2 | ---------------------- | 42 | 45 | 46 | 55 | 5 | |||
| Escherichia coli | ------------T--------- | 51 | 6 | ||||||
| Desulfobotulus sapovorans | ---------------------- | 61 | 4 | ||||||
| Methanococcus thermolithotrophicus | -----G----------U----- | 51 | 6 | ||||||
| S-D-Bact-0338-a-A-18 | 5′GCTGCCTCCCGTAGGAGT3′ | 66.7 | 46 | ||||||
| T1 | 3′CGACGGAGGGCAUCCTCA5′ | 44.5 | 46 | 57 | |||||
| E1 | ------------------ | 54 | 43 | 46 | 62 | 4 | |||
| E2 | ------------------ | —c | 46 | — | |||||
| S-D-Bact-0927-a-A-17b | 5′ACCGCTTGTGCGGGCCC3′ | 76.5 | 46 | 26 | |||||
| S-F-Dsv-0687-a-A-16 | 5′TACGGATTTCACTCCT3′ | 43.8 | 46 | ||||||
| Desulfovibrio africanus | 3′AUGCCUAAAGUGAGGA5′ | 44 | NSd | NS | 19 | ||||
| Geobacter metallireducens | ---------------- | NS | NS | ||||||
| S-*-Dsb-0804-a-A-18 | 5′CAACGTTTACTGCGTGGA3′ | 50.0 | 46 | ||||||
| Desulfobacter latus | 3′GUUGCAAAUGACGCACCU5′ | 46 | 19 | ||||||
| T1 | ------------------ | 39.5 | 46 | 85 | |||||
Probes are named according to the Oligonucleotide Probe Database (2).
These probes were not used in slot-blot hybridization.
—, Td was not determined because the melting profile obtained was straight line and did not show a sigmoid curve.
NS, no signal.
Underlined positions are positions that may cause self-complementality.
Quantitative membrane hybridization.
For quantitative membrane hybridization, denatured RNA samples and a dilution series of pure culture RNA were applied in triplicate to nylon membranes (Magna Charge nylon membrane; Micron Separation Inc., Westboro, Mass.), and hybridization was performed as described previously (48, 55, 64). The concentrations of all samples and reference RNAs were normalized by using the probe S-∗-Univ-907-a-A-22. A known concentration of Escherichia coli native RNA determined spectrophotometrically was used for normalization. The hybridization signals were measured by using a PhosphorImager (Storm 860; Molecular Dynamics, Sunnyvale, Calif.) and analyzed with the ImageQuant software package (Molecular Dynamics). The results were expressed as the fraction of target 16S rRNA relative to total eubacterial rRNA quantified by using probe S-D-Bact-0338-a-A-18.
DNA microarray fabrication.
The DNA microarrays used in this study consisted of miniaturized oligonucleotide arrays in which oligonucleotide probes were individually immobilized within small polyacrylamide gel pads affixed to a glass slide. A matrix of glass-immobilized gel elements (100 by 100 by 20 μm each) spaced 100 μm apart was manufactured with photopolymerization (63) and activated as described previously (33). We have incorporated four new oligonucleotide probes and six previously published probes into the DNA microarray used in this study (Table 1). Approximately 6 nl of individual 1 mM amino-oligonucleotide solutions was applied to each gel pad containing aldehyde groups according to the procedure described previously (63).
RNA fragmentation and labeling.
About 10 to 20 μg of either native or transcribed rRNA was heated at 95°C for 5 min and fragmented by addition of 60 mM MgCl2 (total volume, 20 μl) and incubation at 95°C for 40 min. Phosphatase treatment was performed by addition of 3 μl of 10× alkaline phosphatase buffer (Promega, Madison, Wis.) and 0.2 μl of shrimp alkaline phosphatase (1 U μl−1) (Promega) and heating at 37°C for 30 min. Oxidation of the 3′-end ribosyl moiety was conducted by addition of 6.5 μl of 100 mM sodium periodate and incubation at room temperature for 20 min.
Labeling was carried out by addition of 3.5 μl of 100 mM lissamine rhodamine B ethylenediamine (Molecular Probes, Eugene, Oreg.) and 1.65 μl of 1 M HEPES (pH 7.5) and heating at 37°C for 1 h. Reduction of the Schiff base was conducted by addition of 6.7 μl of 200 mM sodium cyanoborohydride and incubation at room temperature for 30 min. Labeled RNA was precipitated by addition of 15 volumes of 2% lithium perchlorate in acetone and chilling at −20°C for 20 min. After centrifugation at 14,000 rpm for 5 min, RNA pellets were washed twice with acetone, dried at 55°C for 10 min, and suspended in 20 μl of nuclease-free water (43).
Microarray and image analysis.
An aliquot (40 μl) of hybridization solution containing labeled RNA [10 μg of native RNA, 5 μg of transcribed (11-1512)RNA, or 3 μg of transcribed (338-927)RNA], 60% formamide, 0.9 M NaCl, and 20 mM Tris-HCl buffer (pH 8.0) was passed through a 0.22-μm filter (Ultra-MC; Millipore) to remove particulates, then heated at 95°C for 3 min, and held on ice. An aliquot (35 μl) of the hybridization solution was added to a hybridization chamber (Grace Biolabs, Bend, Oreg.), and the hybridization chamber was affixed to the microarray. The microarray was allowed to hybridize at room temperature for at least 16 h in the dark. After the microchamber and hybridization solution were removed, the microarray was soaked in warm washing buffer (20 mM Tris-HCl buffer [pH 8.0], 10 mM NaCl, and 5 mM EDTA [pH 8.0]) at 46°C for 5 min. The microarray was then immediately covered with a chamber filled with 100 μl of washing buffer.
The signal of each probe hybridized with fragmented and labeled rRNA was detected by using a custom-made epifluorescence microscope equipped with a charge-coupled device camera (Princeton Instruments, Princeton, N.J.), and the intensity of each signal was measured at room temperature with WinView software (Princeton Instruments). Exposure times were in the range of 0.1 to 1 s, depending on the signal intensity. When melting curves were determined, the microarray was washed at room temperature for 1 min two times and then covered with the chamber filled with 100 μl of washing buffer. The temperature of the microscope slide was controlled by a thermotable mounted on a stage of the epifluorescence microscope and connected with a thermoelectric temperature controller (LFI-3751; Wavelength Electronics, Inc., Bozeman, Mont.) and a waterbath (Cole Parmer Instruments Co., Chicago, Ill.). Melting profiles for all probes were monitored and recorded at 2°C intervals from 10 to 70°C by increasing the temperature at 1°C per min.
Nucleotide sequence accession numbers.
The partial rRNA gene sequences have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession nos. AB062689, AB062688, and AB062924.
RESULTS AND DISCUSSION
Sulfide production and hydrocarbon degradation in enrichments.
We enriched the marine sediment, which was obtained from Tokyo Bay, by adding toluene (2% [vol/vol] in the carrier phase) or ethylbenzene (2% [vol/vol] in the carrier phase) as a sole electron donor and carbon source under sulfate-reducing, anaerobic conditions. The sulfide production of these enrichment cultures was monitored with respect to growth on hydrocarbon. In the toluene-degrading consortium (TDC), the amount of toluene decreased from 1.53 to 0.72 mmol, associated with 2.92 mmol of sulfide production after 90 days of incubation. In the ethylbenzene-degrading consortium (EDC), the concentration of sulfide increased from 0.39 mmol on day 15 to 1.63 mmol on day 49, then reached 2.14 mmol after 82 days. The amount of ethylbenzene decreased from 0.95 mmol on day 5 to 0.38 mmol on day 82. Attempts to isolate pure cultures from TDC and EDC have so far been unsuccessful.
DGGE analysis and sequencing.
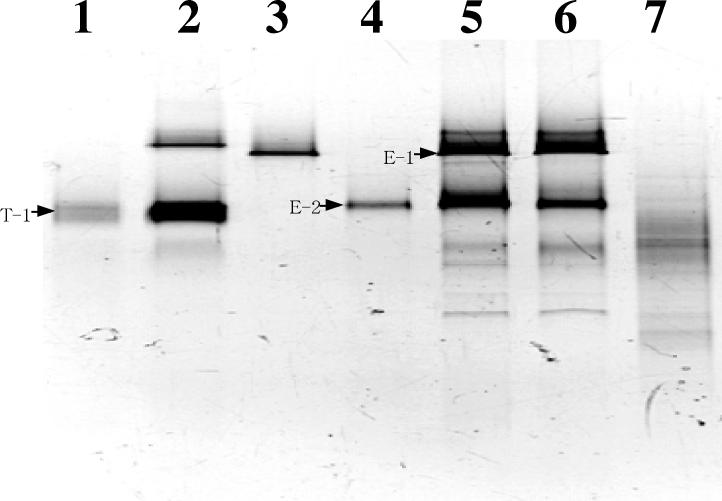
DGGE patterns of TDC at 30 days, EDC at 30 and 60 days, and oil-contaminated sediment are shown in Fig. 1 (lanes 2, 5, 6, and 7, respectively). A few conspicuous bands (T-1 in TDC and E-1 and E-2 in EDC) were observed on the DGGE gel. This result indicated that specific microorganisms were selectively enriched. The DGGE pattern of the DNA amplicon from the oil-contaminated sediment was faint, and no conspicuous bands were detected.
FIG. 1.
DGGE analysis of PCR-amplified 16S rDNA fragments obtained from TDC and EDC, microbial populations in an oil-contaminated sediment at the coastal area of Kuwait, and the major DGGE bands. Inverted image of the DGGE gel stained by ethidium bromide. Lane 1, reamplified band T-1; lane 2, TDC (30 days); lane 3, reamplified band E-1; lane 4, reamplified band E-2; lane 5, EDC (30 days); lane 6, EDC (60 days); lane 7, Kuwaiti sediment. 16S rDNA fragments of TDC, EDC, and oil-contaminated sediment were obtained by PCR amplification of genomic DNA (lanes 2, 5, 6, and 7). Reproducibility of DGGE analysis was confirmed by three replications of PCR.
The bands corresponding to T-1, E-1, and E-2 were not visible in the sediment sample. DNA fragments were excised from the major bands on the DGGE gel (T-1, E-1, and E-2) and reamplified (Fig. 1, lanes 1, 3, and 4, respectively). The sequences of these 16S rRNA fragments (338 to 927, based on E. coli numbering) were determined. The sequence of T-1 was most similar to that of strain oXyS1 (99.8%), which originated from the water phase of an oil tank (28). Strain oXyS1 is affiliated with a relatively deep-branching lineage within the family Desulfobacteriaceae. Growth on toluene was also observed for strain oXyS1.
Even though utilization of ethylbenzene has not previously been detected under mesophilic sulfate-reducing conditions (47, 51), we successfully enriched for sulfate-reducing bacteria by using ethylbenzene as a sole energy and carbon source. DGGE analysis followed by sequencing revealed that the sequence of E-2 was most similar to that of mXyS1 (98.0%), affiliated with the family Desulfobacteriaceae. mXyS1 is a novel type of marine sulfate-reducing bacterium capable of complete anaerobic degradation of m-xylene (28). The sequence of E-1 showed highest similarity (84.6%) to an uncultured soil bacterium, PBS-21, loosely affiliated with a deep-branching lineage within the order Spirochaetales.
These results are consistent with the finding that all known sulfate-reducing bacteria enriched on aromatic compounds are affiliated with the family Desulfobacteriaceae (47). Moreover, Phelps et al. (42) reported that 4 of 12 clones isolated from a sulfate-reducing consortium enriched with benzene belonged to the family Desulfobacteriaceae.
Quantitative membrane hybridization.
In order to determine the abundance of the putative hydrocarbon-degrading bacteria revealed by the DGGE analysis, we designed four oligonucleotide probes based on the sequences of the major DGGE bands (Table 1). In addition, probes S-D-Bact-0338-a-A-18, S-F-Dsv-0687-a-A-16, and S-∗-Dsb-0804-a-A-18 were also used. Probe S-∗-Univ-0907-a-A-22 was used for normalization. The final washing temperature (Tw) for each probe (Table 1) was empirically determined by a temperature of dissociation (Td) study and specificity study as previously described by Zheng et al. (63). Normalized Td curves of these probes were obtained with high reproducibility (Fig. 2). Table 1 lists the targets, the sequence alignments, and the amount of 32P-labeled probes dissociated at each specific wash condition (Tw). Tw for probe S-∗-Univ-0907-a-A-22 corresponded to the temperature at which an almost equal percentage of the duplex between any 16S rRNA target and probe remained intact, but was higher than the temperature causing nonspecific binding.
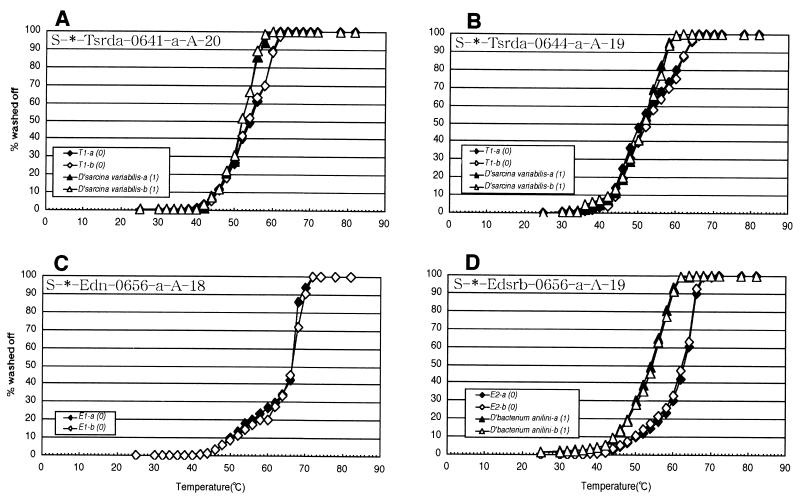
FIG. 2.
Normalized Td (temperature of dissociation) curves determined by using transcribed RNA (338 to 927, E. coli numbering) of target 16S rDNA fragments and native RNA of nontarget microorganisms. Numbers in parentheses indicate the number of mismatches between probe and target sequences. All curves shown are the result of duplicate experiments. Only target RNA was used for probe S-∗-Edn-0656-a-A-18.
Probes S-∗-Tsrda-0641-a-A-20 and S-∗-Tsrda-0644-a-A-19, which target the same region of the 16S rRNA, showed different melting curves. Also, the initial signal intensity of probe S-∗-Tsrda-0641-a-A-20 was 100 times lower than that of probe S-∗-Tsrda-0644-a-A-19. This difference was attributed to self-complementarity (Table 1). For probe S-∗-Tsrda-0641-a-A-20, the Td value for the nontarget organism containing one mismatch was only 2°C lower than that for the target organism (Fig. 2A). Since a Tw achieving complete discrimination between the target and the nontarget containing one mismatch would reduce signal intensity by 90%, 58°C was selected as the Tw. Therefore, the signal of single-mismatch nontargets would be slightly above background (96% washed off) (Table 1).
For probe S-∗-Tsrda-0644-a-A-19, the Td values of the target organism and the nontarget organism with one mismatch were not different. However, the melting curve of the duplex between S-∗-Tsrda-0644-a-A-19 and the target (T-1) had a shoulder at around 53°C (Fig. 2B). Due to this shoulder, it was possible to determine a Tw that would result in reasonable hybridization signals. Probes S-∗-Edn-0656-a-A-18 and S-∗-Edsrb-0656-a-A-19 showed ideal melting curves (Fig. 2C and D). Their Td values were 66 and 63°C, respectively (Table 1). These probes could distinguish between the target organisms and the nontarget reference organisms with one or more mismatches by washing at their respective Td values.
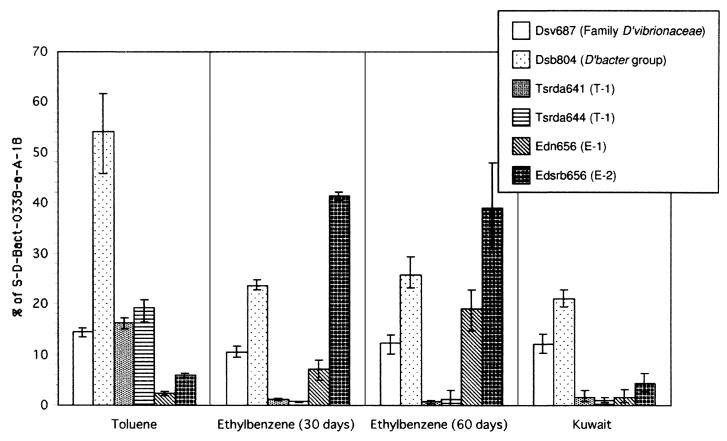
Figure 3 displays the relative fraction of 16S rRNA extracted directly from microbial populations comprising the toluene (TDC)- and ethylbenzene (EDC)-degrading consortia and the oil-contaminated sediment determined by quantitative membrane hybridization. This analysis revealed that T-1 and E-1 and E-2 were members of the TDC and EDC, respectively. This was consistent with the results of DGGE analysis. The signals of the probes for T-1 were negligible in EDC and those for T-1, E-1, and E-2 were negligible in the oil-contaminated sediment.
FIG. 3.
Results of quantitative membrane hybridization signals obtained with specific probes were normalized to the hybridization signal of probe S-D-Bact-0338-a-A-18. The results are expressed as a percentage of the total eubacterial 16S rRNA and represent the mean values of triplicate applications from a single sample. Vertical bars indicate maximum and minimum ratios of specific probes to eubacterial probe (S-D-Bact-0338-a-A-18) calculated by all combinations as follows: aA−1, aB−1, aC−1, bA−1, bB−1, bC−1, cA−1, cB−1, and cC−1, where the lowercase letters represent the signal intensities of each specific probe in triplicate (a, b, and c), and the capital letters represent the signal intensities of eubacterial probe in triplicate (A, B, and C). RNA samples were obtained from TDC, EDC, and oil-contaminated sediment in the coastal area of Kuwait.
Since the dissimilatory, gram-negative sulfate-reducing bacteria can be roughly divided into the families Desulfovibrionaceae and Desulfobacteriaceae (18), we used general probes for these groups to encompass most of the gram-negative sulfate-reducing bacteria mesophilic group. The signals of probe S-F-Dsv-0687-a-A-16 accounted for 11.1 to 14.4% of total eubacterial 16S rRNA in all samples. However, this probe hybridizes to some species of Geobacteriaceae (20), and the contribution of these types in the samples is unknown. The group hybridizing with probe S-∗-Dsb-0804-a-A-18 was abundant in all samples, consistent with the previously reported finding that most aromatic compound-degrading bacteria belong to the family Desulfobacteriaceae (42, 47).
In TDC, probes S-∗-Tsrda-0641-a-A-20 and S-∗-Tsrda-0644-a-A-19 (targeting T-1) gave positive signals and accounted for 16.3 and 19.2% of total 16S rRNA, respectively. There were no significant differences between the results with probes S-∗-Tsrda-0641-a-A-20 and S-∗-Tsrda-0644-a-A-19 in any of the samples. In EDC, the signal intensity of probe S-∗-Edn-0656-a-A-18 increased markedly from 7.2% on day 30 to 19.0% on day 60. The signal with probe S-∗-Edsrb-0656-a-A-19 did not change significantly, from 41.4% to 39.1%, during this period.
The Desulfobacter group was not more abundant in EDC than in TDC. McMahon et al. (36) demonstrated the difference caused by the use of in vitro-transcribed and native RNAs for membrane hybridization. They concluded that transcribed RNA could be used to determine Td values because differences were relatively small. They also mentioned that when in vitro-transcribed rRNA was used as a standard for quantitative hybridization, the population was consistently underestimated. The ratio of T-1, E-1, and E-2 to total bacteria might be underestimated because we used transcribed RNA as a standard of the populations corresponding to the major DGGE bands. Nevertheless, we demonstrated that the combination of DGGE and quantitative membrane hybridization without isolation could be applied more generally to the quantification of slow-growing environmental populations such as hydrocarbon-degrading sulfate-reducing bacteria.
It has been estimated that 6 to 10 million barrels of crude oil were released into the coastal area of Kuwait during the 1991 Gulf War (56). Oil-contaminated sediment from the coast of Kuwait was studied as an example of a bioremediating system inhabited by complex microbial communities. Application of membrane hybridization with the above probes to characterize oil-contaminated marine sediments from the coast of Kuwait revealed the presence of hydrocarbon-degrading sulfate-reducing bacteria. This sediment contained more than 1 mg of oil g−1 (wet weight). Forty percent of total bacteria (total 16S rRNA) were detected with the probes used in this study. The E-2 target population, which may degrade ethylbenzene, comprised 4.3% of rRNA extracted from this sediment. The family Desulfobacteriaceae (probe S-∗-Dsb-0804-a-A-18) accounted for 21.1%. Members of this family are known to degrade the aromatic compounds present in the contaminated sediment. Times series analysis from November 1993 to June 1995 revealed that the asphaltene content decreased from 12 to 2% and the monoaromatic component (containing toluene and ethylbenzene) remained almost constant at 10% during this period (52). These results indicate that the asphaltene component was degraded without accumulation of monoaromatic compounds, suggesting that biodegradation of aromatic compounds was occurring simultaneously.
Application of DNA microarrays.
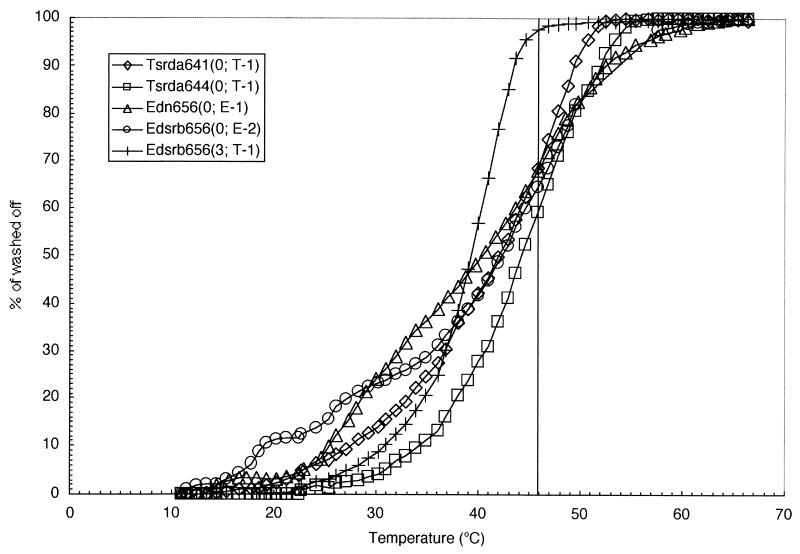
Optimal washing conditions for the microarray used in this study were established by first determining the melting profiles of all probe-target duplexes. The design of the DNA microarray and the numbers of mismatches are listed in Table 2. Nontarget RNAs having three or more mismatches were completely dissociated following two washings at room temperature for 1 min. However, the duplex between probes S-∗-Edsrb-0656-a-A-19 and T-1 was not dissociated under this condition. A single washing temperature of 46°C was established based on the melting profile (Fig. 4 and Table 1). The optimal washing time was determined by evaluating dissociation kinetics for a series of washing times (data not shown). In summary, these studies showed that washing for 5 min was sufficient to eliminate nontargets containing three or more mismatches. However, washing for longer than 5 min reduced signal intensities without improvement in specificity. Thus, a washing protocol of 46°C for 5 min was used for further experiments.
TABLE 2.
Probe names, number of mismatches, and locations on DNA microarray
| Probe | No. of mismatches
|
Location
|
|||
|---|---|---|---|---|---|
| T1 | E1 | E2 | Column | Row | |
| S-*-Tsrda-0644-a-A-19 | 0 | 7 | 4 | 1 | A |
| S-*-Tsrda-0641-a-A-20 | 0 | 6 | 5 | 2 | A |
| S-*-Edn-0656-a-A-18 | 6 | 0 | 6 | 3 | A |
| S-*-Edsrb-0656-a-A-19 | 3 | 7 | 0 | 4 | A |
| S-*-Dsb-0804-a-A-18 | 0 | 3 | 2 | 3 | B |
| S-F-Dsv-0687-a-A-16 | 3 | 5 | 3 | 4 | B |
| S-D-Bact-0338-a-A-18 | 0 | 0 | 0 | 3 | C |
| S-D-Bact-0927-a-A-17 | 0 | 0 | 0 | 4 | C |
| S-*-Univ-0907-a-A-22 | —a | — | — | 3 | D |
| S-*-Univ-1390-a-A-18 | — | — | — | 4 | D |
—, transcribed rRNA (338 to 927, E. coli numbering) T-1, E-1, and E-2 do not contain the target sites.
FIG. 4.
DNA microarray temperature-of-dissociation study for transcribed RNA fragments derived from DGGE bands on DNA microarrays. Numbers in parentheses indicate the number of mismatches between probe and target sequences. All elution curves represent the means of four replicate experiments on one chip. Hybridization to RNA containing more than three mismatches could not be detected at the beginning of the melting curve except for the duplex of S-∗-Edsrb-0656-a-A-19 and T-1. A washing temperature of 46°C (vertical line) was used to achieve suitable mismatch discrimination.
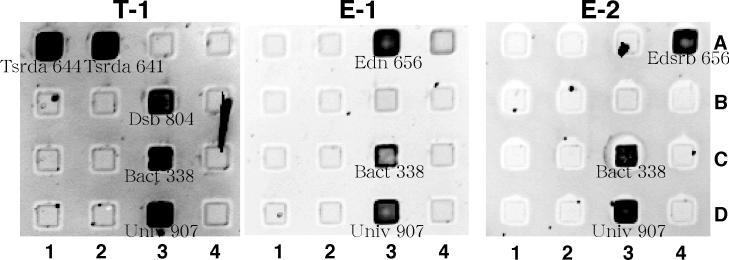
The RNAs transcribed from DGGE fragments hybridized with the complementary probes under the washing condition defined above (Fig. 5). Universal and eubacterial probes gave positive signals for all samples. Since the RNAs of DGGE bands were transcribed from the DNA fragments amplified with the 338f-907r primer pair, probes S-∗-Univ-1390-a-A-18 and S-D-Bact-0927-a-A-17 did not hybridize for lack of target sequences. In contrast, the failure of probe S-F-Dsv-0687-a-A-16 to hybridize is attributed to the lower stability of this short probe under the conditions used in this study (43). The effect of probe length and sequence composition on DNA microarray hybridization is under investigation.
FIG. 5.
Hybridization of lissamine rhodamine-labeled RNA to DNA microarray. RNAs (338 to 927, E. coli numbering) were transcribed from PCR-amplified 16S rDNA fragments from the DGGE gel, fragmented with magnesium, and labeled with lissamine rhodamine. Probes, their locations, and the number of mismatches between each probe and the RNA are shown in Table 2. The DNA microarray was hybridized to in vitro-transcribed 16S rRNA fragments from bands T-1, E-1, and E-2, shown in panels T-1, E-1, and E-2, respectively.
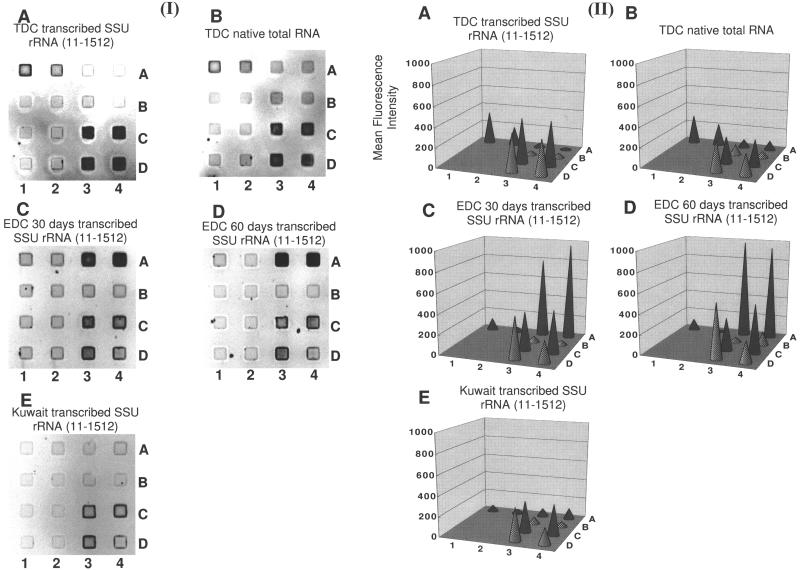
This microarray was then used to characterize enrichment and environmental samples (Fig. 6). The in vitro-transcribed RNA was used to increase the sensitivity of analysis for some samples. The hybridization of the TDC sample with native and in vitro-transcribed RNAs showed comparable patterns, although the native RNA (including 23S and 5S rRNA) generated a higher background than in vitro-transcribed RNA (Fig. 6A and B). Since EDC and the oil-contaminated sediment generally did not yield sufficient native RNA to hybridize to the DNA microarray, transcribed RNA was used to amplify the signal. Each yielded characteristic patterns.
FIG. 6.
Results of DNA microarray hybridization with samples of TDC, EDC, and oil-contaminated sediment from the coastal area of Kuwait. Probes, their locations, and the number of mismatches between each probe and the RNA are shown in Table 2. RNA was prepared by in vitro transcription of 16S rDNA fragments (11 to 1512, E. coli numbering) PCR amplified from genomic DNA. Native RNA was also used for one TDC sample. (I) Negative images of hybridization with a DNA microarray and (II) fluorescent signal intensities for (A) TDC, (B) TDC (native RNA), (C) 30-day EDC, (D) 60-day EDC, and (E) Kuwaiti sediment. Fluorescence intensities were quantified by using WinView and calculated from four replications of each probe on one chip. SSU, small subunit.
The equal variance and significant differences of signal intensities between specific probes and blank were verified by two-tailed F test [F (3, 19; 0.005)] and t test [t (22, 0.01)], respectively. Probes S-∗-Tsrda-0641-a-A-20 and S-∗-Tsrda-0644-a-A-19 gave positive signals for the TDC sample (P < 0.01) (Fig. 6A and B) and probes S-∗-Edn-0656-a-A-18 and S-∗-Edsrb-0656-a-A-19 gave positive signals for EDC (P < 0.01) (Fig. 6C and D). For the Kuwaiti sediment, only the universal and eubacterial probes gave positive signals (P < 0.01) (Fig. 6E). Although the Desulfobacter group (hybridized by probe S-∗-Dsb-0804-a-A-18) accounted for more than 50% of bacterial 16S rRNA as revealed by membrane hybridization (Fig. 3), the signal was not detected (Fig. 6). Possible reasons for this observation include the low Td value of probe S-∗-Dsb-0804-a-A-18 and the length of the transcribed target RNA used in the enrichment and environmental samples.
The Td value of probe S-∗-Dsb-0804-a-A-18 was the lowest of all the probes used in this study (Table 1). This means a lower affinity between the probe and target RNA. In addition, probe specificity on the DNA microarray was checked by using short fragments (338 to 927, E. coli numbering) (Fig. 5), whereas almost full-length transcribed 16S rRNA (11 to 1512, E. coli numbering) was used to compare with native intact 16S rRNA when we applied the DNA microarray to enrichment and environmental samples (Fig. 6). We suggest that the fragmentation of long transcribed 16S rRNA molecules may have been less efficient than for short fragments and that the target site of probe S-∗-Dsb-0804-a-A-18 may have been more difficult to fragment than other regions due to its three-dimensional structure. We need further optimization to design good probes for DNA microarray hybridization.
The unambiguous analysis of natural systems requires an understanding of mismatch discrimination. In particular, single-mismatch discrimination is often difficult to achieve (30, 48, 55, 64). Both probes S-∗-Tsrda-0644-a-A-19 and S-∗-Tsrda-0641-a-A-20 contain one U-G mismatch to D. variabilis located 3 and 6 bases from the 5′ end of the probes, respectively (Table 1). Although the target with one mismatch could not be completely eliminated, the melting profile of the one-mismatch duplex differed from that of the perfect-match duplex. The Td values for the perfect-match duplexes were 42.0 and 44.0°C, 3.5 and 5.5°C higher than those for the one-mismatch duplexes, respectively. This could provide a basis to distinguish between perfect-match and single-mismatch duplexes by further optimization of hybridization and washing condition (e.g., formamide and salt concentration in buffer) (57). Alternatively, monitoring the dissociation kinetics of all array elements simultaneously could also provide improved discrimination.
DNA microarrays have been used mainly as a tool to determine microbial species (7, 15, 27, 34) and to profile gene expression of microorganisms (60). There are few studies of the application of DNA microarrays to analysis of bacterial communities (12, 54). A general objective of our study was to evaluate, in a comparative framework, alternative methods of community analysis. The same microbial communities were characterized by using DGGE, membrane hybridization, and DNA microarray hybridization. All methods were consistent, showing that T-1, E-1, and E-2 existed in our enrichment cultures (Fig. 1, 3, and 6), even though quantification of relative abundance is not possible with DGGE and has yet to be established for DNA microarray hybridization.
Although DNA microarray technology is a rapid and high-throughput format for nucleic acid hybridization, this technique still has limitations. However, technical issues such as sample preparation, sensitivity, and one-mismatch discrimination will be resolved in the near future. For example, the importance of the location of mismatches in probe-target duplexes has been clarified by Urakawa et al. (58). This should be helpful information for the design of probes for microarray hybridization. This study is the first to compare the novel microarray technique and other methods, and it demonstrates that the target populations were qualitatively detected by microarray hybridization. We anticipate that the DNA microarray technology used in this study will ultimately provide a rapid and high-throughput platform for microbial population analysis.
Acknowledgments
We are grateful to Andreas Schramm for critical reading of the manuscript and for valuable discussions. We also thank G. Yershov, A. Kukhtin, and A. Gemmell for their efforts in manufacturing the microchip. We thank S. Takii (Tokyo Metropolitan University) for encouragement and support.
This work was supported by grants from the New Energy and Industrial Technology Development Organization (NEDO) and the Ministry of Education, Culture, Sports, Science and Technology, Japan (no. 12440219) to M.F. and grants from NASA and DARPA to D.A.S.
REFERENCES
- 1.Aeckersberg, F., F. Bak, and F. Widdel. 1991. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156:5-14. [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R. I., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann, R. I., W. Ludwig., and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barsky, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker, S., P. Boger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beller, H. R., D. Grbic-Galic, and M. Reinhard. 1992. Microbial degradation of toluene under sulfate-reducing conditions and the influence of iron on the process. Appl. Environ. Microbiol. 58:786-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beller, H. R., M. Reinhard, and D. Grbic-Galic. 1992. Metabolic by-products of anaerobic toluene degradation by sulfate-reducing enrichment cultures. Appl. Environ. Microbiol. 58:3192-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beller, H. R., A. M. Spormann, P. K. Sharma, J. R. Cole, and M. Reinhard. 1996. Isolation and characterization of a novel toluene-degrading, sulfate-reducing bacterium. Appl. Environ. Microbiol. 62:1188-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borneman, J., M. Chrobak, G. Della Vedova, A. Figueroa, and T. Jiang. 2001. Probe selection algorithms with applications in the analysis of microbial communities. Bioinformatics 17(Suppl. 1):S39-48. [DOI] [PubMed] [Google Scholar]
- 13.Britton, L. N. 1984. Microbial degradation of aliphatic hydrocarbons, p. 89-129. In T. D. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, New York, N.Y.
- 14.Chee, M., R. Yang, E. Hubbell, A. Berno, X. C. Huang, D. Stern, J. Winkler, D. J. Lockhart, M. S. Morris, and S. P. A. Fodor. 1996. Accessing genetic information with high-density DNA arrays. Science 274:610-614. [DOI] [PubMed] [Google Scholar]
- 15.Cho, J. C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 17.Cord-Ruwisch, R. 1985. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4:33-36. [Google Scholar]
- 18.Devereux, R., M. Delaney, F. Widdel, and D. A. Stahl. 1989. Natural relationships among sulfate-reducing eubacteria. J. Bacteriol. 171:6689-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 20.Devereux, R., M. R. Winfrey, J. Winfrey, and D. A. Stahl. 1996. Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment. Syst. FEMS Microbiol. Ecol. 20:23-31. [Google Scholar]
- 21.El-Fantroussi, S. 2000. Enrichment and molecular characterization of a bacterial culture that degrades methoxy-methyl urea herbicides and their aniline derivatives. Appl. Environ. Microbiol. 66:5110-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry, N. K., J. K. Fredrickson, S. Fishbain, M. Wagner, and D. A. Stahl. 1997. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl. Environ. Microbiol. 63:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galushko,A., D. Minz, B. Schink, and F. Widdel. 1999. Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulfate-reducing bacterium. Environ. Microbiol. 1:415-420. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, D. T., and V. Subramanian. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-294. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, New York, N.Y.
- 26.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 27.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harms, G., K. Zengler, R. Rabus, F. Aeckersberg, D. Minz, R. Rosselló-Mora, and F. Widdel. 1999. Anaerobic oxidation of o-xylene, m-xylene, and homologous alkylbenzenes by new types of sulfate-reducing bacteria. Appl. Environ. Microbiol. 65:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hristova, K. R., M. Mau, D. Zheng, R. I. Aminov, H. R. Gaskins, and L. Raskin. 2000. Desulfotomaculum genus- and subgenus-specific 16S rRNA hybridization probes for environmental studies. Environ. Microbiol. 2:143-159. [DOI] [PubMed] [Google Scholar]
- 31.Jørgensen, B. B. 1982. Mineralization of organic matter in the sea bed — the role of sulphate reduction. Nature 296:643-645. [Google Scholar]
- 32.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khrapko, K. R., P. Lysov Yu, A. A. Khorlin, I. B. Ivanov, G. M. Yershov, S. K. Vasilenko, V. L. Florentiev, and A. D. Mirzabekov. 1991. A method for DNA sequencing by hybridization with oligonucleotide matrix. DNA Seq. 1:375-388. [DOI] [PubMed] [Google Scholar]
- 34.Liu, W. T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a nonequilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 35.Maidak, B. L., N. Larsen, M. J. McCaughey, R. Overbeek, G. J. Olsen, K. Fogel, J. Blandy, and C. R. Woese. 1994. The Ribosomal Database Project. Nucleic Acids Res. 22:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMahon, K. D., D. A. Stahl, and L. Raskin. 1998. A comparison of the use of in vitro-transcribed and native RNA for the quantification of microorganisms in the environment. Microb. Ecol. 36:362-371. [DOI] [PubMed] [Google Scholar]
- 37.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran, M. A., V. L. Torsvik, T. Torsvik, and R. E. Hodson. 1993. Direct extraction and purification of rRNA for ecological studies. Appl. Environ. Microbiol. 59:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muyzer, G., S. Hottenträger, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA — a new molecular approach to analyze the genetic diversity of mixed microbial communities, p. 3.4.4/1-3.4.4/23. In A. D. L. Akkermans, J. D. Van Elsas, and F. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Nakagawa, T., S. Sato, Y. Yamamoto, and M. Fukui. 2002. Successive changes in community structure of an ethylbenzene-degrading sulfate-reducing consortium. Water Res., in press. [DOI] [PubMed]
- 42.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 43.Proudnikov, D., and A. Mirzabekov. 1996. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 24:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purdy, K. J., T. M. Embley, S. Takii, and D. B. Nedwell. 1996. Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl. Environ. Microbiol. 62:3905-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purdy, K. J., D. B. Nedwell, T. M. Embley, and S. Takii. 1997. Use of 16S rRNA-targeted oligonucleotide probes to investigate the occurrence and selection of sulfate-reducing bacteria in response to nutrient addition to sediment slurry microcosms from a Japanese estuary. FEMS Microbiol. Ecol. 24:221-234. [Google Scholar]
- 46.Rabus, R., R. Nordhaus, W. Ludwig, and F. Widdel. 1993. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl. Environ. Microbiol. 59:1444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabus, R., M. Fukui, H. Wilkes, and F. Widdel. 1996. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl. Environ. Microbiol. 62:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rueter, P., R. Rabus, H. Wilkes, F. Aeckersberg, F. A. Rainey, H. W. Jannasch, and F. Widdel. 1994. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulphate-reducing bacteria. Nature 372:455-458. [DOI] [PubMed] [Google Scholar]
- 52.Sato, S., A. Matsumura, Y. Urushigawa, M. Metwally, and S. Al-Muzaini. 1998. Type analysis and mutagenicity of petroleum oil extracted from sediment and soil samples in Kuwait. Environ. Int. 24:67-76. [Google Scholar]
- 53.Service, R. F. 1998. Microchip arrays put DNA on the spot. Science 282:396-399. [DOI] [PubMed] [Google Scholar]
- 54.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorhaug, A. 1992. The environmental future of Kuwait, p. 69-72. In A. K. Al-Shatti and J. M. Harrington (ed.), Proceedings of the International Symposium on the Environmental and Health Impacts of the Kuwait Oil Fires. University of Birmingham, Edgbaston, England.
- 57.Tilssen, P. 1993. Hybridization with nucleic acid probes, part I, p. 35-41. Elsevier Science, Amsterdam, The Netherlands.
- 58.Urakawa, U., P. A. Noble, S. El Fantroussi, J. J. Kelly, and D. A. Stahl. 2002. Single-base-pair discrimination of terminal mismatches by by using oligonucleotide microarrays and neural network analysis. Appl. Environ. Microbiol. 68:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, D. G., J. B. Fan, C. J. Siao, A. Berno, P. Young, R. Sapolsky, G. Ghandour, N. Perkins, E. Winchester, J. Spencer, L. Kruglyak, L. Stein, L. Hsie, T. Topaloglou, E. Hubbell, E. Robinson, M. Mittmann, M. S. Morris, N. Shen, D. Kilburn, J. Rioux, C. Nusbaum, S. Rozen, T. J. Hudson, R. Lipshutz, M. Chee, and E. S. Lander. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077-1082. [DOI] [PubMed] [Google Scholar]
- 60.Wei, Y., J. M. Lee, D. R. Smulski, and R. LaRossa. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, M. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. IV. Springer-Verlag, New York, N.Y.
- 62.Wilson, K. 1990. Miniprep of bacterial genomic DNA. p. 2.4.1-2.4.2 In F. M. Ausubel et al. (ed.), Short protocols in molecular biology, 2nd ed. John Wiley and Sons, New York, N.Y.
- 63.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probe for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]