Abstract
By the use of Escherichia coli DH1 harboring cphA from Synechocystis sp. strain PCC6803, large-scale production of cyanophycin at 30- and 500-liter culture volumes was established. Transcription of cphA was controlled by the thermosensitive cI857 repressor, which enabled induction of cphA by a simple temperature shift in the culture fluid. Maximum cyanophycin cell content of up to 24% (wt/wt) of cellular dry matter was obtained by induction in the early exponential growth phase and cultivation of the cells in terrific broth complex medium. Synthesis of cyanophycin was found to be strongly dependent on the presence of complex components, and in mineral salts medium the cells synthesized and accumulated cyanophycin only if Casamino Acids were added. Cultivations were done at the 500-liter scale, allowing the provision of cell mass for the preparation of cyanophycin at the kilogram scale. Isolation of cyanophycin was achieved by a new acid extraction procedure which allowed large-scale purification of the polyamide from whole cells.
Cyanophycin (multiarginyl-poly[l-aspartic acid]) is a protein-like polymer which consists of equimolar amounts of aspartic acid and arginine arranged as a polyaspartic acid backbone, to which arginine residues are linked to the β-carboxyl group of each aspartate by its α-amino group (24). In nature cyanophycin is produced by most but not all cyanobacteria as a temporary nitrogen reserve material during the transition of cells from the exponential phase to the stationary phase (15). At neutral pH and physiological ionic strength, cyanophycin is insoluble and deposited in the cytoplasm as membraneless granules (13).
Cyanophycin is of biotechnological interest because purified cyanophycin can be chemically converted into a polymer with a reduced arginine content (11), which might be used like polyaspartic acid as a biodegradable substitute for synthetic polyacrylate in various technical processes (22). In addition, cyanophycin might also be of interest for other applications if the unknown physical and material properties of this polymer are revealed. Because of the low polymer content, and the slow growth of cyanobacteria, resulting in only low cell densities, cyanobacteria are not suitable for large-scale production of cyanophycin (3), and sufficient amounts of cyanophycin were hitherto not available.
The polymerization reaction is catalyzed by only one enzyme, which is referred to as cyanophycin synthetase (CphA) (27). The cphA genes from Anabaena variabilis ATCC 29413, Anabaena sp. strain PCC7120, Synechocystis sp. strain PCC6803, Synechocystis sp. strain PCC6308, Synechococcus elongatus, and Synechococcus sp. strain MA19 were cloned and expressed in Escherichia coli (1, 4, 7, 19, 27). More recently, heterologous expression of cphA was also demonstrated at a small scale in recombinant strains of Ralstonia eutropha, Corynebacterium glutamicum, and Pseudomonas putida (3). Whereas in cyanobacteria the molecular mass of the polymer strands ranged from 25 to 100 kDa (23), the polymer from recombinant strains harboring cphA as well as in vitro-synthesized polymer exhibited a much lower range (25 to 30 kDa) and polydispersity. Furthermore, it was found that the polymer isolated from recombinant strains contained lysine as an additional amino acid constituent (3, 27).
Due to the wide knowledge of its metabolism and available genetic tools, E. coli is one of the most commonly used bacterial hosts for the production of recombinant proteins (14). Several expression systems have been developed for technical-scale production of recombinant proteins in E. coli based on the regulated trp, lac, or lambda PL promoter (9).
In this study, we describe the cultivation of recombinant E. coli strains harboring cphA from Synechocystis sp. strain PCC6803 at the 500-liter scale for the production of cyanophycin. Since the previously described method for the purification of cyanophycin (24) is not applicable to a large-scale, a simplified method for isolation of the polymer at the technical scale was elaborated.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and preparation of cell extracts for analytical purposes.
The bacterial strains and plasmids used in this study are listed in Table 1. The E. coli strains were cultivated at 30 or 37°C in Erlenmeyer flasks in complex media (Luria-Bertani [LB] or terrific broth [TB]) (19) or in a mineral salts medium (18) in the presence of 100 μg of ampicillin or 100 μg of chloramphenicol ml−1as indicated in the text. The flasks were incubated on a Pilotshake RC-4/6-W horizontal shaker (Kühner AG, Birsfelden, Switzerland) at 150 rpm and at an amplitude of 5 cm.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| K-12 | DSM 498 | |
| DH1 | F−endA1 gyrA96 hsdR17 (rK− mK+) recA1 supE thi-1 λ− | 8 |
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 ΔlacU169 (φ80 lacZΔM15) | 8 |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE λ− Δ(lac-proAB) [F′ traΔ36 proAB lacIqZΔM15] | 26 |
| SMH50 | supE44 endA1 hsdR17 gyrA96 thi-1 relA1 Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 17 |
| TG1 | Δ(lac-pro) supE thiE hsdD5/F′ [traD36 proA+B+ lacIqlacZΔM15] | 6 |
| Top10 | F−araD139 Δ(ara leu)7697 ΔlacX74 galU galK rpsL deoR φ80dlacZΔM15 endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | Invitrogen (San Diego, Calif.) |
| Plasmids | ||
| pMa/c5-914 | Apr Cmrc1857ts λ PL/PR, translational initiation region | Personal gift from T. Wagnera |
| pMa/c5-914::cphA | pMa/c5-914 carrying 2.6-kb PCR product from Synechocystis sp. strain PCC6803 genomic DNA harboring cphA | This study |
Institut für Molekulare Genetik der Georg-August-Universität, Göttingen, Germany.
Growth was monitored by measuring the turbidity at 600 nm. Cells were harvested in the early stationary phase by centrifugation (10 min, 2,800 × g, 4°C), washed twice with 50 mM Tris-HCl buffer (pH 8.2), and resuspended with 2 ml of buffer per g of fresh cell mass. The cells were disintegrated by sonification for 1 min per ml of cell suspension by using a Sonoplus sonifier (Bandelin Electronic, Berlin, Germany). Broken cell debris and membrane particles were removed by centrifugation (1 h, 100,000 × g, 4°C). The supernatant was desalted on NAP5 columns (Pharmacia Biotech, Freiburg, Germany) and served as the soluble cell fraction.
Cultivation at the 30-liter scale.
A Biostat UD30 stainless steel reactor (B. Braun Biotech International, Melsungen, Germany), which had a total volume of 42 liters (28 cm inner diameter and 71 cm height) and a d/D value relation (relation of stirrer diameter to vessel diameter) of 0.375, was used for cultivations at the 30-liter scale. This bioreactor was equipped with three stirrers, each containing six paddles and a Funda-Foam mechanical foam destroyer (B. Braun Biotech International, Melsungen, Germany). In addition, ports were used for sterilizable probes to measure dissolved oxygen (pO2) (model 25; Mettler Toledo GmbH, Steinbach, Switzerland), pH (model Pa/25; Mettler-Toledo GmbH), foam (model L300/Rd. 28; B. Braun Biotech International), temperature (pt 100 electrode; M. K. Juchheim GmbH, Fulda, Germany), and optical density at 850 nm (model CT6; Sentex/Monitek Technology Inc.). The operations were controlled and recorded by a digital control unit in combination with the MFCS/win software package (B. Braun Biotech International).
Carbon dioxide and oxygen concentrations in the spent gas leaving the bioreactor were measured with a URAS 10 P NDIR spectrophotometer (Mannesmann, Hartmann and Braun, Frankfurt, Germany) or a Magnos 6 G oxygen analyzer (Mannesmann, Hartmann and Braun), respectively. Cultivations were done at 30 or 37°C and at a pO2 range of 0 to 100% saturation in the medium, which was controlled by agitation rates between 100 and 800 rpm and aeration rates of 0.4 and 1.0 vvm (volume per volume × minute). The pH in the medium was held between 7.0 and 7.2 by controlled addition of 4 N HCl or NaOH. Foam was removed by a mechanical foam destroyer; if this was not sufficient, the antifoam agent Silikon Antischaum Emulsion SLE (Wacker, Darwin Vertriebs GmbH, Ottobrunn, Germany) was added. Small samples withdrawn from the culture fluid for analytical purposes were separated into a cell pellet and a cell-free supernatant by 15 min of centrifugation at 3,500 × g.
Cultivation at the 500-liter scale.
Cultivations at the 500-liter scale were performed with a Biostat D650 stainless steel bioreactor (B. Braun Biotech International), which had a total volume of 650 liters (64 cm inner diameter and 198 cm height) and the same d/D value as the Biostat UD30 described above. All other equipment was as described above for the Biostat UD30 bioreactor. Fermentations were done as described above or in the Results section; however, the corresponding tip speeds of the stirrer blades were between 1.3 and 5.0 m s−1, with stirrer speeds between 100 and 400 rpm.
Cell harvest from 30- and 500-liter scale cultivations.
Cells were harvested by centrifugation at 4°C in a CEPA type Z41 or type Z61 continuous centrifuge (Carl Padberg Zentrifugenbau GmbH, Lahr, Germany), respectively.
Analysis of ammonium, acetate, and glycerol.
The concentration of ammonium was determined in cell-free supernatants by employing a gas-sensitive ammonium electrode (type 152303000; Mettler Toledo GmbH) or ammonium test bars (Merck AG, Darmstadt, Germany). The concentrations of glycerol and acetic acid in cell-free supernatants were measured enzymatically by using the corresponding test kits (Roche Diagnostics GmbH, Mannheim, Germany).
Cyanophycin synthetase assay.
The enzyme activity of the cyanophycin synthetase was measured in the soluble cell fraction by employing a radiometric enzyme assay as described by Aboulmagd et al. (2).
Small-scale purification of cyanophycin.
Cyanophycin was isolated in small scale from the recombinant cells by the procedure described by Simon and Weathers (24) for Anabaena cylindrica.
Large-scale isolation of cyanophycin from bulk cell mass.
For isolation of cyanophycin from cells obtained from cultivations at the 500-liter scale, a simplified acid extraction procedure was developed. The cellular dry mass was suspended in tap water to give a final concentration of 0.1 g ml−1. Under agitation and pH control, the suspension was acidified to pH 1 by addition of concentrated HCl. After being stirred for 6 h at room temperature, the suspension was centrifuged for 1 h at 3,500 × g. While the supernatant was kept for further use (see below), the cell pellet was resuspended in 0.1 N HCl and stirred for 1 h. After centrifugation, the pellet was discarded, and the supernatant was added to the supernatant obtained from the first centrifugation. The pH of the combined supernatants was then adjusted to 7 to 7.5 by addition of solid or 10 N NaOH. After centrifugation, the pellet, which contained crude cyanophycin, was washed once with water, resuspended in 0.1 N HCl, and agitated for 1 h. The suspension was centrifuged, and the supernatant was neutralized by addition of NaOH. After centrifugation, the pellet was washed twice with water, frozen at −30°C, and lyophilized in a BETA 1-16 type freeze-dryer (Christ Gefriertrocknungsanlagen, Osterode, Germany).
Determination of amino acid constituents of cyanophycin.
The amino acid composition of cyanophycin was determined by high-pressure liquid chromatography analysis of ortho-phthaldialdehyde (OPA)-modified amino acids. Purified cyanophycin was treated for 15 h at 95°C with 6 N HCl. After lyophilization, the material was dissolved in starting eluent (37 mM sodium acetate [pH 7.0], 26% [vol/vol] methanol). The solution was clarified by 5 min of centrifugation at 10,000 × g. To 460 μl of this solution, 200 μl of 0.5 M sodium borate (pH 9.5) and 100 μl of OPA reagent (100 mg of ortho-phthaldialdehyde, 9 ml of methanol, 1 ml of 0.5 M sodium borate [pH 9.5], 100 μl of 2-mercaptoethanol) were added. The reaction was stopped after exactly 200 s by addition of 60 μl of 0.75 N HCl. To 100 μl of this reaction mixture was added 400 μl of starting eluent. Then 20 μl of this solution was applied to a reversed-phase column (0.46 by 12.5 cm; RP18 Techsphere ODS-2; Kontron Instruments, Neufahrn, Germany) equilibrated with starting eluent. OPA-amino acids were eluted with a methanol gradient (26 to 75%, vol/vol) at 40°C at a flow rate of 1.0 ml min−1 and monitored fluorimetrically at 330 and 450 nm (excitation and emission) with a model 1046A fluorescence detector (Hewlett Packard, Waldbronn, Germany). Calibration was done with chromatographically pure amino acids (Kollektion AS-10 from Serva Feinbiochemica, Heidelberg, Germany).
Determination of carbohydrate, protein, and nucleic acid impurities and water content in cyanophycin.
Quantification of carbohydrate was done by following the colorimetric reaction of cyanophycin samples with Anthrone reagent with glucose as the standard as described by Hanson and Philips (10). Protein and nucleic acid content was determined UV spectrophotometrically in clear solutions of cyanophycin preparations in 100 mM HCl by using the following equations: (i) protein, milligrams per milliliter = 1.55 A280 − 0.76 A260 (25), and (ii) nucleic acids, milligrams per milliliter = 0.05 A260 (20). The water content of freeze-dried cyanophycin was determined by the coulometric Karl Fischer method (16).
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in 11.5% (wt/vol) polyacrylamide gels as described by Laemmli (12). Proteins and cyanophycin were stained with Serva Blue R. Protein concentration was determined by the procedure of Bradford (5).
RESULTS
Development of a stable expression system.
The cyanophycin synthetase gene (cphA) from Synechocystis sp. strain PCC6803 was inserted into the expression vector pMa/c5-914. In the resulting plasmid, pMa/c5-914::cphA, transcription was controlled by the λ PL promoter and the temperature-sensitive cI857 repressor. Expression of cphA during large-scale cultivation was achieved by a shift of the cultivation temperature from 30 to 37°C. An increase to higher temperatures (i.e., 42°C), which is usually recommended in laboratory protocols for this induction system (21), was not used in order to reduce heat shock response.
Several E. coli strains (Table 1) were transformed with pMa/c5-914::cphA and tested for stability. Due to the light-scattering cyanophycin inclusions, recombinant strains expressing cphA formed characteristic whitish colonies which could be easily distinguished from the pale wild-type colonies. Whereas transformants of E. coli DH1 produced only whitish colonies even after several transfers in new complex medium under producing conditions (37°C), others, including E. coli strains Top10, DH5α, JM109, and SMH50, also generated various portions of wild-type colonies, indicating plasmid instability. Therefore, E. coli DH1(pMa/c5-914::cphA) was used for cultivations at the larger scale.
Cultivation of E. coli DH1(pMa/c5-914::cphA) at the 30-liter scale in mineral salts medium.
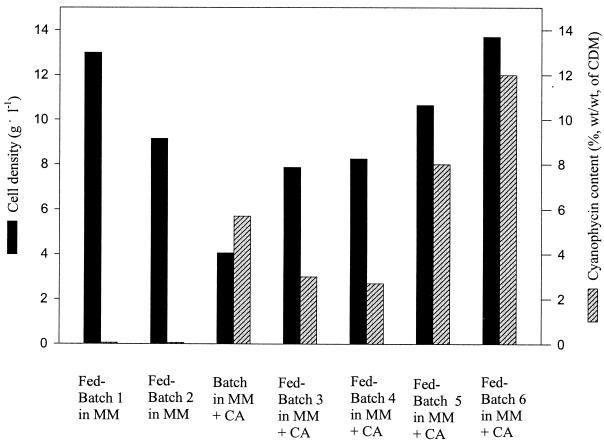
Various batch or fed-batch cultivations of E. coli DH1(pMa/c5-914::cphA) in mineral salts medium with glycerol or glucose as the carbon sources and in the presence or absence of Casamino Acids were done. Only trace amounts of cyanophycin were produced in medium without additional Casamino Acids (see fed batches 1 and 2 in Fig. 1). In order to provide sufficient amino acids as precursors for biosynthesis of cyanophycin, further cultivations were performed after addition of 1% (wt/vol) Casamino Acids (Fig. 1, batch cultivation and fed batches 3 to 6).
FIG. 1.
Different cultivations of E. coli DH1(pMa/c5-914::cphA) at the 30-liter scale in mineral salts medium (MM). Fed-batch cultivations 3 to 6 were done after addition of 1% (wt/vol) Casamino Acids (CA), as indicated. Glucose was used as the carbon source at an initial concentration of 2.5% (wt/wt) in fed batches 1 and 3; all other cultivations were performed with glycerol. The initial concentration of glycerol was 4% (wt/vol) except in fed batch 6, where 2% (wt/vol) was used. Induction was achieved by a temperature shift at the exponential growth phase except in the batch cultivation and fed batch 4, where the temperature was adjusted to 37°C for the beginning of cultivation. CDM, cellular dry matter.
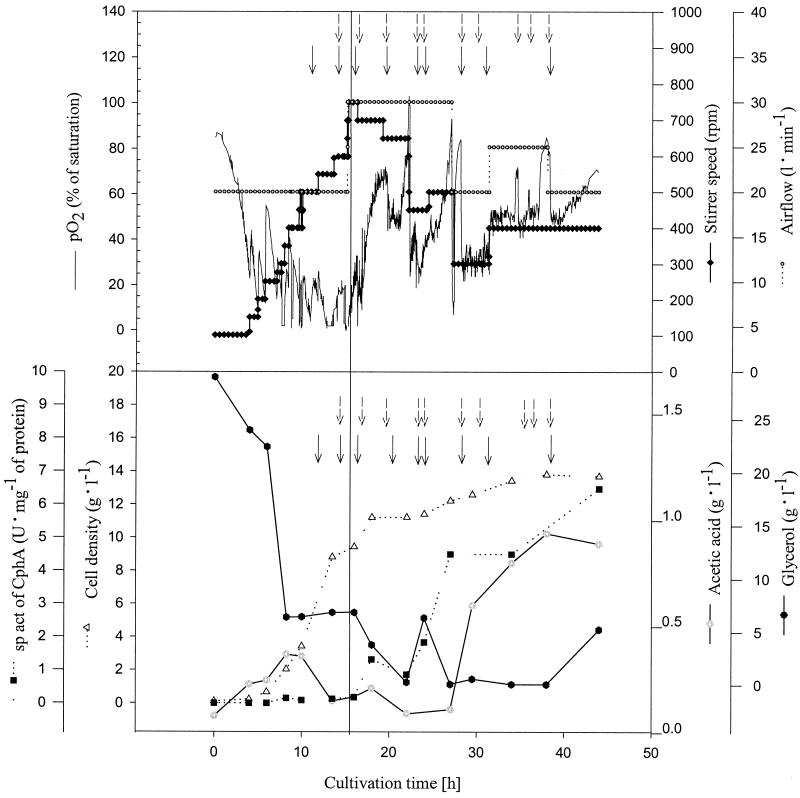
Under optimized conditions (i.e., temperature shift at early exponential growth phase and reduced initial concentration of glycerol), the cells accumulated cyanophycin up to 12% (wt/wt) of the cellular dry matter (CDM). The time course of fed-batch 6, which yielded the highest cell density and cyanophycin content, is shown in Fig. 2. After 44 h, a cell density of 12 g of CDM liter−1 was obtained with a cyanophycin content of 12% (wt/wt). The highest specific activity of 6.5 U (mg of protein)−1 of the cyanophycin synthetase was found at the end of the fermentation (Fig. 2). As can be seen from the time course of the specific activity, no significant expression of cphA occurred until the temperature was raised to provoke induction, illustrating the suitability of the expression system for controlled cyanophycin production. Acetic acid accumulated up to 1 g liter−1 in the cell-free supernatants at the end of the cultivation.
FIG. 2.
Fed-batch fermentation of E. coli DH1(pMa/c5-914::cphA) at the 30-liter scale in semidefined medium. The cultivation was done in a 30-liter stirred-tank reactor containing 26 liters of mineral salts medium with 2% (vol/vol) glycerol, 1 g of ammonium sulfate liter−1, and 1% (vol/vol) Casamino Acids at the beginning of the fermentation. The cultivation parameters were pH 7.0, aeration at 0.8 to 1.0 vvm, and agitation at 100 to 750 rpm. Aeration and agitation were adjusted according to the oxygen demand of the culture. The temperature was raised from 30 to 37°C after 15 h of cultivation time for induction of cphA, as indicated by the solid line. During the fermentation, 0.25 to 0.5 g of glycerol per liter of medium (dashed arrows) and 0.5 g of NH4Cl liter−1 (solid arrows) were added at the time points indicated. Activity of cyanophycin synthetase was determined as described in Materials and Methods. The availability of the carbon source was estimated by monitoring the CO2 content in the exhaust gas and the dissolved oxygen (pO2) level in the medium.
Cultivation of E. coli DH1(pMa/c5-914::cphA) at the 30-liter scale in complex media.
In order to increase both cell density and cyanophycin content, E. coli DH1(pMa/c5-914::cphA) was grown as a batch culture in TB complex medium. The temperature shift for induction of cphA was done at various periods during the time course of the cultivation. The time point of induction was very important for the final amount of product. Induction in stationary phase resulted in a cell density of 7.9 g liter−1 and a cyanophycin content of only 3.3% (wt/wt), whereas a temperature shift in the late exponential phase led to a content of 7.3% (wt/wt) and a cell density of 8.0 g liter−1. Best yields were obtained when the temperature shift was done at the beginning of the exponential growth phase, resulting in cyanophycin contents between 21.0 and 24.0% (wt/wt). The cell densities varied between 6.7 and 8.3 g liter−1 in three independent experiments.
Cultivation of E. coli DH1(pMa/c5-914::cphA) at the 500-liter scale.
Due to the good stability of the expression system in E. coli DH1 in contrast to other strains tested (Table 1), a scale-up to the 500-liter culture volume was performed. Upscaling was facilitated by the similarity of the 30-liter bioreactor to the 500-liter bioreactor used in this study with regard to construction and diameter-to-height ratio as well as stirrer diameter-to-vessel diameter ratio. In contrast to the cultivations at the 30-liter scale, airflow was kept at 0.15 vvm to avoid foam formation and stirrer speed was adjusted manually to avoid insufficient supply of oxygen at the exponential growth phase.
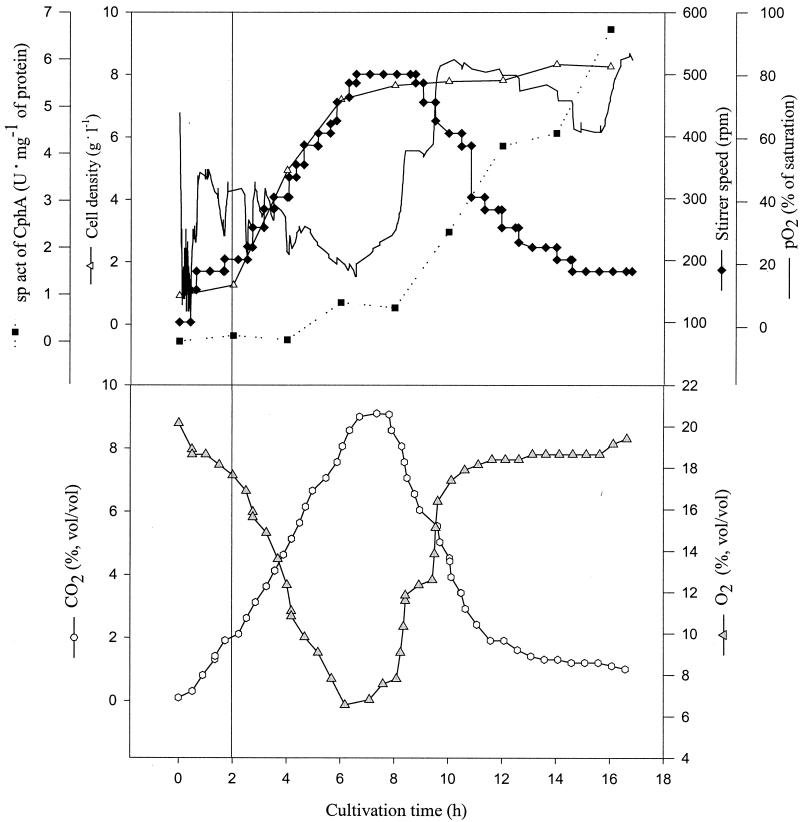
Four cultivation experiments were done at the 500-liter scale. With the exception of one cultivation experiment, reproducible cell densities between 6.6 and 7.7 g liter−1 with cyanophycin contents of between 17 and 21% (wt/wt) were obtained. A fourth cultivation experiment at this scale gave a slightly lower cell density and a cyanophycin content of only 11% (wt/wt). The time course of one representative fermentation at the 500-liter scale is shown in Fig. 3. After 16 h, a cell density of 7.7 g of CDM liter−1 was obtained. Approximately 3.6 kg of dry cellular mass was obtained after centrifugation and freeze-drying of the cells with a cyanophycin content of 21% (wt/wt). Thus, the cells obtained from a single cultivation experiment contained a total of 750 g of cyanophycin.
FIG. 3.
Batch fermentation of E. coli DH1(pMa/c5-914::cphA) at the 500-liter scale in complex medium. The cultivation was done in a 500-liter stirred-tank reactor containing 430 liters of TB medium. Fermentation parameters were pH 7.0, aeration at 0.15 vvm, and agitation at 100 to 500 rpm. The temperature was raised from 30 to 37°C after 2 h of cultivation, as indicated by the solid line. Activity of cyanophycin synthetase was determined as described in Materials and Methods. O2 and CO2, oxygen and carbon dioxide concentrations, respectively, in the exhaust gas.
Large-scale purification of cyanophycin by acid extraction of whole cells.
The protocol for the purification of cyanophycin at an analytical scale as described by Simon and Weathers (24) starts with the disruption of the cells at neutral pH by ultrasonification in the presence of Triton X-100, which is followed by collection of the cyanophycin-containing debris by centrifugation and five washing steps to remove some other cellular components and the detergent. Purification of the crude cyanophycin-containing pellet is then achieved by (i) dissolving the pellet at pH 1.0, (ii) high-speed centrifugation, (iii) precipitation of cyanophycin in the supernatant at neutral pH, and (iv) high-speed centrifugation. Steps i to iv are usually repeated once. After being washed with water, the product is usually lyophilized.
The simplicity of the pH-dependent solubilization-precipitation steps on the one hand was promising for upscaling this part of the purification protocol. On the other hand, simple upscaling of the first part of the protocol is complicated by the time-consuming and laborious cell disruption and washing steps. Therefore, attempts were undertaken to develop a simple and time-saving modification of the method of Simon and Weathers (24).
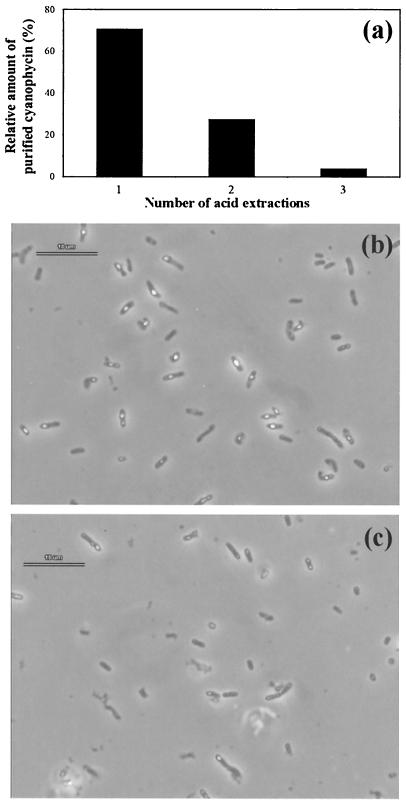
Cell disruption by ultrasonification could be omitted by stirring whole cells in diluted HCl at pH 1.0 for 6 h at room temperature. Higher pHs or shorter incubation times reduced the final yield of cyanophycin (data not shown). Though the cells appeared almost intact after such treatment, as evaluated by microscopic inspection, the occurrence of intracellular cyanophycin granules was greatly reduced (Fig. 4). This indicated that an exogenous pH of 1 both dissolved cyanophycin inside the cells and extracted the dissolved polymer from the cells. Approximately 69% of the cyanophycin was extracted from the cells by this first step. The amount of polymer remaining in the cells could be further reduced by repeating the acid extraction step. After the second acid extraction, approximately 97% of the cyanophycin could be isolated, and only 3% remained in the cells (Fig. 4c). Therefore, two subsequent acid extractions are sufficient and allow an economically feasible isolation of the polymers (see Materials and Methods). The acid extraction of whole cells made not only the mechanical cell disruption unnecessary, but also several centrifugation steps which have to be done according to the conventional procedure to collect and wash the crude cyanophycin (24). Attempts to replace all other centrifugations by filtration through cotton wool, mull, and paper or glass fiber filters resulted in insufficient separations (details not shown).
FIG. 4.
Effect of the number of extraction steps on the release of cyanophycin from cells. (a) Relative amounts of purified cyanophycin obtained after different numbers of acid extractions of the cells. (b) Cells before starting acid treatment. (c) Cells after first acid extraction. Prior to microscopic inspection, the cell-containing samples were neutralized by the addition of NaOH. Bars (b and c), 10 μm.
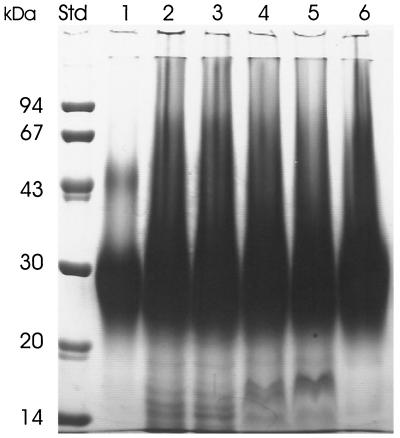
As can be seen in Fig. 5, the molecular size distribution and polydispersity of the cyanophycin prepared by the new acid extraction procedure were comparable to those of cyanophycin obtained by the method of Simon and Weathers (24). In addition, both cyanophycin preparations were visually almost free of contaminating proteins.
FIG. 5.
SDS-PAGE of cyanophycin purified by different purification protocols. Lanes 1 to 6 each contained 75 μg of SDS-denatured cyanophycin preparations. Lane 1, purified by the method of Simon and Weathers (24); lanes 2 to 6, purified by the acid extraction procedure; lanes 2 to 5, different lots of preparations obtained after only one acidification-neutralization step; lane 6, preparation of cyanophycin obtained after two acidification-precipitation steps as described in Materials and Methods. The sizes of molecular mass standard proteins (lane Std) are provided at the left. Cyanophycin and protein bands were visualized by staining with Serva Blue R.
A quantitative comparison of the yield and the purity of the product from both the conventional and the acid extraction procedure is given in Table 2. The acid extraction procedure obviously yields less cyanophycin with a lower content of other cellular components (i.e., protein and nucleic acids) compared to the established method. Both effects are probably due to the elimination of the cell disruption step. Ultrasonic disruption of the cells releases not only the entire amount of cyanophycin from the cells but also various undesired components. In addition, the molar ratio of the three amino acid constituents of both preparation differed in the arginine content, which is slightly lower after acid extraction. This might be due to the long incubation under acidic conditions, which is known to cause some release of arginyl residues from the polymer (24). Traces of long-chain fatty acids were detected by gas chromatography-mass spectrometry analysis of the product after acid extraction (data not shown), whereas no fatty acids were found after the procedure of Simon and Weathers (24), which is probably due to the additional Triton treatment in the latter protocol.
TABLE 2.
Quantitative analysis of cyanophycin preparations obtained by the acid extraction procedure or by the method described by Simon and Weathers (24)a
| Parameter (units) | Acid extraction | Simon and Weathers procedure |
|---|---|---|
| Relative cyanophycin yieldb (%) | 75 ± 5 | 100 |
| Molar ratio, Asp:Arg:Lys | 1.0:0.47 ± 0.05:0.32 ± 0.03 | 1.0:0.62 ± 0.06:0.31 ± 0.03 |
| Asp + Arg + Lys content (%, wt/wt) | 81.2 ± 5.0 | 87.7 ± 6.0 |
| Water (%, wt/wt) | 10.6 ± 1.0 | 9.5 ± 0.96 |
| Proteins (%, wt/wt) | 1.3 ± 0.3 | 3.7 ± 0.5 |
| Nucleic acids (%, wt/wt) | 0.10 ± 0.02 | 0.3 ± 0.02 |
| Carbohydrates (%, wt/wt) | 0.03 ± 0.01 | 0.03 ± 0.01 |
The cell dry mass for both purifications was derived from the same cultivation. Data represent the means of three determinations ± SD.
The amount of cyanophycin which was obtained by the established method of Simon and Weathers (24) was set to 100%.
DISCUSSION
Until now, cyanophycin could only be obtained at the laboratory scale for analytical purposes. The aim of this study was to establish an efficient biotechnological process for the production of cyanophycin at a technical scale, allowing the isolation of several hundred grams or even kilogram amounts of cyanophycin in order to reveal the physical and material properties of this polymer. Therefore, the cyanophycin synthetase gene (cphA) was inserted into a vector which allowed induction of cphA by a simple temperature shift and was stably maintained in the host. E. coli DH1(pMa/c5-914::cphA) exhibited best stability of heterologous expression. With this system, expensive agents such as isopropyl-β-d-thiogalactopyranoside for the induction of product synthesis could be avoided. It was shown that the time point of temperature shift seemed to be crucial for product formation. Best yields were obtained when the induction was performed during the early exponential growth phase.
Cultivation experiments in different media revealed that cyanophycin synthesis in the recombinant E. coli strains used in this study was dependent on complex nutrient components such as Casamino Acids. This indicates that these strains need the provision of amino acids or small peptides which can serve as substrates or primers for cyanophycin synthetase and cyanophycin formation (2, 3). In order to minimize process costs, cultivation in mineral medium is desirable. Further investigations using metabolically engineered strains of E. coli are required to obtain cyanophycin production in mineral salts medium.
The acid extraction procedure presented here abolished a bottleneck in the biotechnological production of cyanophycin. In contrast to the labor-intensive, costly, and time-consuming method described by Simon and Weathers (24), which is useful at the laboratory scale, the novel acid extraction method of whole cells is simple and time- and cost-saving. Thus, it offers the possibility of purifying cyanophycin from bulk cell mass at the large scale. The minor disadvantage in yield is compensated for by a lower content of cellular impurities in the final product. Further attempts will be undertaken to reduce the remaining centrifugation steps. In addition, the application of this method to other gram-negative and gram-positive cyanophycin-producing recombinant strains (3) will be examined.
Acknowledgments
This work was financially supported by the Bundesministerium für Verbraucherschutz, Ernährung und Landwirtschaft (00NR125).
The technical assistance of Katja Kemper during large-scale purification of cyanophycin is gratefully acknowledged.
REFERENCES
- 1.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Purification of Synechocystis sp. strain PCC6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Environ. Microbiol. 67:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aboulmagd, E., I. Voβ, F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2:1338-1342. [DOI] [PubMed] [Google Scholar]
- 4.Berg, H., K. Ziegler, K. Piotukh, K. Baier, W. Lockau, and R. Volker-Engert. 2000. Biosynthesis of the cyanobacterial reserve polymer multi-liter-arginyl-poly-liter-aspartic acid (cyanophycin). Mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 267:5561-5570. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Carter, P., H. Bedouelle, and G. Winter. 1985. Improved oligonucleotide site-directed mutagenesis using M 13 vectors. Nucleic Acids Res. 13:4431-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hai, T., F. B. Oppermann-Sanio, and A. Steinbüchel. 1999. Purification and characterization of cyanophycin synthetase from the thermophilic Synechococcus sp. MA19. FEMS Microbiol. Lett. 181:229-236. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 9.Hannig, G., and S. C. Makrides. 1998. Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol. 16:54-60. [DOI] [PubMed] [Google Scholar]
- 10.Hanson, R. S., and J. A. Phillips. 1981. Chemical composition, p. 328-364. In P. Gerhardt (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 11.Joentgen, W., T. Groth, A. Steinbüchel, T. Hai, and F. B. Oppermann. 1998. Polyaspartic acid homopolymers and copolymers: biotechnical production and use thereof. International patent application WO 98/39090.
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lawry, N. H., and R. D. Simon. 1982. The normal and induced occurrence of cyanophycin inclusion bodies in several blue-green algae. J. Phycol. 18:391-399. [Google Scholar]
- 14.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 15.Mackerras, A. H., N. M. De Chazal, and G. D. Smith. 1990. Transient accumulation of cyanophycin in Anabaena cylindrica and Synechocystis 6308. J. Gen. Microbiol. 136:2057-2065. [Google Scholar]
- 16.Margolis, S. A. 1997. Sources of systematic bias in the measurement of water by the coulometric and volumetric Karl Fischer methods. Anal. Chem. 69:4864-4871. [DOI] [PubMed] [Google Scholar]
- 17.Messing, J., R. Crea, and P. H. Seeberg. 1981. A system for shotgun DNA sequencing. Nucleic Acids Res. 9:309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, K., M. Rischke, S. Sato, and H. Märkl. 1997. Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Appl. Microbiol. Biotechnol. 48:597-601. [DOI] [PubMed] [Google Scholar]
- 19.Oppermann-Sanio, F. B., T. Hai, E. Aboulmagd, F. F. Hezayen, S. Jossek, and A. Steinbüchel. 1999. Biochemistry of microbial polyamide metabolism, p. 185-193. In A. Steinbüchel (ed.), Biochemical principles and mechanisms of biosynthesis and biodegradation of polymers. Wiley-VCH, Weinheim, Germany.
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Schmidt, M., K. R. Babu, N. Khanna, S. Marten, and U. Rinas. 1998. Temperature-induced production of recombinant human insulin in high-cell density cultures of recombinant Escherichia coli. J. Biotechnol. 68:71-83. [DOI] [PubMed] [Google Scholar]
- 22.Schwamborn, M. 1998. Chemical synthesis of polyaspartates: a biodegradable alternative to currently used polycarboxylate homo- and copolymers. Polym. Degrad. Stabil. 59:39-45. [Google Scholar]
- 23.Simon, R. D. 1976. The biosynthesis of multi-liter-arginyl-poly(liter-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422:407-418. [DOI] [PubMed] [Google Scholar]
- 24.Simon, R. D., and P. Weathers. 1976. Determination of the structure of the novel polypeptide containing aspartic acid and arginine which is found in cyanobacteria. Biochim. Biophys. Acta 420:165-176. [DOI] [PubMed] [Google Scholar]
- 25.Warburg, O., and W. Christian. 1941. Isolierung und Kristallisierung des Gärungsferments Enolase. Biochem. Z. 310:384-421. [Google Scholar]
- 26.Yannisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp8 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 27.Ziegler, K., A. Diener, C. Herpin, R. Richter, R. Deutzmann, and W. Lockau. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-liter-arginyl-poly-liter-aspartate (cyanophycin). Eur. J. Biochem. 254:154-159. [DOI] [PubMed] [Google Scholar]