Abstract
Live Lactobacillus casei is present in fermented dairy products and has beneficial properties for human health. In the human digestive tract, the resident flora generally prevents the establishment of ingested lactic acid bacteria, the presence of which is therefore transient. The aim of this work was to determine if L. casei DN-114 001 survives during transit and how this bacterium behaves in the digestive environment. We used the human flora-associated (HFA) mouse model. L. casei DN-114 001 was genetically modified by the introduction of erm and lux genes, encoding erythromycin resistance and luciferase, respectively. For this modified strain (DN-240 041), light emission related to luciferase expression could easily be detected in the contents of the digestive tract. When inoculated into the digestive tract of HFA mice, L. casei (DN-240 041) survives but is eliminated with the same kinetics as an inert transit marker, indicating that it does not establish itself. In pure culture of L. casei, luciferase activities were high in the exponential and early stationary growth phases but decreased to become undetectable 1 day after inoculation. Viability was only slightly reduced even after more than 5 days. After transit in HFA mice, luciferase activity was detected even when 5-day-old L. casei cultures were given to the mice. In culture, the luciferase activity could be restored after 0.5 to 7 h of incubation in fresh medium or milk containing glucose, unless protein synthesis was inhibited by the addition of chloramphenicol or rifampin. These results suggest that in HFA mice L. casei DN-240 041, and thus probably L. casei DN-114 001, is able to initiate new protein synthesis during its transit with the diet. The beneficial properties of L. casei-fermented milk for human health might be related to this protein synthesis in the digestive tract.
Fermented milk containing live lactic acid bacteria shows probiotic effects (8, 35): yogurt with living Lactobacillus bulgaricus and Streptococcus thermophilus is far more efficient to prevent lactose intolerance than the same product with heat-killed bacteria (33). Live Lactobacillus casei reduces diarrhea (26, 27) and appears to modify the digestive microflora (9, 18) and to enhance the immune system during its transit in the digestive tract (DT) (25, 28). The question raised here is how the probiotic bacterium ingested with food survives and adapts to the environmental change from fermented milk to the DT.
The animal model we used is, from a bacteriological point of view, close to the human DT: germfree mice associated with a human flora (2, 20). The bacterial genera that are dominant in humans remain dominant in such mice, in contrast with conventional mice in which lactobacilli are dominant (12).
To study adaptation of L. casei in the DT, we developed a genetic approach successfully used with other lactic acid bacteria, Lactococcus lactis and S. thermophilus (6, 10, 11). It consists of transcriptional coupling of known promoters of the bacteria with the luciferase genes from Vibrio harveyi or Photorhabdus luminescens (13, 14). Using this approach it was demonstrated (6) that L. lactis can respond to a dietary stimulus (malate) in the DT and that S. thermophilus was able to produce beta-galactosidase during its transit in the DT (10).
The purpose of this work was to characterize survival during intestinal transit of L. casei DN-114 001 and a possible change(s) in L. casei DN-114 001 physiology during the environmental shift from fermented milk to the DT of human flora-associated (HFA) mice.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are presented in Table 1. Escherichia coli strain DH5α was grown aerobically at 37°C in Luria-Bertani medium. L. casei strain DN-240 041 was derived from the strain DN-114 001 (CNCM number, I-1518). The L. casei strains were grown at 37°C in MRS medium (7) or glucose-milk medium (GM medium; 174 g of milk powder/liter of water, autoclaved for 15 min at 110°C, 5% glucose). When required, the concentrations of antibiotics used were 50 μg of ampicillin or 100 μg of erythromycin per ml to select E. coli transformants and 5 μg of erythromycin or 5 μg of chloramphenicol per ml for L. casei. Chloramphenicol (ICN) and rifampin (Merck, Darmstadt, Germany) were used at 100 μg/ml for inhibition of translation and transcription studies, respectively. Thermoresistant Bacillus subtilis spores were used as transit markers and activated at 60°C in G-spore medium according to the method described for enumeration (4).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−lacZΔM15 Δ(lacZYA-argF)U169 deoR recA endA hsdR17 phoA supE λ−thi-1 gyrA relA | Life Technologies |
| Lactic acid bacteria | ||
| JIM4930 | L. lactis strain IL1403 containing the plasmid pJIM2530 | 32 |
| DN-114 001 | L. casei Danone strain | |
| DN-240 013 | L. casei strain DN-114 001 containing the plasmid pJIM2530 | This work |
| DN-240 034 | L. casei strain DN-114 001 containing the plasmid pDN13 | This work |
| DN-240 041 | DN-114 001, Emr, lacTp::luxAB on pDN26 integrated in the chromosome | This work |
| Plasmids | ||
| pBluescript | Ampr, M13ori pBR322ori | Stratagene |
| pGEM-T | Ampr, M13ori pBR322ori, linear T-overhang vector | Promega |
| pDN2 | Ampr, 423-bp fragment of L. casei containing lacTp cloned in pGEM-T | This work |
| pmj763 | Cmr, pSH71ori | 21 |
| pJIM2279 | Emr, derivative of pIL252 with copF::linker | 32 |
| pJIM2530 | Emr, derivative of pJIM2279 with luxAB genes from V. harveyi | 32 |
| pDN13 | Emr, derivative of pJIM2279 with luxAB genes from P. luminescens | This work |
| pVE6007 | Cmr, thermosensitive derivative of pGKV12 | 22 |
| pDN20 | Emr, ori (pWV01 ΔrepA) with luxAB genes from P. luminescens, integrative vector | This work |
| pDN26 | Emr, derivative of pDN20 carrying lacTp::luxAB | This work |
DNA manipulations.
Purification of genomic DNA from L. casei was performed with the DNeasy tissue kit (Qiagen, Courtaboeuf, France). Restriction endonucleases, T4 DNA polymerase, T4 DNA ligase, and Taq DNA polymerase were obtained from Roche Diagnostics (Roche Diagnostics GmbH, Mannheim, Germany) and were used according to the instructions of the manufacturers. L. casei was transformed by electroporation with a Gene-Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) as previously described (31). Plasmid DNA extractions were performed by a method adapted from that of Sambrook et al. (34).
Plasmid and strain constructions.
In order to test different lux genes, a plasmid similar to pJIM2530 (32) but carrying lux genes from P. luminescens was constructed as follows. luxAB genes from P. luminescens were obtained by EcoRI and BamHI digestion of the plasmid pmj763. The resulting fragment of 2.35 kb was cloned into pBluescript. This plasmid was digested with SacI and ligated with pJIM2279 also cut by SacI. This large hybrid plasmid was then digested with BclI and BssHII, and blunt ends were created with T4 DNA polymerase and religated in order to place lux genes under the control of the promoter of the plasmid resolvase as in plasmid pJIM2530.
A transcriptional fusion of the L. casei DN-114 001 lacTEGF operon promoter with the lux genes from P. luminescens was constructed. The sequences of the lacTEGF operons from the strains L. casei 64H (1) and ATCC 393 pLZ15− (17), available in GenBank (accession no. U21391 and Z80834, respectively), were used to design primers OFF21 (5′-GGCATCGTCGATGAGCCAA-3′) and OFF25 (5′-GCCTAATTAAATAGTCACAATCCAC-3′). The PCR fragment of 423 bp obtained with the primers OFF21 and OFF25 and containing the lac promoter (lacTp) lacks the stem-and-loop structure involved in the antiterminator (LacT)-dependent induction in the presence of lactose. The promoter region was cloned into pGEM-T, generating plasmid pDN2. pDN2 was digested with SalI and ligated with pDN20, an integrative vector carrying the luxAB genes from P. luminescens, also digested with SalI. The resulting plasmid was cut with PvuII, leading to the transcriptional fusion of the constitutive lactose promoter, lacTp, and the luxAB genes. This plasmid, called pDN26, was used to introduce the construction into the L. casei DN-114 001 strain. DN-114 001 was first transformed with pVE6007, a thermosensitive plasmid (22) used as helper plasmid to provide the replicase for pDN26 replication. The strain was then transformed with pDN26 which could replicate in the presence of pVE6007 at a permissive temperature (30°C). A shift to a restrictive temperature (42°C) inhibiting plasmid pVE6007 replication allowed the insertion of pDN26 into the chromosome by homologous recombination. Integrants were analyzed by PCR with primers OFF43 (5′-GGTCATTAAAACCGGTCGCGTG-3′), located upstream of the lacTp, and OFF44 (5′-CCCAGATTAACCAATCGC-3′), located in the luxA gene, to check for correct insertion into the chromosome.
Growth curves and luminescence measurements.
GM medium was inoculated at a 1/10 dilution with exponentially growing L. casei culture (optical density at 600 nm of around 0.3 in a Jenway 6400 spectrophotometer [Felsted, United Kingdom]). Cultures were grown at 37°C and samples were collected at regular intervals for measurement of luminescence and enumeration of bacteria. The luciferase activity was measured immediately after the addition of 5 μl of decanal (Sigma, Detroit, Mich.) to 1 ml of a broth culture. Light emission was measured in a luminometer (LB9501; Berthold, Germany). The values obtained were expressed as micro-relative light units (μRLU) per CFU or RLU per optical density unit at 600 nm.
HFA mice.
Germfree C3He/J mice were reared in sterile Texler-type isolators (La Cahlène, Vélizy, France) fitted with a rapid transfer system in an environmentally controlled room (21°C) with a 12-h light-dark cycle. Mice were given free access to irradiated food (UAR, Villemoisson, France) and sterilized water. To obtain HFA mice, a breeding stock of donors was prepared: one healthy human volunteer donated fecal material used as inoculums over 3 intervals of 3 weeks in germfree animals. Germfree animals for the experiments received 0.5 ml of a 1/10 fecal dilution from donor mice associated with human flora three times in a 3-week interval.
Experimental design.
One hundred milliliters of GM medium containing erythromycin was inoculated with an overnight culture of L. casei DN-240 041 and maintained at 37°C for 5 days. Inoculum size was chosen such that the culture was saturated after 1 day and little initial luciferase activity was detected. The viability of bacteria and the regeneration of the luciferase activity (see below) were studied daily in culture and in the DT of HFA mice.
Regeneration of luciferase activity. (i) In culture.
Culture samples were diluted (1/10) in prewarmed medium containing erythromycin and incubated at 37°C. The experiment was carried out with and without addition of chloramphenicol or rifampin to the medium. Samples were collected at different times. Enumerations and luminescence measurements were performed until 12 h after dilution.
(ii) In DT.
Mice received 0.5 ml of a mix of 4 ml of L. casei culture and 1 ml of B. subtilis spore solution (108 CFU of each per ml) at time zero by oral administration. Feces were collected individually 6 h later. Enumerations and luminescence measurements were performed with feces diluted to 1/10 in saline buffer.
Fecal elimination of L. casei in HFA mice.
Each mouse received, by oral administration, 0.5 ml of a mix containing L. casei culture and spores as described above. Fecal samples were collected from different animals every 2 h after inoculation until 24 h. Enumerations of L. casei and spores were immediately performed as described above.
RESULTS
Genetic constructions and luciferase expression in GM broth. (i) Construction of L. casei with luciferase activity.
Several vectors have been previously developed with the lux genes isolated from V. harveyi and used successfully in L. lactis (5, 6, 13). One of these, pJIM2530, was introduced into L. casei DN-114 001. The resulting strain, DN-240 013, exhibited very poor luciferase activity (3,500 RLU per optical density unit) in contrast to the level of luciferase activity obtained with the same plasmid introduced in L. lactis IL-1403, strain JIM4930 (500,000 RLU per optical density unit). lux genes isolated from another microorganism, P. luminescens, were tested in L. casei. For this purpose, plasmid pDN13 was constructed in such a way that the lux genes from P. luminescens were in the same genetic context (vector, promoter) as the lux genes from V. harveyi in pJIM2530. Plasmid pDN13 was introduced in L. casei strain DN-114 001 and luciferase activity was measured for the resulting strain, DN-240 034. This strain exhibited a 90-fold increase in luciferase activity compared to strain DN-240 013. lux genes from P. luminescens were therefore used to construct a lacTp::luxAB transcriptional fusion.
This fusion was integrated in the DN-114 001 chromosome by Campbell-like recombination using a two-step procedure previously described for L. lactis (15) involving the use of a thermosensitive helper plasmid. The resulting strain, used in this study, was named DN-240 041.
(ii) Growth and luciferase expression in GM broth.
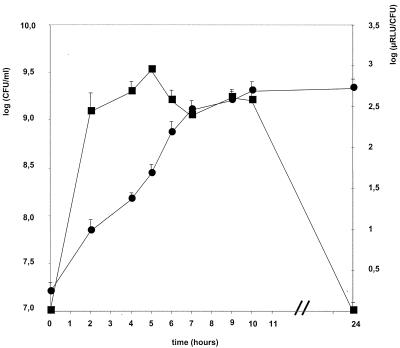
Growth of strain DN-240 041 in GM broth was assessed by enumeration of CFU (Fig. 1). Strain DN-240 041 exhibited a generation time of about 1 h. Moreover, growth of modified strain DN-240 041 was similar to the growth of its parental strain, DN-114 001 (data not shown). When the strain DN-240 041 was grown in GM broth, luciferase expression was high during the exponential phase and remained high until the culture reached a density of 109 CFU/ml (6 h). Then luciferase activity gradually decreased and no luciferase activity was detected in the late stationary phase. No luciferase activity was observed after 1 day of incubation at 37°C.
FIG. 1.
Growth and luciferase expression of L. casei DN-240 041 GM broth. Cultures of L. casei DN-240 041 were grown at 37°C and samples were removed at timed intervals for bacterial enumeration (circles) and luminescence measurement (squares). The luciferase activity was measured immediately after the addition of 5 μl of decanal to 1 ml of culture.
Influence of an environmental shift from GM to the DT. (i) L. casei survival during intestinal transit in HFA mice.
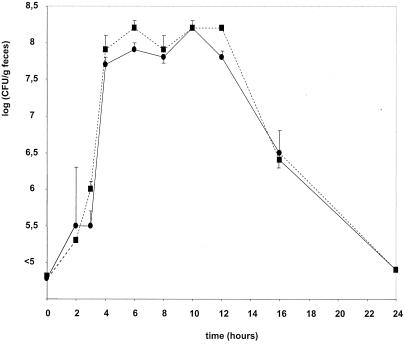
An experiment was carried out to determine the kinetics of L. casei fecal excretion after a single oral inoculation into HFA mice. Thermoresistant B. subtilis spores were added to a fresh L. casei culture before inoculation and served as an inert intestinal transit marker (Fig. 2). The number of L. casei bacteria and spores in the feces reached a plateau of 108 CFU/g between 4 and 12 h after inoculation. Then the levels of both bacterial species decreased to reach 106 CFU/g after 14 h and levels below 105 CFU/g 1 day after inoculation. The ratio between bacteria and spores remained constant during the elimination period. In the following experiments, fecal samples were collected 6 h after inoculation.
FIG. 2.
Fecal elimination kinetics of L. casei DN-240 041. Mice received 0.5 ml of a mix containing L. casei culture (about 108 CFU/ml) and B. subtilis spores (same quantity) (transit markers) by oral administration. Fecal samples were collected every 2 h after inoculation until 24 h. Bacteria (circles) and spores (squares) were analyzed in triplicate; fecal samples from each mouse were plated on MRS agar with erythromycin and on G-spore medium to enumerate L. casei DN-240 041 and B. subtilis, respectively.
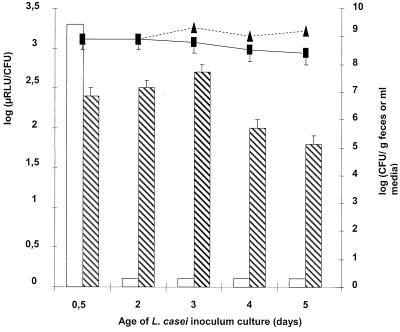
The effect of the age of the initial culture in GM broth on the survival during intestinal transit was analyzed. The viability of L. casei remained constant during the 5-day incubation period in GM broth at 37°C. The same viability of bacteria was found in fecal samples collected 6 h after inoculation (Fig. 3).
FIG. 3.
Regeneration of luciferase activity in vivo from GM culture. Mice received 0.5 ml of a mix of L. casei (108 to 109 CFU/ml) culture and spores (same quantity) of B. subtilis (transit markers) daily at 10 a.m. by oral administration. Feces were collected 6 h later. Enumerations (E) and luminescence (L) measurements were performed with inoculum (E, triangles; L, empty columns) and feces (E, squares; L, hatched columns) diluted to 1/10.
(ii) Luciferase activity after transit.
Experiments were designed to determine whether L. casei shows metabolic activity during transit. Measurement of luciferase activity during the DT transit is the way chosen to estimate if L. casei is able to synthesize protein during transit in the DT.
When L. casei was cultured in GM medium, 12 h after inoculation the luciferase activities were high in pure culture and in HFA mouse feces (Fig. 3). When the culture was older than 1 day, however, the luciferase activity was not detectable in the inoculum. The acidity is not the main reason for the drop of luciferase activity since a shift to neutral pH did not enhance the activity except for in the 2-day culture (data not shown). After passage through the DT, luciferase activities were enhanced 30- to 100-fold (Fig. 3). The reinitiation of luciferase activities in the DT occurred while no bacterial growth was observed.
(iii) Restart of growth and luciferase activity of L. casei in culture.
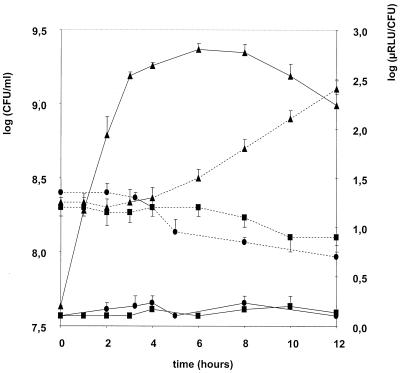
Experiments in pure culture were carried out to determine if reinitiation of luciferase activity is linked to new protein synthesis. Stationary overnight cultures in GM medium reinitiated growth and showed luciferase activity very quickly after dilution (1/10) in fresh medium (data not shown). For older cultures the reinitiation of growth was delayed, although the number of viable cells directly after dilution was the same as at the beginning of the stationary phase. Figure 4 illustrates this with L. casei strain DN-240 041 cultivated for 3 days in GM medium: cultures were diluted in prewarmed medium and kept at 37°C. The luciferase activity increased very rapidly, whereas multiplication was delayed for 4 h. Under the same conditions during the first 4-h period, chloramphenicol or rifampin addition blocked luciferase activity but did not affect L. casei viability.
FIG. 4.
Regeneration of luciferase activity in pure culture of L. casei. Three-day-old culture samples were diluted (1/10) in prewarmed GM medium containing erythromycin (triangles) and incubated at 37°C. Further experiments were carried out with a supplement of chloramphenicol (squares) or rifampin (circles) in the medium. Enumerations (dotted lines) and luminescence (solid lines) measurements were performed until 12 h after dilution.
After the 4-h delay period, bacterial growth became visible in GM medium, and luciferase activities were similar to those observed in Fig. 1. Chloramphenicol or rifampin additions reduced viability after the 4-h delay.
DISCUSSION
A previous approach using a modified L. lactis strain (5, 6, 13) gave us the ability to evaluate how this lactic acid bacterium behaves during transit in HFA mice. No effect of L. lactis as a probiotic on human health was reported. In this work the approach was adapted to L. casei, which is known as a probiotic for humans. It represents the first successful attempt to express luciferase genes in L. casei. Luciferase from P. luminescens shows a higher activity in L. casei than luciferase from V. harveyi. This may be due to a higher stability of this enzyme at high temperature since the optimal temperature for the P. luminescens luciferase activity is 45°C compared to 30°C for the V. harveyi enzyme (19). In pure cultures, L. casei (DN-240 041) shared similar features with L. lactis (6) since luciferase activity was high in the exponential growth phase and reduced in the late stationary phase.
The HFA mouse model reflects the human situation concerning the antagonistic effect against lactic acid bacteria: L. casei did not multiply but survived well as demonstrated by the fact that the ratio between the bacteria and spores used as transit markers remained constant. This observation is consistent with what has been observed in humans where different lactic acid bacteria were progressively eliminated from the DT (16, 23, 37). We determined the transit time based on fecal excretion of L. casei: a maximum was observed between 4 and 12 h postinoculation. This period was not precisely estimated in humans since fecal emission is more rare than in mice. Bouhnik et al. (3) observed that the level of bifidobacterium decreases 1 day after product consumption. Pochart et al. (29) observed that the transit time was 2 h postingestion by using in vivo ileal perfusion. In this HFA model we chose a 6-h delay between inoculation and fecal sample collection for further experiments.
When exponential-phase cultures were used to inoculate mice, the luciferase activity per CFU measured in the inoculum was recovered in the feces. In contrast, when old stationary-phase cultures were used to inoculate mice, the luciferase activity per CFU observed in the corresponding fecal samples was high while it was not detectable in the inoculum. These data suggested that L. casei DN-240 041, which transited the DT without any detectable multiplication, underwent a physiological change resulting in the reinitiation of luciferase activity.
We have studied, in pure culture, the restart of luciferase activity with such aged L. casei cultures: during the first 4 h after dilution into fresh culture medium luciferase activity increased to a large extent while no multiplication could be observed. Such reinitiation of luciferase activity preceding L. casei multiplication was blocked by rifampin or chloramphenicol. These antibiotics are known to block transcription (24) and translation (30), respectively, and we therefore postulate that the reinitiation of luciferase activity corresponded to the synthesis of new proteins. The protein(s) could be LuxAB itself, another protein(s) needed for the enzymatic reaction (36), or protein synthesis affecting the global physiology of the cell.
Based on our studies, performed in pure culture with chloramphenicol and rifampin, where reinitiation of luciferase activity was shown to be linked to the synthesis of a new protein(s), the reinitiation of luciferase activity observed after transit suggests that L. casei not only survives in the DT but also synthesizes new protein during its transit in the DT. The environmental shift from fermented milk to DT may induce protein synthesis in L. casei to adapt to the new conditions. This feature is observed without bacterial division. To our knowledge, this is the first study to suggest that lactic acid bacteria in transit in human flora synthesize proteins. The promoter used in the luciferase construct could influence this result. Other promoters may be more or less active as previously observed with the same luciferase reporter gene (10). This work opens a perspective for comparison of different L. casei promoters in HFA mice.
In previous work dealing with L. lactis, it was shown that bacteria adapted to diet stimulation (6), but the experiments were carried out in germfree mice. Here we investigated HFA mice. The presence of autochthonous microflora exerted an antagonistic effect on L. casei growth, but the bacterium was still able to adapt to the environment during transit. This feature is of importance for probiotic bacteria that are supposed to exert in the DT functions beneficial for human health. The next step will be to determine which functions are induced in the bacteria during digestive transit.
Acknowledgments
We thank P. Renault for advice in genetic constructions.
REFERENCES
- 1.Alpert, C. A., and U. Siebers. 1997. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the Bg1G family of transcriptional antiterminators. J. Bacteriol. 179:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andremont, A., P. Raibaud, and C. Tancrede. 1983. Effect of erythromycin on microbial antagonisms: a study in gnotobiotic mice associated with a human fecal flora. J. Infect. Dis. 148:579-587. [DOI] [PubMed] [Google Scholar]
- 3.Bouhnik, Y., P. Pochart, P. Marteau, G. Arlet, I. Goderel, and J. C. Rambaud. 1992. Fecal recovery in humans of viable Bifidobacterium sp. ingested in fermented milk. Gastroenterology 102:875-878. [DOI] [PubMed] [Google Scholar]
- 4.Contrepois, M. 1969. Utilisation d'une technique microbiologique pour la mesure de la vitesse de transit des microparticules dans le tractus digestif des ruminants. C. R. Acad. Sci. 268:1757-1759. [Google Scholar]
- 5.Corthier, G., C. Delorme, S. D. Ehrlich, and P. Renault. 1997. Assessment of bacterial physiology in the digestive tract by use of luciferase gene as promoter probe. Adv. Exp. Med. Biol. 418:831-832. [DOI] [PubMed] [Google Scholar]
- 6.Corthier, G., C. Delorme, S. D. Ehrlich, and P. Renault. 1998. Use of luciferase genes as biosensors to study bacterial physiology in the digestive tract. Appl. Environ. Microbiol. 64:2721-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of Lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 8.de Roos, N. M., and M. B. Katan. 2000. Effects of probiotic bacteria on diarrhea, lipid metabolism, and carcinogenesis: a review of papers published between 1988 and 1998. Am. J. Clin. Nutr. 71:405-411. [DOI] [PubMed] [Google Scholar]
- 9.Djouzi, Z., C. Andrieux, M. C. Degivry, C. Bouley, and O. Szylit. 1997. The association of yogurt starters with Lactobacillus casei DN 114.001 in fermented milk alters the composition and metabolism of intestinal microflora in germ-free rats and in human flora-associated rats. J. Nutr. 127:2260-2266. [DOI] [PubMed] [Google Scholar]
- 10.Drouault, S., J. Anba, and G. Corthier. 2002. Streptococcus thermophilus is able to produce a β-galactosidase active during its transit in the digestive tract of germ-free mice. Appl. Environ. Microbiol. 68:938-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducluzeau, R. 1969. Influence of the zoological species on the microflora of the gastrointestinal tract. Rev. Immunol. Ther. Antimicrob. 33:345-383. [PubMed] [Google Scholar]
- 13.Eaton, T. J., C. A. Shearman, and M. J. Gasson. 1993. The use of bacterial luciferase genes as reporter genes in Lactococcus: regulation of the Lactococcus lactis subsp. lactis lactose genes. J. Gen. Microbiol. 139:1495-1501. [DOI] [PubMed] [Google Scholar]
- 14.Frackman, S., M. Anhalt, and K. H. Nealson. 1990. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J. Bacteriol. 172:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godon, J. J., C. J. Pillidge, K. Jury, C. A. Shearman, and M. J. Gasson. 1995. Molecular analysis of the Lactococcus lactis sex factor. Dev. Biol. Stand. 85:423-430. [PubMed] [Google Scholar]
- 16.Goldin, B. R., S. L. Gorbach, M. Saxelin, S. Barakat, L. Gualtieri, and S. Salminen. 1992. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig. Dis. Sci. 37:121-128. [DOI] [PubMed] [Google Scholar]
- 17.Gosalbes, M. J., V. Monedero, C. A. Alpert, and G. Perez-Martinez. 1997. Establishing a model to study the regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol. Lett. 148:83-89. [DOI] [PubMed] [Google Scholar]
- 18.Guerin-Danan, C., C. Chabanet, C. Pedone, F. Popot, P. Vaissade, C. Bouley, O. Szylit, and C. Andrieux. 1998. Milk fermented with yogurt cultures and Lactobacillus casei compared with yogurt and gelled milk: influence on intestinal microflora in healthy infants. Am. J. Clin. Nutr. 67:111-117. [DOI] [PubMed] [Google Scholar]
- 19.Hill, P. J., C. E. Rees, M. K. Winson, and G. S. A. B. Stewart. 1993. The application of the lux genes. Biotechnol. Appl. Biochem. 17:3-14. [PubMed] [Google Scholar]
- 20.Hirayama, K. 1999. Ex-germfree mice harboring intestinal microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Exp. Anim. 48:219-227. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, M. F. 1995. Highly bioluminescent Streptococcus thermophilus strain for the detection of dairy-relevant antibiotics in milk. 44:405-412. [DOI] [PubMed]
- 22.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marteau, P., P. Pochart, Y. Bouhnik, S. Zidi, I. Goderel, and J. C. Rambaud. 1992. Survival of Lactobacillus acidophilus and Bifidobacterium sp. in the small intestine following ingestion in fermented milk. A rational basis for the use of probiotics in man. Gastroenterol. Clin. Biol. 16:25-28. [PubMed] [Google Scholar]
- 24.McClure, W. R., and C. L. Cech. 1978. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253:8949-8956. [PubMed] [Google Scholar]
- 25.Paubert-Braquet, M., G. X.-H., C. Gaudichon, N. Hedef, A. Serikoff, C. F. Bouley, B. Bonavida, and P. Braquet. 1995. Enhancement of host resistance against Salmonella typhimurium in mice fed a diet supplemented with yogurt or milks fermented with various Lactobacillus casei strains. Int. J. Immunother. 11:153-161. [Google Scholar]
- 26.Pedone, C. A., C. C. Arnaud, E. R. Postaire, C. F. Bouley, and P. Reinert. 2000. Multicentric study of the effect of milk fermented by Lactobacillus casei on the incidence of diarrhoea. Int. J. Clin. Pract. 54:568-571. [PubMed] [Google Scholar]
- 27.Pedone, C. A., A. O. Bernabeu, E. R. Postaire, C. F. Bouley, and P. Reinert. 1999. The effect of supplementation with milk fermented by Lactobacillus casei (strain DN-114 001) on acute diarrhoea in children attending day care centres. Int. J. Clin. Pract. 53:179-184. [PubMed] [Google Scholar]
- 28.Perdigon, G., E. Vintini, S. Alvarez, M. Medina, and M. Medici. 1999. Study of the possible mechanisms involved in the mucosal immune system activation by lactic acid bacteria. J. Dairy Sci. 82:1108-1114. [DOI] [PubMed] [Google Scholar]
- 29.Pochart, P., P. Marteau, Y. Bouhnik, I. Goderel, P. Bourlioux, and J. C. Rambaud. 1992. Survival of bifidobacteria ingested via fermented milk during their passage through the human small intestine: an in vivo study using intestinal perfusion. Am. J. Clin. Nutr. 55:78-80. [DOI] [PubMed] [Google Scholar]
- 30.Porse, B. T., C. Rodriguez-Fonseca, I. Leviev, and R. A. Garrett. 1995. Antibiotic inhibition of the movement of tRNA substrates through a peptidyl transferase cavity. Biochem. Cell. Biol. 73:877-885. [DOI] [PubMed] [Google Scholar]
- 31.Posno, M., R. J. Leer, N. van Luijk, M. J. F. van Giezen, P. T. H. M. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renault, P., G. Corthier, N. Goupil, C. Delorme, and S. D. Ehrlich. 1996. Plasmid vectors for gram-positive bacteria switching from high to low copy number. Gene 183:175-182. [DOI] [PubMed] [Google Scholar]
- 33.Rizkalla, S. W., J. Luo, M. Kabir, A. Chevalier, N. Pacher, and G. Slama. 2000. Chronic consumption of fresh but not heated yogurt improves breath-hydrogen status and short-chain fatty acid profiles: a controlled study in healthy men with or without lactose maldigestion. Am. J. Clin. Nutr. 72:1474-1479. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schaafsma, G. 1996. State of the art concerning probiotic strains in milk products. Nutr. Newsl. 5:23-24. [Google Scholar]
- 36.Stewart, G. S., and P. Williams. 1992. lux genes and the applications of bacterial bioluminescence. J. Gen. Microbiol. 138:1289-1300. [DOI] [PubMed] [Google Scholar]
- 37.Yuki, N., K. Watanabe, A. Mike, Y. Tagami, R. Tanaka, M. Ohwaki, and M. Morotomi. 1999. Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: selective isolation from faeces and identification using monoclonal antibodies. Int. J. Food Microbiol. 48:51-57. [DOI] [PubMed] [Google Scholar]