Abstract
GacS/GacA comprises a two-component regulatory system that controls the expression of secondary metabolites required for the control of plant diseases in many pseudomonads. High mutation frequencies of gacS and gacA have been observed in liquid culture. We examined whether gacS/gacA mutants could competitively displace the wild-type populations on roots and thus pose a threat to the efficacy of biological control. The survival of a gac mutant alone and in competition with the wild type on roots was examined in the biological control strain Pseudomonas aureofaciens 30-84. In this bacterium, GacS/GacA controls the expression of phenazine antibiotics that are inhibitory to plant pathogenic fungi and enhance the competitive survival of the bacterium. Wheat seedlings were inoculated with strain 30-84, and bacteria were recovered from roots after 21 days in sterile or nonsterile soil to check for the presence of gacS or gacA mutants. Although no mutants were detected in the inoculum, gacS/gacA mutants were recovered from 29 out of 31 roots and comprised up to 36% of the total bacterial populations. Southern hybridization analysis of the recovered gacA mutants did not indicate a conserved mutational mechanism. Replacement series analysis on roots utilizing strain 30-84 and a gacA mutant (30-84.gacA) or a gacS mutant (30-84.A2) demonstrated that although the mutant population partially displaced the wild type in sterile soil, it did not do so in natural soil. In fact, in natural soil final rhizosphere populations of wild-type strain 30-84 starting from mixtures were at least 1.5 times larger than would be predicted from their inoculation ratio and generally were greater than or equal to the population of wild type alone despite lower inoculation rates. These results indicate that although gacS/gacA mutants survive in natural rhizosphere populations, they do not displace wild-type populations. Better survival of wild-type populations in mixtures with mutants suggests that mutants arising de novo or introduced within the inoculum may be beneficial for the survival of wild-type populations in the rhizosphere.
GacS/GacA global two-component regulatory systems are comprised of a membrane-bound environmental sensor, GacS, and a transcriptional response regulator, GacA. GacA was first described as a global activator of antibiotics and cyanide production in Pseudomonas fluorescens (15). GacS, initially called LemA, was identified by its role in lesion manifestation by Pseudomonas syringae pv. syringae strain B728a (13). GacS/GacA homologs have since been identified in many gram-negative bacteria (reviewed in reference 11).
The GacS/GacA system controls the expression of genes required for the synthesis of secondary metabolites with antimicrobial activity in many plant-associated fluorescent Pseudomonas species. Secondary metabolites contributing to biological control that are regulated directly or indirectly by this system include the antibiotics 2,4-diacetylphloroglucinol, 2-hexy-5-propyl-resorcinol, phenazines, pyoluteorin, and pyrrolnitrin, as well as hydrogen cyanide, chitinase, and exoproteases (1, 3, 4, 6, 8, 15). Many of these compounds also play a role in stress resistance, ecological fitness, and rhizosphere competence (2, 18, 19, 27).
It has been observed that gacS and gacA mutants occur at high frequencies in culture (2, 3, 6, 15, 22). In one study, gacS/gacA mutants were found in 192 separate cultures and comprised up to 61% of the total bacteria in each culture (6). The presence of gacS/gacA mutants in equally high proportions in the rhizosphere could compromise the use of fluorescent pseudomonads as biological control agents. Previous work suggested that media conditions altered the occurrence of mutants in liquid culture (2, 6). Given the importance of secondary metabolites and fitness traits controlled by this system for biological control, efficacy could still be reduced if mutations arising de novo in the rhizosphere or introduced with the inoculum were able to displace the wild-type bacterium in the rhizosphere.
We used the root-colonizing biological control bacterium Pseudomonas aureofaciens strain 30-84 to examine whether gacS/gacA mutants displace the wild-type population in the rhizosphere, thereby posing a threat to the efficacy of biological control. Production of phenazine antibiotics by P. aureofaciens 30-84 is responsible for approximately 90% of the control of take-all disease of wheat by this bacterium (21). Phenazines also contribute to the persistence of strain 30-84 on roots in natural soil (18). We previously characterized mutations in gacA and gacS from strain 30-84 (3) and demonstrated that mutations in either gene have identical phenotypes (phenazine−, exoprotease−, hydrogen cyanide−, and loss of pathogen inhibition). In this report we provide the sequence of gacS from strain 30-84 and report on the isolation and characterization of gacS/gacA mutants from the wheat rhizosphere. We used replacement series experiments on roots to examine the competitive survival of the wild type and a gac mutant alone and in mixtures in sterile and natural soil.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
Bacterial strains and plasmids are described in Table 1. A spontaneous rifampin-resistant derivative of strain 30-84 was used in all studies (21). Strain 30-84 and its derivatives were grown at 28°C in Luria-Bertani (LB) medium containing 5 g of NaCl per liter (17) or AB minimal broth (23). Strains were also grown on LB agar, pigment production medium-D agar (30), King's medium B (KMB) agar (14), and skim milk agar (Difco Laboratories, Detroit, Mich.). Antibiotics used when appropriate included rifampin (50 μg/ml), tetracycline (50 μg/ml), and kanamycin (50 μg/ml). Triparental matings with Escherichia coli strains DH5α and HB101 (containing pRK2013) were described previously (21).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| P. aureofaciens | ||
| 30-84 | Phz+ Rifr wild type | W. W. Bockus |
| 30-84Z | Phz− RifrphzB::lacZ genomic fusion | 20 |
| 30-84.gacA | Phz− Rifr Phz− RifrgacA::Kmr of strain 30-84 | 3 |
| 30-84SGS1 | Phz− Rifr spontaneous gacS mutant of strain 30-84 | This study |
| 30-84SGA2 | Phz− Rifr spontaneous gacA mutant of strain 30-84 | This study |
| 30-84.A2 | Phz− Rifr Kmr spontaneous gacS mutant of strain 30-84 containing Tn5 in an unidentified locus | 3 |
| E. coli | ||
| DH5α | F−recA1 endA1 hsdR17 supE44 ti-1 gyrA96 relA1 Δ(argF-lacZYA) 1169 φ80lacZ ΔM15 | Gibco-BRL |
| HB101 | F−hsdS20 (rB− mB−) supE44 recA1 ara14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-5 | Gibco-BRL |
| Plasmid | ||
| pLAFR3 | IncP1 Tetr cos+ rlx+ | 25 |
| pRK415 | IncP1 Tetr cos+ rlx+ | |
| pRK2013 | IncP1 tra oriE1 Kmr | 5 |
| pRKgacSB03 | An approximately 3-kb PCR product containing gacS of strain 30-84 cloned into the BamHI site of pRK415 | This study |
| pLSP259 | pLAFR3 containing phzI, phzR, and the phenazine biosynthetic locus on a 20.4-kb EcoRI fragment of strain 30-84 chromosomal DNA | 20 |
| pLSP19-21 | pLAFR3 containing a 23-kb EcoRI fragment of 30-84 genomic DNA that contains the 3′ end of gacS | This study |
| pSTC121 | pLAFR3 containing a 4.5-kb EcoRI-PstI gacA fragment of pSTC110 | 3 |
| pSTC140 | pUC18 containing a 2.2-kb KpnI/PstI gacA fragment of pSTC121 | 3 |
Cloning and nucleotide sequence analysis of P. aureofaciens 30-84 gacS.
The gacS nucleotide sequence was determined from a partial gacS genomic subclone and a full-length gacS-containing PCR product. The partial subclone was identified by colony blot hybridization and was sequenced by subcloning and primer walking. The full-length PCR clone was amplified by using Elongase polymerase enzyme mix (Gibco-BRL, Gaithersburg, Md.) from strain 30-84 chromosomal DNA utilizing primers designed from the flanking regions of the Pseudomonas chlororaphis strain 06 gacS (accession number AF192795). A BamHI restriction site was designed on the end of each primer to accommodate cloning into pRK415. The primers gacSBam_F (5′ CGGGATCCGACAGATACGGGATTCATTAG 3′) and gacSBam_R (5′ CGGGATCCCGATAGTTCTGGCTGCTGAAGAGA 3′) were synthesized by Gibco-BRL. The purified PCR product was sequenced directly prior to cloning. All sequencing was performed at the University of Arizona Biotechnology Center with an Applied Biosystems automatic sequencer (model 373A, version 1.2.1). DNA sequence analyses were performed with the University of Wisconsin Genetics Computer Group software package (version 9.1). The gacS PCR product was cloned into the BamHI restriction site of pRK415 to generate plasmid pRKgacSB03.
Isolation of mutants from wheat roots.
Wheat seeds (cv. Fielder) were surface disinfested as described previously (29). Disinfested seeds were pregerminated on LB agar for 3 days to ensure sterility before planting. The seedlings were suspended for 1 min in cultures of either strain 30-84 or strain 30-84Z (phzB::lacZ) grown for 24 to 30 h with shaking in LB broth (ca. 1010 CFU/ml). Each culture was used to treat 20 seedlings. Seedlings were sown in 24- by 100-mm glass tubes that contained 55 cm3 of sterile sand and 5 ml of sterile 1/3 Hoaglands solution (macronutrients only) (12). The seedlings were covered with 1 cm of sterile sand and were placed in a Conviron growth chamber (22°C/18°C light/dark cycle, 25% relative humidity, 12-h light/dark cycles). Plants were grown for 19 to 21 days. Plants were aseptically harvested, and bacteria were isolated via sonication (twice for 30 s) from the entire excised root mass in 2 ml of phosphate-buffered saline, pH 7.0. Bacterial populations were determined by serial dilution in phosphate-buffered saline and plating on LB or LB containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to differentiate wild-type and mutant bacteria counts. Orange colonies from the strain 30-84 cultures and blue colonies (on X-Gal) from the strain 30-84Z cultures were counted as wild type. White colonies from both cultures were counted as gac-like mutants. White colonies were selected randomly and tested for hyperfluorescence and the loss of exoprotease production by patching onto KMB agar or skim milk agar, respectively. When present, a single white mutant from each plant was selected for further characterization.
Characterization of mutants.
Complementation analysis was performed on each mutant to determine the nature of the mutation. Each mutant was conjugated with three plasmids: pRKgacSB03 and pSTC121 containing gacS and gacA from 30-84, respectively, and pLSP259 containing the entire phenazine biosynthetic operon (phzFABCD) plus phzI and phzR (Table 1). Complementation was indicated by restoration of orange pigmentation to 30-84 mutant strains and β-galactosidase activity to 30-84Z mutant strains.
Southern hybridization.
EcoRI/PstI-digested chromosomal DNA from 30-84 derivative strains was probed with the 1.2-kb HincII/HindIII restriction fragment of pSTC140, which contained the 30-84 gacA gene. The probe was labeled with digoxigenin, and homology was detected as described in the Genius System User's Guide for Membrane Hybridization (version 3.0) (Boehringer-Mannheim, Indianapolis, Ind.).
Growth curves.
Relative growth rates of wild-type strain 30-84 and gacS and gacA mutants 30-84SGS1 and 30-84SGA2, respectively, were determined in rich and minimal media. Single colonies of each strain were inoculated into 2 ml of LB broth and AB minimal broth and were grown overnight at 28°C. The optical density at 620 nm of the cultures was measured, and the cell densities were normalized before 200 μl of each culture was added to 20 ml of the same medium (LB or AB minimal). The cultures were grown with shaking and were sampled at ca. 1-h intervals. Absorbance at 620 nm was determined and plotted versus time. At the last sampling, final population densities were determined via serial dilution plating.
Replacement series.
The relative competitive fitness of wild-type strain 30-84 and the gacA mutant strain 30-84.gacA in the wheat rhizosphere was determined by mixing strains for growth on plants in a replacement series. Wheat seeds (cv. Penewawa) were surface disinfested and pregerminated as described above. Prior to planting of the seedlings, each was dipped for 1 min into one of five bacterial mixtures containing specific proportions of wild-type 30-84 (gac+) and strain 30-84.gacA (gac mutant) (ratios of 1:0, 0.9:0.1, 0.5:0.5, 0.1:0.9, and 0:1, respectively). The bacterial mixtures were generated from 150 ml of LB overnight cultures of each strain. The cultures were normalized to an optical density at 620 nm of 1.1 (ca. 1010 CFU/ml) prior to mixing, and the ratios were confirmed via serial dilution plating. Each starting culture was serially diluted and plated on six sets of pigment production medium-D agar plates (3 plates/set) to screen for the presence of preexisting gac mutants. Each dilution plate contained ca. 103 bacteria. The inoculated seedlings were planted as described above. However, instead of sterile sand, soil taken from a wheat field in southern Arizona was used. Two soil treatments were performed: sterilized soil which consisted of field soil that was autoclaved for 45 min, allowed to sit at room temperature 24 h, and then reautoclaved, and natural field soil. Seedlings were allowed to grow for 14 days under the conditions described above. Bacteria were isolated from the roots by sonication in phosphate buffer solution (0.01 M, pH 7.4), and the roots were dried overnight at 65°C and dry weights were recorded. The bacterial suspensions were serially diluted and plated onto KMB agar amended with rifampin. Rough colony morphology and orange pigmentation indicated strain 30-84, and smooth, white, highly fluorescent colonies indicated strain 30-84.gacA. Strain 30-84 colonies, strain 30-84.gacA colonies, and total colonies were recorded and standardized to CFU/gram of dry weight of roots. The reported results are the means of eight replicates (separate plants) of each treatment. The replacement series were performed twice with similar results. The results of one experiment are shown. In a second set of replacement series experiments with the wild type and the kanamycin-resistant gacS mutant (30-84.A2), seedlings were allowed to grow 1, 5, 10, and 20 days. Additionally, one set of seedlings was planted and harvested immediately to provide inoculation population sizes (day 0). Bacteria were isolated as above but were serially diluted and plated onto LB agar amended with rifampin and rifampin plus kanamycin. Dilution plates for the wild-type-alone treatment (1.0:0, gac+:gac mutant) were carefully examined for spontaneous mutants (colonies with the gac mutant phenotype and insensitivity to kanamycin) at each census. Experiments were repeated twice, and reported results are the means of three replicates from one experiment. Data for days 10 and 20 are similar to those for day 14 in the previous experiments and are not shown.
Nucleotide sequence accession number.
The nucleotide sequence of P. aureofaciens 30-84 gacS has been deposited in the GenBank database under accession no. AY037869.
RESULTS
Cloning and sequencing P. aureofaciens 30-84 gacS.
Oligonucleotide primers from DNA flanking the P. chlororaphis 06 gacS gene (accession no. AF192795) were used to amplify a 3-kb region of the P. aureofaciens chromosome. The cloned PCR product (on pRK415gacSB03) restored wild-type levels of phenazine production to strain 30-84.A2, a gacS mutant (3), indicating that the gacS gene was functional. The nucleotide sequence of gacS was determined by direct sequencing of the PCR mix and from cosmid pLSP19-21, which contained approximately 70% of the 3′ end of gacS. The 30-84 gacS was 96% identical to gacS of P. chlororaphis 06. The predicted amino acid sequence was 98.8% identical to the predicted GacS sequence of P. chlororaphis and was highly similar to other GacS amino acid sequences (data not shown).
Characterization of gacS/gacA mutants isolated from wheat roots.
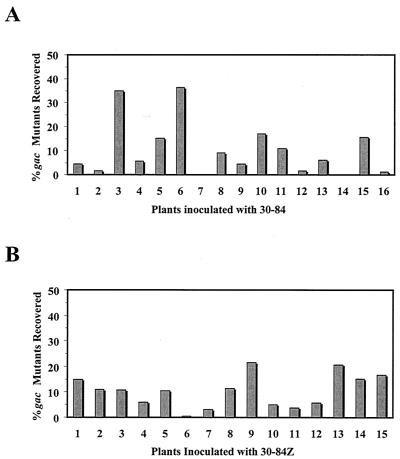
Cultures of strain 30-84 or the phenazine reporter strain 30-84Z (phzB::lacZ) were used to inoculate pregerminated surface-disinfested wheat seeds. Initial populations of strains 30-84 and 30-84Z were 9.7 × 108 and 7.2 × 108 CFU/ml, respectively. No gacS/gacA mutants were identified from either inoculum as determined by dilution plating. We calculated the level of detection of mutants to be less than 0.01% on the basis of examination of over 10,000 colonies on dilution plates from 10 separate samples of the initial wild-type strain 30-84 inoculum. Treated seedlings were planted in sterile sand and were incubated for 3 weeks. The plants were harvested, and total bacterial populations (averages of 2 × 107 and 2.5 × 107 CFU/ml) were isolated from the roots of 16 plants inoculated with strain 30-84 and 15 plants inoculated with strain 30-84Z, respectively. White colonies indicative of the loss of phenazine production were observed from 14 out of 16 plants inoculated with strain 30-84 and from 15 of 15 plants inoculated with strain 30-84Z. Therefore, possible gacS/gacA mutants were recovered from 97% (29 out of 31) of the inoculated plants and comprised up to 36% of the total bacterial population isolated from each root (Fig. 1). The fact that mutants were isolated from roots colonized by strains 30-84 and 30-84Z in approximately equal frequency suggests that phenazine production does not affect the formation of gacS/gacA mutations. To preclude the characterization of siblings, a single white mutant was chosen from each of the 29 plants for further characterization.
FIG. 1.
Percentage of gac mutants in the total bacterial populations recovered from wheat roots. Pregerminated wheat seedlings were inoculated with cultures of P. aureofaciens strain 30-84 (A) or 30-84Z (B). Neither culture contained detectable gac mutants prior to inoculation (detection limit, ∼0.01%).
All 29 mutants failed to express the phenazine biosynthetic operon, were deficient in the production of exoprotease (on skim milk agar), and were hyperfluorescent (on KMB), consistent with reported gac mutant phenotypes (3). The nature of the mutations was characterized by complementation analysis. Functional copies of gacS (on pRK415gacSB03), gacA (on pSTC121), and the phenazine biosynthetic operon (phzFABCD) plus the quorum-sensing regulators phzI and phzR (on pLSP259) were introduced in trans into the mutants. None of the mutants was complemented by pLSP259, indicating that none was mutated in either the quorum-sensing system, which controls phenazine production, or the biosynthetic region. Of the 14 mutants derived from the wild type, 6 were complemented only by gacA while the other 8 were complemented only by gacS (data not shown). Of the 15 mutants derived from strain 30-84Z, 5 carried mutations in gacA while 10 contained mutations in gacS. Thus, all 29 independently isolated mutants from wheat roots were mutated only in gacA or gacS.
Southern hybridization analysis of chromosomal DNA from the gacA mutants described above revealed at least three classes of gacA mutations. Of the 11 gacA mutants isolated, 9 contained a band hybridizing to gacA which comigrated with the wild-type gacA gene, consistent with the occurrence of point mutations or small deletions or insertions. One mutant, 30-84SGA2, appeared to harbor a 200-bp insertion, while another, 30-84SGA4, showed a 3-kb deletion (data not shown). None of the mutants reverted to the wild-type phenotype even after multiple serial platings (data not shown).
Growth of the wild type and gac mutants in liquid culture.
Growth curves were performed on strain 30-84 and the mutants 30-84SGS1 (gacS mutant) and 30-84SGA2 (gacA mutant) in both rich and minimal media. In LB broth, both mutants entered the exponential growth phase earlier than the wild type (within 1 h, versus 3 h for the wild type). In AB broth, the mutants and strain 30-84 entered exponential growth at similar times. There was no difference in doubling times between mutants and wild type in either medium (1 h in LB versus 1.3 h in AB, respectively). These results indicated that the gacS/gacA mutants had an early growth advantage compared to the wild type under rich nutrient conditions.
Replacement series analysis of gac+ and gac mutant 30-84 strains.
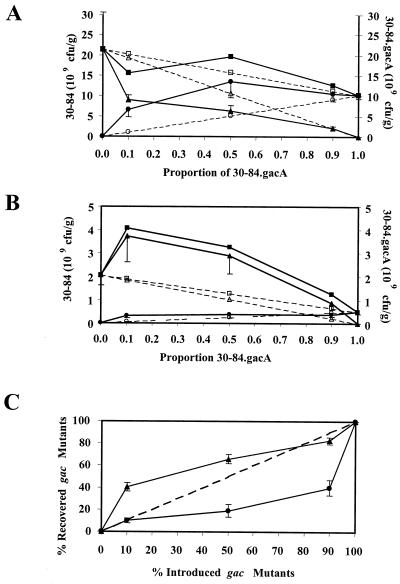
To determine whether gac mutants have a competitive advantage on plant roots, replacement series analysis was performed with wild-type 30-84 (gac+) and 30-84.gacA (gacA mutant, Kmr) strains. The wild-type and gac mutant strains were inoculated in specific ratios (1:0, 0.9:0.1, 0.5:0.5 0.1:0.9, and 0:1) on wheat roots grown in sterile (Fig. 2A) and nonsterile soils (Fig. 2B) for 14 days. Populations recovered from roots inoculated by each strain alone indicated the carrying capacity of the rhizosphere for each strain under these conditions. In sterile soil after 14 days, the rhizosphere population of the wild-type strain alone (gac+:gac mutant, 1:0) was twofold larger than the population of the gac mutant strain alone (gac+:gac mutant, 0:1). However, after 14 days in natural soil wild-type populations on roots reached levels fourfold larger than those of the gac mutants. Final rhizosphere populations of strain 30-84 and the gac mutant were 10- and 20-fold smaller, respectively, in natural versus sterile soil.
FIG. 2.
Replacement series between P. aureofaciens strains 30-84 and 30-84.gacA after 14 days in sterile (A) or natural (B) soil. Observed population sizes are represented by solid lines (filled symbols), and expected population sizes are represented by dashed lines (open symbols). Wild-type strain 30-84 populations are represented by triangles, 30-84.gacA mutants populations are represented by circles, and total populations are represented by squares. Expected values were calculated on the basis of population sizes of each strain inoculated alone and the proportion of the wild type and mutants in the original inoculum for each treatment. Each point represents the mean of eight replicates and includes positive standard error bars. (C) Comparison of the percentage of 30-84.gacA mutants in the mixture with that of the wild type introduced onto and recovered from roots grown in sterile (triangles) and nonsterile soil (circles) after 14 days. Expected values (dashed lines) are the same as the proportion introduced in each treatment. Each point represents the mean of eight replicates.
To assess the competitive fitness of the gac mutants and wild-type strains in mixtures, rhizosphere populations were determined after 14 days. The observed population sizes after 14 days were compared to predicted population sizes that were calculated on the basis of the final population of each strain alone and the proportion of each strain in the initial mixture. If there was no interaction (competition) between the two strains, the recovered populations would be expected to be in the same proportion as that when it was introduced. In sterile soil, final mutant (gac mutant) populations in mixtures were larger and wild-type populations were smaller than would be predicted, indicating a competitive advantage for gac mutants in the rhizosphere in sterile soil (Fig. 2A). However, in natural soil the wild-type populations were recovered from roots at higher levels than predicted (Fig. 2B). Interestingly, the wild-type populations were at least 1.5 times larger when coinoculated with the gac mutant than their predicted sizes on the basis of their inoculation ratio. Wild-type populations originating from mixtures where 30-84 composed at least 50% of the inoculum were larger than or equal to the population of the wild type alone despite lower inoculation rates. In contrast, gac mutants were recovered from roots at similar levels regardless of their proportion in the initial inoculum (Fig. 2B), suggesting that better survival of the wild type was not due simply to competitive displacement of the mutant.
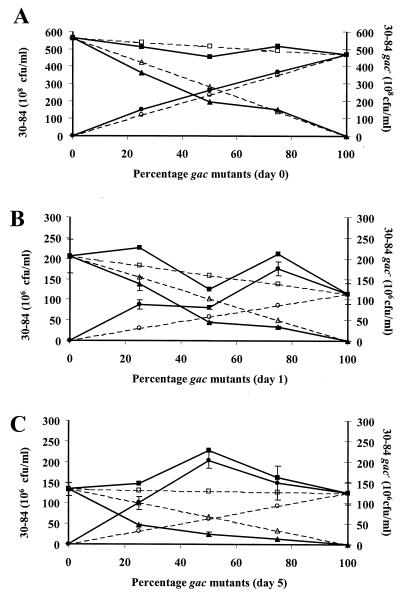
The effect of the indigenous microbial community on the survival of gac mutants can be determined by comparing the percentage of gac mutants recovered from roots in sterile versus nonsterile soil (Fig. 2C). In sterile soil, the percentage of gac mutants recovered was generally greater than the percentage introduced, whereas the opposite was observed in natural soil. In separate experiments, we examined gac mutants and wild-type rhizosphere populations over shorter time intervals in sterile soil to determine how quickly mutants became prominent on roots. After 24 h in sterile soil, gac mutant populations are larger and wild-type rhizosphere populations are generally smaller than what would be predicted (Fig. 3A and B). By day 5, the competitive advantage of the gac mutants in sterile soil is well established (Fig. 3C). We were able to differentiate our kanamycin-resistant strain 30-84.A2 (gac mutant) from spontaneous mutants arising de novo or introduced within the wild-type inoculum. Despite the lack of detectable kanamycin-sensitive mutants in our original inoculum, kanamycin-sensitive mutants were present on plant roots treated with the wild type alone in every experiment. By 10 to 14 days, kanamycin-sensitive mutants comprised less than 1, 3, 26, and 35% of the wild-type population on roots in sterile soil (four experiments) and less than 1 and 17% of the wild-type population on roots in natural soil (two experiments).
FIG. 3.
Replacement series between P. aureofaciens strains 30-84 and 30-84.A2 in sterile soil after inoculation (A), after 1 day (B), and after 5 days (C). Observed population sizes are represented by solid lines, and expected population sizes are represented by dashed lines. Wild-type strain 30-84 populations are represented by triangles, 30-84.A2 mutant populations are represented by circles, and total populations are represented by squares. Expected values were calculated on the basis of population sizes of each strain inoculated alone and the proportion of the wild type and mutants in the original inoculum for each treatment. Each point represents the mean of three replicates and includes standard error bars.
DISCUSSION
Mutations in gacS or gacA have been shown to occur at a high frequency in a diverse range of pseudomonads (2, 3, 6, 15, 22). Evidence from multiple studies has demonstrated that the GacA/GacS system controls the expression of genes required for the synthesis of secondary metabolites and other traits required for successful colonization and biological control (reviewed in reference 11). Further, evidence from independent studies has demonstrated that adequate establishment of bacterial populations and production of these metabolites on roots is important for successful control of pathogenic fungi (reviewed in reference 26). Since this global two-component regulatory system controls multiple traits required for successful biological control of plant pathogens in strain 30-84, the occurrence of such mutants could reduce the efficacy of this biological control agent if mutants could competitively displace wild-type strains in the rhizosphere. Previous work demonstrated that mutations in these genes accumulate during culture but that their formation can be limited by utilizing low-nutrient conditions (6). However, there is little information regarding the rate of occurrence of gacS or gacA mutations or their success in the rhizosphere.
Comparisons of 14-day rhizosphere populations composed of 100% wild type to those composed of 100% gac mutants in sterile and natural soils suggest that a functional Gac system confers a survival advantage, especially in the presence of the indigenous rhizosphere microflora. These results are consistent with previous work with strain 30-84 describing the competitive advantages conferred by phenazine production in natural versus sterile soil (18). Work with gacS/gacA mutants of strain P. fluorescens CHA0 also demonstrated that the rhizosphere survival of mutants was similar to that of the wild type in sterile bulk soil but was diminished in natural soil (19). Interestingly, survival of mutants was no different than that of the wild type in the rhizosphere of several natural soils in their study.
Replacement series analysis was used to assess the competitive fitness of the gac mutants and wild-type strains in mixtures. In sterile soil, final gac mutant populations in mixtures were larger and wild-type gac+ populations were smaller than would be predicted, indicating a competitive advantage for gac mutants under these conditions. This competitive advantage was evident by 24 h and was fully established within 5 days. Mutations in gacS/gacA lead to increased production of fluorescent compounds, possibly pyoverdin siderophores in strain 30-84 (3) and other pseudomonads (6, 24). Reduced energy expenditure due to the lack of secondary metabolite production and increased production of fluorescent siderophores or other beneficial compounds are all plausible hypotheses for the competitive success of gac mutants relative to the wild type in the absence of the indigenous rhizosphere competitors.
In contrast to sterile soil, wild-type gac+ populations in natural soil were larger than predicted, and gac mutants accounted for less than 40% of the total population and reached a constant population size regardless of their inoculation rate. These results indicate that gac mutants can survive and compete with the wild type in sterile soil, but in a rhizosphere in competition with other microorganisms they do not displace the wild-type population. In fact, in natural soil final rhizosphere populations of wild-type 30-84 starting from mixtures were at least 1.5 times larger than would be predicted from their inoculation ratio. Surprisingly, final populations of the wild type originating from 90:10 and 50:50 mixtures with mutants were larger than or equal to the population of 30-84 inoculated alone, despite their lower inoculation rate. These results demonstrate that the presence of gac mutants promote the survival of wild-type populations in the rhizosphere in the presence of the indigenous microflora.
Given the increased fitness of the wild type in mixtures with gac mutants and the constancy of the mutant population size in the rhizosphere, it is intriguing to speculate whether gacS/gacA mutants may be a normal component of healthy P. aureofaciens 30-84 populations in the rhizosphere. If so, what selective pressure might be responsible for maintaining them? One hypothesis is that enhanced siderophore production by these mutants could improve the fitness of a mixed community by enhancing the ability of both member strains to acquire and sequester sufficient iron to limit the growth of competing microorganisms. We have also observed that high inoculation densities of wild-type strain 30-84 are somewhat phytotoxic to plant roots, whereas similar inoculum densities of gac mutants produce healthier plant roots. The role of the plant in the increased success of the wild-type and mutant populations over the 100% wild-type populations remains to be determined.
In our experiments, mixed wild-type and mutant inoculum contained potentially larger numbers of mutants than would be expected in careful inoculum preparation. However, mixed populations also occurred on our plants treated with wild-type inoculum alone. Despite the lack of detectable mutants in our wild-type inoculum, by 10 to 14 days mutants accounted for up to 35 and 17% of the rhizosphere population in sterile and natural soils, respectively. These data suggest that undetected mutants introduced within the original inoculum or arising de novo on roots can survive and proliferate on plant roots. Although we do not rule out the possibility of mutants in our initial inoculum, several lines of evidence support the idea that some of the mutations we observed occurred during growth in the rhizosphere. First, in all experiments the number of mutants present in our initial inoculum was below our estimated level of detection of 0.01%. Second, no mutants were observed on plants prior to day 5 in sterile soil, where they have a competitive advantage. Third, the percentage of mutants recovered per plant in a single experiment was highly variable (for example, 0 to 36%; Fig. 1). Thus, the occurrence of mutants was not evenly distributed, as would be expected if all the mutations had occurred in the original inoculum. This suggests that mutations occurred on different plants after different lengths of time. Mutants that occurred early after inoculation would be analogous to the jackpot cultures described by Luria and Delbruck (16). Finally, Southern analysis of chromosomal DNA fragments from gacA mutants isolated from roots revealed three likely mutation types (insertion, deletion, and point mutation). Together these results suggest that gacS/gacA mutants isolated from the wheat rhizosphere of plants treated only with wild-type inoculum not only originated from the inoculum but also arose on plant roots.
Conserved mechanisms in generating reversible mutations in two-component systems similar to gacS/gacA have been reported. For example, Pseudomonas tolaasii, the causative agent of mushroom brown blotch, inversely regulates the expression of virulence factors and motility via the reversible insertion of a 661-bp duplication within pheN, the sensor-kinase member of a two-component system (9). Agrobacterium tumefaciens C58 utilizes the plasmid-borne VirA/VirG two-component system to regulate vir gene expression. Upon exposure to high acetosyringone levels, virA or virG become inactivated via insertion of IS426, a 1.3-kb insertion element (7). However, in contrast to characteristics of these systems we observed (i) no single mechanism of gacS/gacA mutation and (ii) no revertants back to the wild-type phenotype in P. aureofaciens strain 30-84. Similar results were observed for P. fluorescens CHA0 (2).
These experiments are some of the first to address the relative competitive survival of gacS/gacA mutants and wild-type strains in the plant rhizosphere. Replacement series experiments are an elegant way to compare the competitive fitness of two species under various environmental conditions (10, 28). Treatments of test strains introduced alone demonstrate the carrying capacity of the roots for each strain in the absence of influence from the other and have typically been used to infer relative rhizosphere competence. However, replacement series of specific proportions of each strain provide information on the relative competitive fitness of each strain in mixture. Our experiments demonstrated that gacS/gacA mutants can survive in natural rhizosphere populations, but these mutants do not displace wild-type populations and therefore are unlikely to reduce the efficacy of biological control by competitive displacement of wild-type strain 30-84. Better survival of the wild type in mixtures with mutants suggests the intriguing possibility that mutants arising de novo or introduced within the inoculum may be a normal, beneficial component of rhizosphere populations of strain 30-84. However, a better understanding of the ecological role of gacS/gacA mutants will require a more comprehensive understanding of the phenotypes controlled by these systems, the signals perceived by these systems, and the mechanisms for generating mutations in these genes.
Acknowledgments
We thank Patricia Figuli and Cheryl Whistler for technical assistance and Cheryl Whistler for critical review of early drafts of the manuscript.
This work was supported by U.S. Department of Agriculture NRICGP grants 98-02129 and 2001-02684.
REFERENCES
- 1.Aarons, S., A. Abbas, C. Adams, A. Fenton, and F. O'Gara. 2000. A regulatory RNA (PrrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J. Bacteriol. 182:3913-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull, C. T., B. Duffy, C. Voisard, G. Défago, C. Keel, and D. Haas. 2001. Characterization of spontaneous gacS and gacA regulatory mutants of Pseudomonas fluorescens biocontrol strain CHA0. Antonie Leeuwenhoek 79:327-336. [DOI] [PubMed] [Google Scholar]
- 3.Chancey, S. T., D. W. Wood, and L. S. Pierson III. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbell, N., and J. E. Loper. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for Gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy, B. K., and G. Défago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortin, C., C. Marquis, E. W. Nester, and P. Dion. 1993. Dynamic structure of Agrobacterium tumefaciens Ti plasmids. J. Bacteriol. 175:4790-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaffney, T. D., S. T. Lam, J. Ligon, K. Gates, A. Frazelle, J. Di Maio, S. Hill, S. Goodwin, N. Torkewitz, A. M. Allshouse, H. Kempf, and J. O. Becker. 1994. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol. Plant-Microbe Interact. 7:455-463. [DOI] [PubMed] [Google Scholar]
- 9.Han, B., A. Pain, and K. Johnstone. 1997. Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol. Microbiol. 25:211-218. [DOI] [PubMed] [Google Scholar]
- 10.Harper, J. L. 1977. Population biology of plants. Academic Press, London, United Kingdom.
- 11.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 12.Hoagland, D. R., and K. I. Arnon. 1950. The water-culture method for growing plants without soil. University of California Experiment Station circular 347, revised edition. University of California Experiment Station, Davis, Calif.
- 13.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on beans is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 28:301-307. [PubMed] [Google Scholar]
- 15.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Défago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luria, S., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resisitance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Mazzola, M., R. J. Cook, L. S. Thomashow, D. M. Weller, and L. S. Pierson. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58:2616-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Défago. 1994. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierson, L. S., III, V. D. Keppenne, and D. W. Wood. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierson, L. S., III, and L. S. Thomashow. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant-Microbe Interact. 5:330-339. [DOI] [PubMed] [Google Scholar]
- 22.Rich, J. J., T. G. Kinscherf, T. Kitten, and D. K. Willis. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the gacS sensor in Pseudomonas syringae. J. Bacteriol. 176:7468-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schleif, R. F., and P. C. Wensink. 1981. Practical methods in molecular biology. Springer-Verlag, New York, N.Y.
- 24.Schmidli-Sacherer, P., C. Keel, and G. Défago. 1997. The global regulator GacA of Pseudomonas fluorescens CHA0 is required for suppression of root diseases in dicotyledons but not in gramineae. Plant Pathol. 46:80-90. [Google Scholar]
- 25.Staskawicz, B., D. Dahlbreck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomashow, L. S., and D. M. Weller. 1996. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites, p. 187-235. In G. Stacey and N. Keen (ed.), Plant-microbe interactions, vol. 1. Chapman & Hall, New York, N.Y.
- 27.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators of GacS and GacA influence accumulation of the stationary-phase sigma factor σs and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, M., and S. E. Lindow. 1994. Ecological similarity and coexistence of epiphytic ice-nucleating (Ice+) Pseudomonas syringae strains and a non-ice nucleating (Ice−) biological control agent. Appl. Environ. Microbiol. 60:3128-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood, D. W., F. Gong, M. M. Aykin, P. Williams, and L. S. Pierson III. 1997. N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 179:7663-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood, D. W., and L. S. Pierson III. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]