Abstract
Mealybugs (Hemiptera, Coccoidea, Pseudococcidae), like aphids and psyllids, are plant sap-sucking insects that have an obligate association with prokaryotic endosymbionts that are acquired through vertical, maternal transmission. We sequenced two fragments of the genome of Tremblaya princeps, the endosymbiont of mealybugs, which is a member of the β subdivision of the Proteobacteria. Each of the fragments (35 and 30 kb) contains a copy of 16S-23S-5S rRNA genes. A total of 37 open reading frames were detected, which corresponded to putative rRNA proteins, chaperones, and enzymes of branched-chain amino acid biosynthesis, DNA replication, protein translation, and RNA synthesis. The genome of T. princeps has a number of properties that distinguish it from the genomes of Buchnera aphidicola and Carsonella ruddii, the endosymbionts of aphids and psyllids, respectively. Among these properties are a high G+C content (57.1 mol%), the same G+C content in intergenic spaces and structural genes, and similar G+C contents of the genes encoding highly and poorly conserved proteins. The high G+C content has a substantial effect on protein composition; about one-third of the residues consist of four amino acids with high-G+C-content codons. Sequence analysis of DNA fragments containing the rRNA operon and adjacent regions from endosymbionts of several mealybug species suggested that there was a single duplication of the rRNA operon and the adjacent genes in an ancestor of the present T. princeps. Subsequently, in one mealybug lineage rpS15, one of the duplicated genes, was retained, while in another lineage it decayed. These results extend the diversity of the types of endosymbiotic associations found in plant sap-sucking insects.
Mealybugs (Hemiptera, Coccoidea, Pseudococcidae) are plant sap-sucking insects which have a novel symbiotic association (20, 21, 47, 48). Within the body cavity of the insect is a large multicellular structure called a bacteriome that is made up of cells called bacteriocytes. Within the bacteriocytes are host-derived vesicles containing the gram-negative primary endosymbiont Tremblaya princeps (48; M. Thao, P. J. Gullan, and P. Baumann, submitted for publication). This organism is a member of the β subdivision of the Proteobacteria (35;Thao et al., submitted). Remarkably, T. princeps may harbor within its cells other gram-negative bacteria (secondary endosymbionts) belonging to the γ subdivision of the Proteobacteria (18, 48; Thao, submitted). Recently, using 16S-23S ribosomal DNA (rDNA) sequences, we have examined the evolutionary relationships of T. princeps from 22 species of mealybugs and the secondary endosymbionts from 12 of these species. The results suggest that the symbiotic association between T. princeps and mealybugs is a result of a single infection of an insect host 100 to 200 million years ago (Thao et al., submitted). In contrast to this result, it appears that infection of T. princeps with different precursors of the secondary endosymbionts occurred multiple times and that following infection there was cotransmission of both endosymbionts (Thao et al., submitted).
Mealybugs are members of the suborder Sternorrhyncha (Hemiptera), which contains a number of other plant sap-sucking insect families, including psyllids and aphids (20, 21). The diet of these insects is high in carbohydrates and low in essential amino acids. All of these insects harbor endosymbionts within bacteriocytes; there is good evidence that in aphids one of the functions of the endosymbionts is synthesis of essential amino acids for the host (16, 37). Evolutionary studies of both psyllids and aphids suggest that the endosymbiotic association is a consequence of a single infection of an ancient ancestor, followed by vertical evolution of the endosymbiont and the host (7, 32, 46). In these organisms, as well as in mealybugs, the endosymbionts are transmitted maternally.
The 641-kb genome of the endosymbiont of aphids (Buchnera aphidicola) has been sequenced (40); its G+C content is 26.3 mol%, which is at the lower end of the range of G+C contents of free-living bacteria (24). The sequence of 37 kb of DNA from the endosymbiont of psyllids (Carsonella ruddii) has recently been determined (12). The genome of this endosymbiont has an unusual organization, and the G+C content of its DNA is 19.9 mol%, a value lower than that of any known prokaryote (12). Both B. aphidicola and C. ruddii are members of the γ subdivision of the Proteobacteria. Because T. princeps is a member of the β subdivision and because of the higher G+C content of its 16S-23S rDNA, we decided to determine the nucleotide sequence of a fragment of its genome and compare the results with the data obtained for C. ruddii and B. aphidicola. The results indicate that T. princeps differs from these two endosymbionts in a number of properties.
MATERIALS AND METHODS
General methods.
Standard molecular biology methods were used in this study (3). Additional methods have been described elsewhere (7, 12; Thao et al., submitted). These methods include isolation of total mealybug DNA, restriction enzyme and Southern blot analyses, and cloning into λZAP (Stratagene, La Jolla, Calif.). The nucleotide sequence of T. princeps DNA was determined at the University of Arizona (Tucson) LSME sequencing facility. In addition to the T3 and T7 primers, custom-made oligonucleotide primers were also designed for sequencing. For sequence determination of most of the DNA fragments, a double-stranded DNA nested deletion kit (Pharmacia, Piscataway, N.J.) was used. The reaction mixtures and the PCR conditions used have been described previously (12).
Most of the sequence data were for T. princeps from the mealybug Dysmicoccus brevipes. Additional sequence data were for T. princeps from Melanococcus albizziae, Planococcus citri, Maconellicoccus australiensis, and Maconellicoccus hirsutus. Both species of Maconellicoccus lacked a secondary endosymbiont (Thao et al., submitted). The sources of these mealybugs will be described elsewhere (Thao et al., submitted).
General approach.
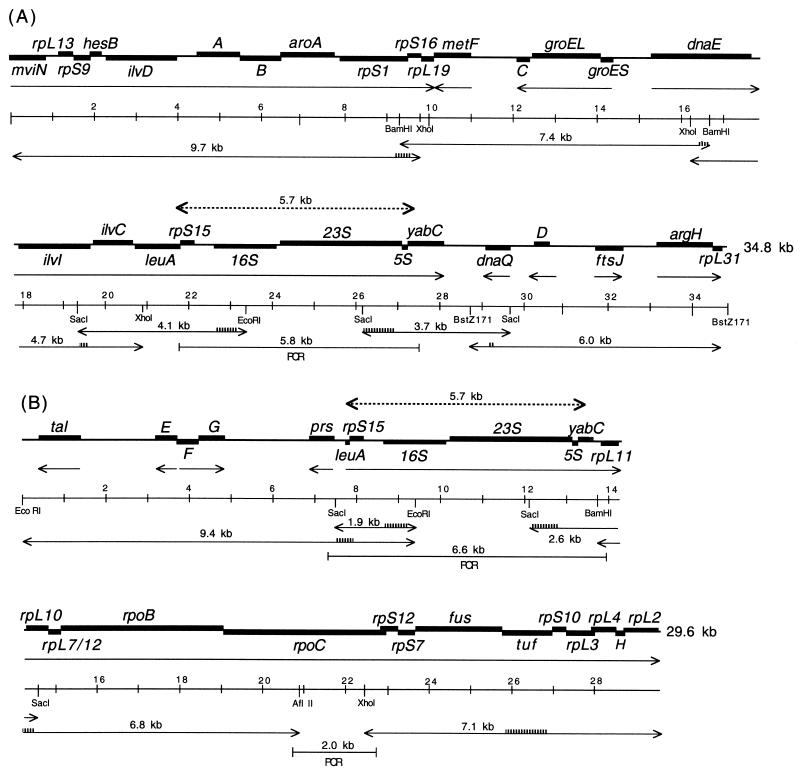
We have obtained the sequence of a 4-kb 16S-23S rDNA-containing DNA fragment from T. princeps of D. brevipes (Thao et al., submitted). In previous studies of B. aphidicola (6) and C. ruddii (12) only one copy of the rRNA genes was detected by restriction enzyme and Southern blot analyses. By using the strategy used for C. ruddii (12), we hoped to extend the sequence upstream and downstream of the 16S-23S rDNA of T. princeps from D. brevipes. Restriction enzyme and Southern blot analyses performed with probes for 16S rDNA indicated the presence of two SacI-EcoRI fragments at 4.1 kb (Fig. 1A) and 1.9 kb (Fig. 1B), which is consistent with the presence of two copies of 16S-23S rDNA genes. Similar analyses with a probe for 23S rDNA indicated the presence of two SacI fragments at 3.7 kb (Fig. 1A) and 2.6 kb (Fig. 1B). These fragments were cloned into λZAP (Stratagene) and sequenced. Subsequently, the sequences of overlapping DNA fragments upstream or downstream of these DNA fragments were determined. In brief, the methods used consisted of finding a convenient restriction site within the sequenced DNA fragment, obtaining a DNA fragment that served as a probe, and performing a restriction enzyme and Southern blot analysis. Restriction enzyme-digested DNA fragments of the appropriate size (4.7 to 9.7 kb) were eluted from agarose gels, cloned into λZAP, and sequenced (Fig. 1) (12). The nucleotide sequences of the primers used for making the probes are available upon request.
FIG. 1.
Genetic maps of DNA fragments from T. princeps from D. brevipes. The thick lines indicate structural genes; the arrows with one arrowhead indicate the direction of transcription; the arrows with two arrowheads indicate overlapping DNA fragments that were cloned into λZAP and sequenced; the striped lines on arrows with two arrowheads indicate the positions of probes; the dashed lines with two arrowheads indicate an identical 5.7-kb sequence; and the lines with brackets indicate DNA fragments obtained by PCR.
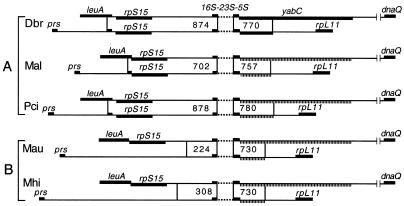
FIG. 2.
Genetic map of the upstream and downstream portions of the duplicated regions containing 16S and 23S rDNAs. A and B are two different clusters established on the basis of the evolutionary relationships of T. princeps (Thao et al., submitted). The numbers in the boxes indicate the lengths of fragments having identical nucleotide sequences from the 16S (upstream) and 23S (downstream) oligonucleotide primers used in the PCRs. The dashed lines indicate missing sequence; the striped lines indicate pseudogenes. T. princeps was obtained from the following mealybug species: D. brevipes (Dbr), M. albizziae (Mal), P. citri (Pci), M. australiensis (Mau), and M. hirsutus (Mhi).
Sequences upstream and downstream of 16S-23S rDNA.
The leuA-16S DNA (Fig. 2) was amplified by PCR by using the following oligonucleotide primers: leuA (XbaI; 5′-GTA TCT AGA GGN ATH CAY CAR GAY GGN G-3′) and U16S (5′-GCC GTM CGA CTW GCA TGT G-3′) containing an EcoRI site (Pci and Mau [Fig. 2]) or a BamHI site (Pci and Mau [Fig. 2]) added to the 5′ end. Similarly, the prs-16S DNA (Fig. 2) was amplified by using prs5 (KpnI; 5′-GTA GGT ACC GCT WRA GTG GAG GTC CAT TGC-3′) or prs6 (KpnI; 5′-GTA GGT ACC GAT ATC CTG CGC GCK AGT C-3′) and primer U16S containing an added EcoRI site (Pci, Mau, and Mhi [Fig. 2]) or a BamHI site (Mal [Fig. 2]). The 23S-dnaQ DNA (Fig. 1A and 2) was amplified by using the following oligonucleotide primers: B23S (5′-GTT TGG CAC CTC GAT GTC G-3′) containing an EcoRI site (Pci [Fig. 2]) or a BamHI site (Mal, Mau, and Mhi [Fig. 2]) at the 5′ end and dnaQ-JH (XbaI; 5′-GTA TCT AGA GTN YTN GAY ACN GAR ACN ACN G-3′) (Pci and Mhi [Fig. 2]) or dnaQ-A (XbaI; 5′-GTA TCT AGA GCA GAG GTG GGG TGC GTG GAG-5′) (Mal and Mau [Fig. 2]). Similarly, the DNA from 23S-rpL11 (Fig. 2) was amplified by using primer B23S-JH and primer rpL-11 (XbaI; 5′-GTA TCT AGA GTN GCN GCR TTR AAN GCY TTA SA-3′). After digestion with the appropriate restriction enzymes, the inserts were cloned into pBluescript (Stratagene), and the nucleotide sequences were determined.
Analysis of the DNA.
We used GeneJockey II (Biosoft, Ferguson, Mo.) to identify open reading frames (ORFs) and Blast searches (National Center for Biotechnology Information, Bethesda, Md.) to identify proteins with amino acid sequence similarity. Alignment of amino acids was performed by using Gap (Genetics Computer Group, Madison, Wis.). In comparative studies sequences of B. aphidicola (accession no. AF000398), Neisseria meningitidis (AE002098), and C. ruddii (AF274444, AF291051, and AF211141) were also included.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences of the fragments obtained in this study are as follows: T. princeps from D. brevipes, AF481102 and AF481103; T. princeps from M. albizziae, AF481907, AF481911, AY090468, and AY090468; T. princeps from P. citri, AF481908, AF481912, AY079511, and AY079513; T. princeps from M. australiensis, AF481909, AF481913, AY090469, and AY090470; and T. princeps from M. hirsutus, AF481910, AF481914, AY079512, and AY079514.
RESULTS
General properties of T. princeps DNA.
We sequenced two T. princeps DNA fragments, one of which is 34,806 nucleotides (nt) long (Fig. 1A) and one of which is 29,559 nt long (Fig. 1B) (total, 64,365 nt). Searches of databases (in December 2001) identified 37 ORFs as corresponding to known genes; 36 of these ORFs were found in N. meningitidis, the nearest relative having a fully sequenced genome (Thao et al., submitted), and 1 ORF (yabC) was found in the Escherichia coli genome. The two DNA fragments had an identical 5.7-kb region, which contained the genes for 16S, 23S, and 5S rDNA (Fig. 1). The total G+C content of the two fragments was 57.1 mol%. There are three large gene clusters that are transcribed from left to right consisting of mviN-rpL19 and dnaE-yabC (Fig. 1A), as well as rpS15-rpL2 (Fig. 1B); the remaining genes are transcribed in either direction.
General properties of the ORFs.
A list of the T. princeps genes together with their G+C contents and percentages of amino acid sequence identity to N. meningitidis homologs is presented in Table 1. The G+C contents of the genes ranged from 54.2 mol% (rpL11) to 59.9 mol% (rpS10). The proteins having the most conserved sequences are Tuf, RpS12, and GroEL (75.3, 74.8, and 71.7% amino acid identity to N. meningitidis proteins, respectively). The least conserved proteins are Tal, DnaQ, and Prs (21.6, 26.3, and 28.4% amino acid identity to N. meningitidis proteins, respectively). Eight ORFs were detected that could not be readily equated with genes in the databases (ORF-A to -H) (Fig. 1). The putative proteins encoded by these ORFs contained 343 (ORF-A), 333 (ORF-B), 67 (ORF-C), 120 (ORF-D), 174 (ORF-E), 165 (ORF-F), 208 (ORF-G), and 85 (ORF-H) amino acids.
TABLE 1.
Genes found in T. princeps
| Gene | Protein | G+C content (mol%) | % Amino acid identitya |
|---|---|---|---|
| Pentose phosphate nonoxidative branch | |||
| tal | Transaldolase | 54.5 | 21.6 |
| Amino acid biosynthesis | |||
| Glutamate family | |||
| argH | Arginosuccinate lyase | 59.3 | 43.6 |
| Aromatic amino acid family | |||
| aroA | 5-Enolpyruvylshikimate-3-phosphate synthase | 57.3 | 29.3 |
| Branched-chain family | |||
| ilvI | Acetolactate synthase III (subunit), valine sensitive | 56.4 | 35.3 |
| ilvC | Ketol-acid reductoisomerase | 55.6 | 55.5 |
| ilvD | Dihydroxyacid dehydrase | 57.3 | 39.0 |
| leuA | α-Isopropylmalate synthase | 57.9 | 43.1 |
| Purine ribonucleotide biosynthesis | |||
| prs | Phosphoribosylpyrophosphate synthetase | 58.2 | 28.4 |
| Central intermediary metabolism | |||
| metF | 5,10-Methylenetetrahydrofolate reductase | 56.8 | 31.5 |
| Chaperones | |||
| groEL | Chaperone, Hsp 60 | 57.6 | 71.7 |
| groES | Chaperone, Hsp 10 | 55.3 | 57.9 |
| Cell division | |||
| ftsJ | Cell division protein | 55.2 | 30.6 |
| rRNA | |||
| rrf | 5S rRNA | 56.9 | 58.8b |
| rrl | 23S rRNA | 55.8 | 75.8b |
| rrs | 16S rRNA | 55.3 | 80.6b |
| Ribosomal proteins | |||
| rpS1 | Ribosomal protein S1 (rpsA) | 56.6 | 42.8 |
| rpS7 | Ribosomal protein S7 (rpsG) | 57.8 | 43.2 |
| rpS9 | Ribosomal protein S9 (rpsI) | 59.7 | 54.3 |
| rpS10 | Ribosomal protein S10 (rpsJ) | 59.9 | 32.0 |
| rpS12 | Ribosomal protein S12 (rpsL) | 59.0 | 74.8 |
| rpS15 | Ribosomal protein S15 (rpsO) | 52.7 | 39.3 |
| rpS16 | Ribosomal protein S16 (rpsP) | 57.7 | 40.3 |
| rpL2 | Ribosomal protein L2 (rplB) | 58.1 | 53.6 |
| rpL3 | Ribosomal protein L3 (rplC) | 58.4 | 35.8 |
| rpL4 | Ribosomal protein L4 (rplD) | 58.8 | 32.2 |
| rpL7/12 | Ribosomal protein L7/12 (rplL) | 54.9 | 43.3 |
| rpL10 | Ribosomal protein L10 (rplJ) | 54.5 | 38.1 |
| rpL11 | Ribosomal protein L11 (rplK) | 54.2 | 40.7 |
| rpL13 | Ribosomal protein L13 (rplM) | 55.2 | 35.8 |
| rpL19 | Ribosomal protein L19 (rplS) | 57.8 | 30.0 |
| rpL31 | Ribosomal protein L31 (rpmE) | 58.8 | 27.0 |
| DNA replication | |||
| dnaE | DNA polymerase III, α-subunit | 58.0 | 38.4 |
| dnaQ | DNA polymerase III, ɛ-subunit | 59.0 | 26.3 |
| Protein translation | |||
| fus | Elongation factor G | 55.7 | 57.6 |
| tuf | Elongation factor Tu | 54.6 | 75.3 |
| RNA synthesis | |||
| rpoB | RNA polymerase, β-subunit | 57.2 | 36.3 |
| rpoC | RNA polymerase, β′-subunit | 57.8 | 43.7 |
| Miscellaneous | |||
| hesB | Putative protein | 58.7 | 31.2 |
| mviNd | Putative virulence protein | 57.3 | 30.0 |
| yabC | Putative protein | 59.1 | 25.0c |
Unless otherwise indicated, amino acids were compared to the amino acids of N. meningitidis proteins.
Compared to rRNAs.
Compared to the amino acids of E. coli protein.
Partial sequence.
Amino acid composition and codon usage.
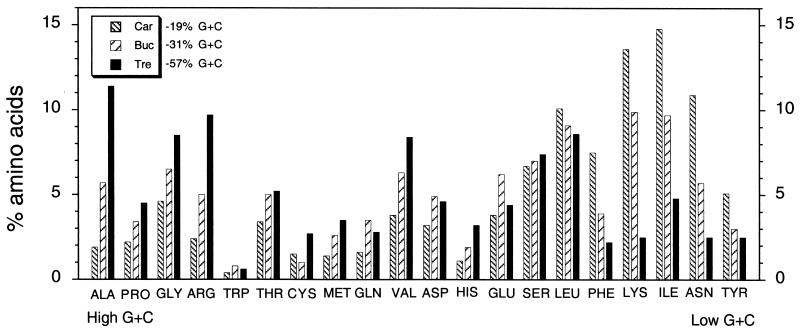
The G+C content of the coding regions has an influence on the codon usage and amino acid composition of proteins (42). A comparison of the amino acid compositions and the G+C contents of codons of proteins of C. ruddii, B. aphidicola, and T. princeps is presented in Fig. 3. T. princeps differs from the other endosymbionts in having substantially greater alanine, glycine, and arginine contents and, to a lesser extent, a greater proline content (codons with a high G+C content) than B. aphidicola and C. ruddii (34.1, 20.6, and 11.1%, respectively, of the total proteins). Similarly, T. princeps differs from B. aphidicola and C. ruddii in having the lowest content of phenylalanine, lysine, isoleucine, asparagine, and tyrosine (codons with a high A+T content) (14.5, 32.2, and 51.9%, respectively, of the total proteins). These results correlate with the G+C contents of the DNAs which encode these proteins (Fig. 3).
FIG. 3.
Amino acid compositions and A+T and G+C contents of codons of proteins of C. ruddii (Car), B. aphidicola (Buc), and T. princeps (Tre). The G+C contents are the G+C contents for the total coding regions. The data for C. ruddii are from reference 12; the data for T. princeps are from this study; and the data for B. aphidicola are data for the genes present in C. ruddii and T. princeps (40).
Protein size.
Prs and LeuA of T. princeps are reduced in size, having 61 and 75% of the amino acid content, respectively, of N. meningitidis proteins. Prs appears to have a 92-amino-acid deletion following amino acid 181 of the N. meningitidis protein. The smaller size of T. princeps LeuA is due to truncation at the C terminus. DnaQ of N. meningitidis has 470 amino acids, while the E. coli enzyme has 246 amino acids. The size of T. princeps DnaQ (203 amino acids) resembles the size of the E. coli enzyme; the reduction in the size of T. princeps DnaQ is also due to truncation at the C terminus. The sizes of the remaining 36 proteins of T. princeps range from 86 to 117% of the sizes of the homologous N. meningitidis proteins. A summation of the total amino acids of these proteins from T. princeps and N. meningitidis indicates that the amino acid content of the T. princeps proteins is on average 3.7% less than that of the N. meningitidis proteins. Compared to E. coli, a substantial decrease in protein size has been observed in C. ruddii (9.5%) (12), and a smaller decrease has been observed in B. aphidicola (8).
Intergenic spaces.
Of the DNA sequenced, 81.6% corresponded to coding regions for proteins and rRNAs. The G+C content of the intergenic spaces was 57.4 mol%, which is similar to the G+C content of the coding regions (57.0 mol%). There was considerable variation in the sizes of the intergenic spaces. If we excluded the major segments which did not contain identifiable genes (ilvD-aroA, metF-groEL, yabC-dnaQ, dnaQ-ftsJ, and tal-prs), then the intergenic spaces ranged from 2 to 730 nt long (22 cases). There were also overlaps between the start and stop codons of adjacent genes, which ranged from −2 to −185 nt (nine cases). The largest overlap was between rpoC and rpS12 (Fig. 1B).
Duplication of the rRNA genes.
In T. princeps from D. brevipes, there is a 5,689-bp duplication involving the transfer of a copy of this segment from fragment A (Fig. 1A) to fragment B (Fig. 1B). The sequences of these two segments are identical. The excised fragment contains the end of leuA, rpS15-16S-23S-5S, and the beginning of yabC (Fig. 1A). This sequence is inserted downstream of prs and upstream of rpL11 (Fig. 1B). We investigated this gene duplication further by cloning and sequencing DNA fragments upstream of the two copies of the rRNA genes from T. princeps (leuA-16S, prs-16S) and downstream of the two copies of the rRNA genes (23S-dnaQ, 23S-rpL11) from four additional mealybug species (Thao et al., submitted). Figure 2 is a summary of the results. In T. princeps from D. brevipes, M. albizziae, and P. citri the regions of sequence identity between leuA-16S and prs-16S are different lengths (874, 702, and 878 nt), but all begin in similar positions at the end of leuA and all include the end of this gene as well as rpS15. In the different lineages represented by T. princeps from M. australiensis and M. hirsutus, the size of the region of sequence identity is reduced (224 and 308 nt) and the prs-16S DNA fragments lack rpS15. Comparisons of 23S-dnaQ and 23S-rpL11 also indicated a different length for the region of sequence identity (730 to 780 nt) downstream of the 23S rDNA primer. In T. princeps from D. brevipes, following 5S rDNA is an ORF having some similarity to E. coli yabC, a gene having no known function (Fig. 3). In T. princeps from the remaining mealybug species (Fig. 3) this gene appeared to be in the process of degradation, with frameshifts and stop codons resulting in shorter ORFs corresponding to segments of yabC of T. princeps from D. brevipes.
rRNA operon.
Comparisons of sequences upstream of rRNA operons from B. aphidicola from different aphid species indicated that there was conservation of sequences resembling the −35,−10 promoter region (6, 7, 34). A similar comparison of C. ruddii from different psyllid species indicated that there was a lack of conserved sequences upstream of the rRNA operons (12). We used a similar approach with T. princeps (Fig. 2) and did not find conserved sequences resembling a −35,−10 promoter region upstream of the 16S rRNA genes. There was, however, conservation of two sequences; 16 bp upstream of the putative beginning of the 16S rRNA gene was the sequence CCCG, and 22 bp upstream of this sequence was another conserved sequence, AGGCTTTAGGT. The significance of these conserved sequences is not known. No inverted repeats, which are characteristic of rho-independent terminators, were detected in T. princeps following the 5S rDNA gene (Fig. 2). Such repeats have not been detected in C. ruddii (12) but were found in B. aphidicola (6, 7, 34).
DISCUSSION
General properties.
T. princeps has a unique combination of properties that distinguishes it from B. aphidicola and C. ruddii, the two other characterized endosymbionts of plant sap-sucking insects (Table 2). Among these properties are a DNA G+C content of 57.1 mol% and no significant difference between the G+C contents of the coding regions and intergenic spaces (57.0 and 57.4 mol%, respectively). The genes detected in the sequenced DNA segments (Fig. 1) are primarily those encoding housekeeping functions (Table 1), such as transcription (RNA polymerase), translation (rRNA, ribosomal proteins, elongation factors), chaperones (groESL), DNA replication, and cell division. In addition, genes for amino acid and purine ribonucleotide biosynthesis, as well as the pentose phosphate pathway, were detected. These genes have also been found in the sequenced genome of B. aphidicola (40). The order of the genes in the rpL11-rpL2 segment in T. princeps (Fig. 1B) is also found in C. ruddii (12) and N. meningitidis (22). A major difference is the presence of rpL23 in N. meningitidis following rpL4, the absence of this gene in C. ruddii, and its substitution by a gene encoding a putative protein (ORF-H) in T. princeps (Fig. 1B). In B. aphidicola the segments corresponding to T. princeps rpL11-rpoC and rpS12-rpL2 are separated. B. aphidicola has a sigma-32 promoter preceding groES (38); no such sequences were detected in T. princeps upstream of this gene.
TABLE 2.
Genetic characteristics of endosymbionts of plant sap-sucking insectsa
| Organism | Proteo- bacterial group | G+C content of DNA (mol%) | % Coding | G+C content of inter- genic spaces (mol%) | Recognizable −35, −10 region preceding rRNA genes | Inverted repeats following rRNA genes | 3′ end of 16S rDNA contains Shine-Dalgarno complement | Translational coupling the normc | Increased A+T content in poorly conserved regions |
|---|---|---|---|---|---|---|---|---|---|
| T. princeps | β | 57.1 | 81.6 | 57.4 | − | − | + | − | − |
| B. aphidicola | γ | 26.3 | 87.9 | 19.1b | + | + | + | − | ? |
| C. ruddii | γ | 19.9 | >99.9 | − | − | − | + | + |
Interpretations were based on analysis of sequence data.
Decreases of 4 to 9 mol% G+C were found in the intergenic spaces of the genomes of Rickettsia prowazekii, Chlamydia trachomatis, E. coli, Haemophilus influenzae, P. aeruginosa, N. meningitidis, Ralstonia solanacearum, and Clostridium acetobutylicum.
Suggested by the large number of gene pairs in which the initiation codon and the stop codon overlap (12).
Protein sequence conservation and G+C content.
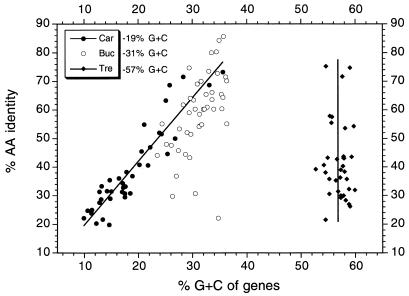
The ages of the endosymbiotic associations of T. princeps, C. ruddii, and B. aphidicola are approximately the same (100 to 230 million years) (7, 33, 36, 46; Thao et al., submitted). It appears that during this time the most radical alterations from a free-living ancestor have occurred in C. ruddii (12). This endosymbiont has a G+C content of 19.9 mol%, has essentially no intergenic spaces, and lacks the complement of a Shine-Dalgarno sequence in the 3′ end of its 16S rRNA. As a consequence of the latter finding, it is probable that long mRNAs are transcribed and translational coupling occurs (12). An additional feature of C. ruddii is a decrease in the G+C content of proteins with poorly conserved amino acid sequences. This is illustrated in Fig. 4, which shows that, as the level of amino acid sequence identity between the C. ruddii and E. coli homologous proteins decreases, the G+C content of the genes also decreases. This feature is absent in T. princeps (Fig. 4). Comparisons of the protein sequences of this endosymbiont with the sequences of homologous proteins of N. meningitidis indicate that, as the amino acid sequence identity decreases, there is little effect on the G+C content of the genes. B. aphidicola appears to occupy an intermediate position between these two extremes, with several poorly conserved genes having a relatively high G+C content (Fig. 4).
FIG. 4.
Comparisons of the levels of amino acid (AA) sequence identity of homologous proteins of C. ruddii and E. coli (Car), B. aphidicola and P. aeruginosa (Buc), and T. princeps and N. meningitidis (Tre) with the G+C contents of the genes of the endosymbiont.
Duplication of the rRNA genes.
Previously it was shown that, on the basis of evolutionary relationships of 16S-23S rDNA, T. princeps could be subdivided into two major clusters (Thao et al., submitted). The endosymbionts of D. brevipes, M. albizziae, and P. citri are representatives of cluster A, while the T. princeps endosymbionts of M. australiensis and M. hirsutus constitute cluster B (Fig. 2). Two copies of the 16S-23S-5S rRNA genes were found in all the T. princeps endosymbionts tested that were representatives of the two clusters (Fig. 2); the new copy was inserted between prs and rpL11. In members of cluster A it was evident that the duplication involved insertion of a segment containing the terminal portion of leuA followed by rpS15-16S-23S-5S rDNA and a portion of yabC (Fig. 1B and 2). In all of these organisms a segment containing an identical sequence was found in both copies, suggesting that concerted evolution resulted in sequence identity (19). There were, however, differences in the lengths of the identical sequences (Fig. 2, cluster A), suggesting that considerable changes occurred in the noncoding regions during evolution of the different lineages. In contrast to these results, cluster B T. princeps lacks rpS15 in the prs-16S rDNA fragments, and the region of sequence identity is consequently considerably reduced. The simplest interpretation of these results is that in the ancestor of the present T. princeps there was a duplication of the 16S-23S-5S rRNA genes. In cluster A rpS15 was preserved in the prs-16S rDNA region, while in the lineage leading to cluster B this region was degraded prior to divergence of M. australiensis and M. hirsutus. The physiological significance of this gene duplication is not understood. B. aphidicola and C. ruddii, the endosymbionts of aphids and psyllids, respectively, have only one copy of the rRNA genes (12, 40). Frequently, organisms that have low growth rates have few copies of the rRNA genes (6, 7, 25). It is possible that the demand for greater protein synthesis in an ancestral T. princeps led to duplication of rRNA genes and resulted in an increase in ribosomes.
Endosymbiosis and G+C content.
Recently, it has been suggested that there is a correlation between the intracellular lifestyle of prokaryotic mutualists and pathogens and low DNA G+C contents (1, 29, 31, 33). In the case of mutualistic endosymbionts this correlation is based on only a few examples. Cossart and Lecuit (15) performed a useful compilation of mammalian pathogens with respect to their intracellular locations. An examination of the G+C contents of these organisms indicates that obligately intracellular pathogens have G+C contents ranging from 29 to 58 mol% (26, 43, 45, 49). Facultatively intracellular pathogens have G+C contents ranging from 34 to 66 mol%, while extracellular pathogens have G+C contents ranging from 25 to 67 mol%. The total range of the G+C contents of prokaryotes is about 25 to 75 mol% (24). T. princeps, the endosymbiont of mealybugs, has a G+C content of 57.1 mol%. The weevil Sitophilus oryzae has an endosymbiont related to members of the Enterobacteriaceae which has a G+C content of 54 mol% (23). B. aphidicola and C. ruddii have G+C contents of 26.3 and 19.9 mol%, respectively (12, 40). Thus, the G+C contents of endosymbionts of insects which are currently known range from 20 to 57 mol%. This information, combined with the ranges for intracellular and facultatively intracellular pathogens (29 to 66 mol%), does not suggest that there is an unequivocal association between a low G+C content and an intracellular lifestyle.
Mutualistic intracellular endosymbionts, as well as some intracellular pathogens, appear to have increased rates of sequence change (29). This has been attributed to the population structure of these organisms and the functioning of Muller's ratchet (29). It has been suggested that the small number of these organisms transmitted to progeny results in reduced purifying selection, accumulation of deleterious mutations, and a reduction in the G+C contents of their DNAs (29). From compilations of the G+C contents of free-living prokaryotes and the properties of these organisms, it is clear that the extremes of G+C content are in themselves not deleterious to an organism. At the lower end of the G+C content range we find such common vigorous soil organisms as the clostridia (25 to 28 mol%) (50). It should also be noted that the mycoplasmas, whose low G+C contents have been attributed to their habitats within plants or animals, are themselves descended from the low-G+C-content gram-positive bacteria (27). At the upper end of the G+C content range are the vigorous and nutritionally highly versatile species Pseudomonas aeruginosa and the actinomycetes (9, 26, 43, 45, 49). Similarly, organisms growing well on minimal media and having G+C contents at the lower and higher ends of the G+C content range (28 and 68 mol%) were found in a taxonomic study of respiratory, gram-negative marine bacteria (5). Although no systematic comparisons have been made, there is no evidence that organisms with extreme G+C contents have functionally deficient enzymes. In the case of the endosymbionts, it appears to be necessary to compare the properties of some of the enzymes of these organisms to their homologous counterparts from related free-living bacteria in order to determine if their evolutionary history has led to decay of the enzymatic function.
One of the results that was interpreted as being consistent with operation of Muller's ratchet in B. aphidicola is the major increase in nonsynonymous substitutions relative to synonymous substitutions as compared to E. coli and Salmonella enterica serovar Typhimurium (14, 29, 33). This was interpreted to reflect a decrease in purifying selection due to the population structure that results in bottlenecks during the transmission of endosymbionts to progeny. Subsequent studies, however, have shown that the ratio of synonymous to nonsynonymous substitutions in the E. coli-S. enterica comparison is the exception and that the ratios in Buchnera are similar to those in other organisms (36).
Due to accelerated evolution and in some cases the decreased G+C content of the rDNA, there may be considerable uncertainty concerning the nearest relatives of some of the endosymbionts (30). This is especially true for C. ruddii (46), but it is somewhat less true for B. aphidicola. In the case of the latter organism, it is clear that members of the Enterobacteriaceae are close relatives and that the branches leading to Aeromonas hydrophila or Vibrio cholerae precede the branching of B. aphidicola and E. coli. Since members of the Enterobacteriaceae, A. hydrophila, and V. cholerae have G+C contents of 38 to 62 mol% (26, 43, 45, 49), it is reasonable to postulate that the G+C content of B. aphidicola (26.3 mol%) is a consequence of a decrease from an ancestor having a higher G+C content (14, 31). An adenine-thymine (AT) pressure is also suggested by the fact that intergenic spaces in B. aphidicola have a lower G+C content than coding regions (Table 2). Although a close ancestor of C. ruddii cannot be established with certainty, the exceptionally low G+C content of this organism also suggests high AT pressure. T. princeps is in marked contrast to C. ruddii and B. aphidicola. In this organism the intergenic spaces and coding regions have similar G+C contents (Table 2), suggesting the absence of AT pressure. Consistent with this interpretation and in contrast to C. ruddii, there is no increase in the A+T content of T. princeps genes coding for proteins that are poorly conserved (Fig. 4).
Determination of the sequence of the B. aphidicola genome and the relatively close relationship of this organism to E. coli have allowed attempts at reconstruction of the evolutionary events that led to the formation of the reduced endosymbiont genome from the genome of a postulated common ancestor (31, 41). These analyses indicated the importance of large deletions that may eliminate genes not essential for endosymbiont function (31). It has been suggested that such deletions may remove genes (or parts of genes) that have an effect on DNA replication-repair fidelity, and the increased A+T content may be the result (28, 31, 33, 51).
Mutualistic endosymbiotic associations between insects and bacteria are widespread and so far have involved members of the γ subdivision of the Proteobacteria (Buchnera, Carsonella, Blochmannia, Wigglesworthia), the β subdivision (Tremblaya), and the Flavobacterium-Bacteroides group (Blattabacterium) (4, 10, 11, 13, 17, 32, 39, 44, 46, 48). Some of these endosymbionts are distantly related, and consequently in principle there is nothing which suggests that members of diverse bacterial groups may not potentially be able to enter into mutualistic associations with insects. One theme that may be common to these associations is the reduction of the genome size (2, 31, 33), and future studies to determine the size of the T. princeps genome will be of interest. Based on the diversity of bacterial types and the stochastic nature of the deletion process, it may be that the endosymbionts have a variety of different genetic properties, as exemplified by the two current extremes (namely, C. ruddii and T. princeps).
Acknowledgments
This work was supported by National Science Foundation awards MCB-9807145 (P. Baumann) and DEB-9978518 (N. A. Moran and P. Baumann) and by the University of California Experiment Station (P. Baumann).
We are grateful to N. A. Moran and H. Ochman for suggestions and discussions and to D. Natale for determining the G+C contents of the intergenic spaces in a number of bacterial genomes.
REFERENCES
- 1.Akman, L., and S. Aksoy. 2001. A novel application of gene arrays: Escherichia coli array provides insight into the biology of the obligate endosymbiont of tsetse flies. Proc. Natl. Acad. Sci. USA 98:7546-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, S. G. E., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263-268. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2001. Current protocols in molecular biology. Wiley, New York, N.Y.
- 4.Bandi, C., G. Damiani, L. Magrassi, A. Grigolo, R. Fani, and L. Sacchi. 1994. Flavobacteria as intracellular symbionts of cockroaches. Proc. R. Soc. Lond. B Biol. Sci. 257:43-48. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, L., P. Baumann, M. Mandel, and R. D. Allen. 1972. Taxonomy of aerobic marine bacteria. J. Bacteriol. 110:402-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann, P., L. Baumann, C. Y. Lai, D. Rouhbakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 49:55-94. [DOI] [PubMed] [Google Scholar]
- 7.Baumann, P., N. A. Moran, and L. Baumann. 2000. Bacteriocyte-associated endosymbionts of insects. In M. Dworkin (ed.), The prokaryotes. [Online.] Springer, New York, N.Y. http://link.springer.de/link/service/books/10125.
- 8.Charles, H., D. Mouchiroud, J. Lobry, I. Goncalves, and Y. Rahbe. 1999. Gene size reduction in the bacterial aphid endosymbiont Buchnera. Mol. Biol. Evol. 16:1820-1822. [DOI] [PubMed] [Google Scholar]
- 9.Chater, K. F., and D. A. Hopwood. 1993. Streptomyces, p. 83-99. In A. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. ASM Press, Washington, D.C.
- 10.Chen, X., S. Li, and S. Aksoy. 1999. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossinia and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J. Mol. Evol. 48:49-58. [DOI] [PubMed] [Google Scholar]
- 11.Clark, J. W., S. Hossain, C. A. Burnside, and S. Kambhampati. 2000. Coevolution between a cockroach and its bacterial endosymbiont: a biogeographical perspective. Proc. R. Soc. Lond. B Biol. Sci. 268:393-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, M. A., L. Baumann, M. L. Thao, N. A. Moran, and P. Baumann. 2001. Degenerative minimalism in the genome of a psyllid endosymbiont. J. Bacteriol. 183:1853-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, M. A., L. Baumann, M. A. Munson, P. Baumann, B. C. Campbell, J. E. Duffus, L. S. Osborne, and N. A. Moran. 1992. The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodoidea) constitute a lineage distinct from the endosymbionts of aphids and mealybugs. Curr. Microbiol. 25:119-123. [Google Scholar]
- 14.Clark, M. A., N. A. Moran, and P. Baumann. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 16:1586-1598. [DOI] [PubMed] [Google Scholar]
- 15.Cossart, P., and M. Lecuit. 2000. Microbial pathogens: an overview, p. 1-27. In P. Cossart, P. Boquet, S. Normark, and R. Rappuoli (ed.), Cellular microbiology. ASM Press, Washington, D.C.
- 16.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 17.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukatsu, T., and N. Nikoh. 2000. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera). Appl. Environ. Microbiol. 66:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futuyama, D. J. 1998. Evolutionary biology, 3rd ed., p. 637-639. Sinauer Associates, Inc., Sunderland, Mass.
- 20.Gullan, P. J., and M. Kosztarab. 1997. Adaptations in scale insects. Annu. Rev. Entomol. 42:23-50. [DOI] [PubMed] [Google Scholar]
- 21.Gullan, P. J., and J. H. Martin. Sternorrhyncha (jumping plant-lice, whiteflies, aphids and scale insects). In R. T. Cardé and V. H. Resh (ed.), Encyclopedia of insects, in press. Academic Press/Elsevier Science.
- 22.Hansmann, S., and W. Martin. 2000. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: influence of excluding poorly alignable sites from analysis. Int. J. Syst. E vol. Microbiol. 50:1655-1663. [DOI] [PubMed] [Google Scholar]
- 23.Heddii, A., H. Charles, C. Khatchadourian, G. Bonnot, and P. Nardon. 1998. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G+C content of an endocytobiotic DNA. J. Mol. Evol. 47:52-61. [DOI] [PubMed] [Google Scholar]
- 24.Ingraham, J. L., and C. A. Ingraham. 1999. Introduction to microbiology, 2nd ed., p. 264. Wadsworth Publishing Co., Belmont, Calif.
- 25.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krieg, N. R., and J. G. Holt (ed.). 1984. Bergey's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md.
- 27.Maniloff, J. 1992. Phylogeny of mycoplasmas, p. 549-559. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. ASM Press, Washington, D.C.
- 28.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 29.Moran, N. A. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran, N. A., and P. Baumann. 2000. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3:270-275. [DOI] [PubMed] [Google Scholar]
- 31.Moran, N. A., and A. Mira. 2001. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. Res. 2:0054.1-0054.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran, N. A., and A. Telang. 1998. Bacteriocyte-associated symbionts of insects: a variety of insect groups harbor ancient prokaryotic endosymbionts. BioScience 48:295-304. [Google Scholar]
- 33.Moran, N. A., and J. J. Wergegreen. 2000. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15:321-326. [DOI] [PubMed] [Google Scholar]
- 34.Munson, M. A., L. Baumann, and P. Baumann. 1993. Buchnera aphidicola (a prokaryotic endosymbiont of aphids) contains a putative 16S rRNA operon unlinked to the 23S rRNA-encoding gene: sequence determination, and promoter and terminator analysis. Gene 137:171-178. [DOI] [PubMed] [Google Scholar]
- 35.Munson, M. A., P. Baumann, and N. A. Moran. 1992. Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol. Phylogenet. Evol. 1:26-30. [DOI] [PubMed] [Google Scholar]
- 36.Ochman, H., S. Elwyn, and N. A. Moran. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 96:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandström, J., and N. Moran. 1999. How nutritionally imbalanced is phloem sap for aphids? Entomol. Exp. Appl. 91:203-210. [Google Scholar]
- 38.Sato, S., and H. Ishikawa. 1997. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J. Bacteriol. 179:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauer, C., E. Stackebrandt, J. Gadau, B. Hölldobler, and R. Gross. 2000. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int. J. Syst. Evol. Microbiol. 50:1877-1886. [DOI] [PubMed] [Google Scholar]
- 40.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Mutualism as revealed at the genomic level: the whole genome sequence of Buchnera sp. APS, an endocellular bacterial symbiont of aphids. Nature (London) 407:81-86. [DOI] [PubMed] [Google Scholar]
- 41.Silva, F. J., A. Latorre, and A. Moya. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17:615-618. [DOI] [PubMed] [Google Scholar]
- 42.Singer, G. A. C., and D. A. Hickey. 2000. Nucleotide bias causes genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17:1581-1588. [DOI] [PubMed] [Google Scholar]
- 43.Sneath, P. H. A., N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.). 1986. Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 44.Spaulding, A. W., and C. D. von Dohlen. 1998. Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha). Mol. Biol. Evol. 15:1506-1513. [DOI] [PubMed] [Google Scholar]
- 45.Staley, J. T., M. P. Bryant, N. Pfennig, and J. G. Holt (ed.). 1989. Bergey's manual of systematic bacteriology, vol. 3. The Williams & Wilkins Co., Baltimore, Md.
- 46.Thao, M. L., N. A. Moran, P. Abbot, E. B. Brennan, D. H. Burckhardt, and P. Baumann. 2000. Cospeciation of psyllids and their prokaryotic endosymbionts. Appl. Environ. Microbiol. 66:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tremblay, E. 1989. Coccoidea endosymbiosis, p. 145-173. In W. Schwemmler and G. Gassner (ed.), Insect endocytobiosis: morphology, physiology, genetics, evolution. CRC Press, Boca Raton, Fla.
- 48.von Dohlen, C. D., S. Kohler, S. T. Alsop, and W. R. McManus. 2001. Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature (London) 412:433-436. [DOI] [PubMed] [Google Scholar]
- 49.Williams, S. T., M. E. Sharpe, and J. G. Holt (ed.). 1989. Bergey's manual of systematic bacteriology, vol. 4. The Williams & Wilkins Co., Baltimore, Md.
- 50.Young, M., and S. T. Cole. 1993. Clostridium, p. 35-52. In A. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. ASM Press, Washington, D.C.
- 51.Zientz, E., F. J. Silva, and R. Gross. 2001. Genome interdependence in insect-bacterium symbioses. Genome Biol. Rev. 2:1032.1-1032.6. [DOI] [PMC free article] [PubMed] [Google Scholar]