Abstract
To assess the extent of genotypic and phenotypic diversity within species of purple nonsulfur bacteria found in aquatic sediments, a total of 128 strains were directly isolated from agar plates that had been inoculated with sediment samples from Haren and De Biesbosch in The Netherlands. All isolates were initially characterized by BOX-PCR genomic DNA fingerprinting, and 60 distinct genotypes were identified. Analyses of 16S rRNA gene sequences of representatives of each genotype showed that five and eight different phylotypes of purple nonsulfur bacteria were obtained from the Haren and De Biesbosch sites, respectively. At the Haren site, 80.5% of the clones were Rhodopseudomonas palustris, whereas Rhodoferax fermentans and Rhodopseudomonas palustris were numerically dominant at the De Biesbosch site and constituted 45.9 and 34.4% of the isolates obtained, respectively. BOX-PCR genomic fingerprints showed that there was a high level of genotypic diversity within each of these species. The genomic fingerprints of Rhodopseudomonas palustris isolates were significantly different for isolates from the two sampling sites, suggesting that certain strains may be endemic to each sampling site. Not all Rhodopseudomonas palustris isolates could degrade benzoate, a feature that has previously been thought to be characteristic of the species. There were differences in the BOX-PCR genomic fingerprints and restriction fragment length polymorphisms of benzoate-coenzyme A ligase genes and form I and form II ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) genes between benzoate-degrading and non-benzoate-degrading genotypes. The ability to distinguish these two Rhodopseudomonas palustris groups based on multiple genetic differences may reflect an incipient speciation event resulting from adaptive evolution to local environmental conditions.
Many species of bacteria in the environment, particularly in soil, are composed of genetically distinct and diverse clones (5, 13, 21, 22, 27, 28, 52). This diversification has resulted from evolutionary processes (4, 28, 30, 35, 41) that occur during adaptive evolution to local conditions in heterogeneous environments that contain a range of ecologically distinct habitats (28). This is in contrast to what is observed with many pathogenic and commensal bacterial species, which often exhibit clonal population structures that are correlated to differences in host environmental specificity and geographic differences (1, 7, 12, 27, 29, 40, 43, 51).
There have been numerous studies to assess the genetic diversity in closely related bacteria by PCR-based genome analysis techniques, including repetitive-sequence-based PCR (rep-PCR), amplified fragment length polymorphism, and random amplified polymorphic DNA techniques. The species studied include human-, animal-, and plant-associated bacteria, such as Escherichia coli (7, 17), Helicobacter pylori (2), Bradyrhizobium sp. (47, 48), Pseudomonas syringae (24, 25), Rhizobium sp. (6, 23), and several species of the genus Xanthomonas (32, 33). These studies were aimed at a better understanding of the epidemiology of pathogens, the host specificity of pathogenic strains, or the competitive strength of agriculturally important microorganisms. In contrast, the genetic diversity of free-living bacterial populations is poorly understood. Studies of 3-chlorobenzoate-degrading strains (13) and fluorescent Pseudomonas strains (5) have provided preliminary insight into the biogeographic distribution of these bacteria in soil. The majority (91%) of the genotypes identified were unique to the sites from which they were isolated, and each genotype was found only in the region from which it was isolated (13). Thus, there was almost no overlap of genotypes between sampling sites or continental regions, indicating that these heterotrophic soil bacteria were not globally mixed but were regionally endemic.
Purple nonsulfur bacteria have a wide range of growth modes and are able to grow under photoheterotrophic, photoautotrophic, and chemoheterotrophic conditions (20). This metabolic versatility is reflected in their wide distribution in natural environments and their isolation from surficial aquatic sediments and activated sludge, as well as from freshwater rivers and lakes (19, 20). Despite their widespread occurrence, the extent of phylogenetic, genotypic, and functional diversity found in purple nonsulfur bacteria is poorly understood.
A study which we conducted on the genetic diversity found in 14 strains of Rhodopseudomonas palustris isolated from various sources showed that all the strains were genetically unique based on BOX-PCR genomic DNA fingerprinting and that there were significant genotypic and phenotypic differences among Rhodopseudomonas palustris strains that were geographically dispersed (Y. Oda, W. G. Meijer, J. L. Gibson, J. C. Gottschal, and L. J. Forney, submitted for publication). While this study yielded insights into the biogeography of this species, the apparent extent of diversity within this species may have been distorted by the fact that the strains studied had been isolated from selective enrichment cultures. In the present study, 128 strains of purple nonsulfur bacteria were isolated from agar plates that had been inoculated with samples from two different sites. The phylogenetic, genotypic, and phenotypic characteristics were determined based on the sequences of 16S rRNA genes, BOX-PCR genomic DNA fingerprints, restriction fragment length polymorphisms (RFLPs) of catabolic genes, and the ability to metabolize (chlorinated) aromatic compounds. The data obtained were used to assess the diversity within species of purple nonsulfur bacteria.
MATERIALS AND METHODS
Media and growth conditions.
Batch cultures of purple nonsulfur bacteria were routinely grown at 30°C in closed screw-cap tubes (16 ml) that contained anoxically prepared LCM medium (46) under anoxic conditions in the light. After the basal LCM medium was autoclaved, 25 ml of sterile 1 M K(NH4)PO4 (pH 7.0) per liter of medium and 2 ml of a filter-sterilized vitamin solution (46) per liter of medium were added. Carbon sources were added from separately autoclaved stock solutions at the concentrations indicated below. To evaluate growth on 2 mM benzoate, 0.1% sodium bicarbonate was added to the medium. Growth was monitored by measuring the optical density at 660 nm. Isolates showing growth on benzoate were further tested for the ability to degrade 1 mM 3-chlorobenzoate in the presence or absence of 1 mM benzoate. Residual benzoate and 3-chlorobenzoate concentrations were determined by reverse-phase high-performance liquid chromatography by using a MicroSpher C18 column (Chrompack, Middelburg, The Netherlands) and methanol-water-acetic acid (50:49.5:0.5, vol/vol/vol) as the eluent and monitoring the effluent with a UV-975 Intelligent UV/VIS detector (Jasco, Tokyo, Japan) at 254 nm.
Isolation of purple nonsulfur bacteria.
Samples used for isolation of bacteria were collected from the top layer (about 0.5 cm) of sediments at two different sites in The Netherlands. To compare bacterial diversity within the sampling sites, two locations (1.5 m apart) from uncontaminated freshwater marsh sediments at Haren (Haren A and Haren B, collected on 12 August 1999) and two locations (5 m apart) from freshwater marsh sediments at De Biesbosch (De Biesbosch A and De Biesbosch B, collected on 16 August 1999) were used. The Haren site is an isolated small marsh with no inlet or outlet, whereas the De Biesbosch site is an area in direct contact with the industrially polluted River Merwede (14). The sediment samples (5.0 g [wet weight] of material) were suspended in 20 ml of anoxically prepared LCM medium in which yeast extract was omitted. The suspensions were sonicated briefly three times for 10 s (B3 sonicator; Branson, Dietzenbach, Germany) and subsequently shaken for 1.5 h at room temperature. After sedimentation, aliquots of the supernatant and a dilution series were spread on LCM medium plates containing 2 mM benzoate and 0.1% sodium bicarbonate. These plates were incubated at 30°C under anoxic conditions (N2 atmosphere) in the light. After approximately 2 weeks of incubation, pigmented colonies were transferred to LCM medium plates containing 15 mM malate and incubated anoxically in the light by using a BBL GasPak anaerobic jar system (Becton Dickinson and Company, Sparks, Md.). Single colonies were restreaked several times on LCM medium plates containing 15 mM malate, and well-separated single colonies were transferred into anoxically prepared LCM liquid medium containing 15 mM malate and incubated in the light. The purity of cultures was checked by microscopic observation and by streaking cultures on LCM medium supplemented with 0.3% peptone, 0.3% yeast extract, and 1.5% agar and then incubating the preparations under oxic conditions in the dark and under anoxic conditions in the light (two 40-W light bulbs at a distance of 20 cm) by using a BBL GasPak anaerobic jar system (Becton Dickinson and Company).
Isolation of DNA.
Genomic DNA was isolated from liquid cultures grown on 15 mM malate under anoxic conditions in the light. Cells (2 ml) were harvested by centrifugation and resuspended in 50 mM EDTA (pH 8.0), and genomic DNA was isolated by using a Wizard genomic DNA purification kit (Promega) according to the manufacturer's instructions. For genomic DNA used for Southern blotting, cells (3 ml) were harvested by centrifugation and resuspended in 750 μl of TE buffer (10 mM Tris-HCl, 0.1 mM EDTA; pH 8.0). Lysozyme (final concentration, 2.5 mg/ml) and RNase (final concentration, 20 μg/ml) were added, and the suspension was incubated for 1 h at 37°C. After addition of 1% (wt/vol) sodium dodecyl sulfate and incubation for an additional 1 h at 50°C, proteinase K (final concentration, 100 μg/ml) was added, and the mixture was incubated with gentle agitation for 2 h at 50°C. The suspension was extracted twice with phenol-chloroform (1:1) and once with chloroform, and then the DNA was precipitated from the aqueous phase by addition of 2 volumes of absolute ethanol. The DNA was collected by centrifugation, washed in 70% ethanol, and resuspended in 100 to 200 μl of TE buffer.
BOX-PCR genomic DNA fingerprinting and computer-assisted cluster analysis of genomic fingerprints.
Aliquots of genomic DNA from various isolates were used as templates to generate rep-PCR genomic fingerprints with the BOX A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′) (34). PCR were carried out in 25-μl reaction mixtures containing Gitschier buffer [83 mM (NH4)2SO4, 335 mM Tris-HCl (pH 8.8), 33.5 mM MgCl2, 33.5 μM EDTA, 150 mM β-mercaptoethanol], 4 μg of bovine serum albumin per ml, 10% (vol/vol) dimethyl sulfoxide, each deoxynucleoside triphosphate at a concentration of 1.25 mM, 2 μM BOX A1R primer, 2 U of Taq DNA polymerase (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands), and 50 ng of DNA. The temperature profile was as follows: initial denaturation at 95°C for 2 min; 35 cycles of 94°C for 3 s, 92°C for 30 s, 50°C for 1 min, and 65°C for 8 min; and final extension at 65°C for 8 min. PCR products were separated by electrophoresis on 1.5% agarose gels at 70 V and 4°C for 19 h. After the agarose gels were stained with ethidium bromide, images of the gels were digitized by using an ImageMaster VDS system (Amersham Pharmacia Biotech Benelux) and stored as TIFF files. Computer-assisted analysis of genomic fingerprints was performed by using the GELCOMPARE software program (version 4.1; Applied Maths, Kortrijk, Belgium). Similarity matrices of whole densitometric curves of the gel tracks were calculated by using the pairwise Pearson's product-moment correlation coefficient (r value) (31). Cluster analysis of similarity matrices was performed by the unweighted pair group method using arithmetic averages (39). BOX-PCR genomic fingerprint patterns having r values of more than 0.8 were considered to be patterns of the same genotype. BOX-PCR genomic fingerprints of Rhodopseudomonas palustris strains AP1, BIS3, BIS6, BIS10, BIS11, BIS14, BIS17, BIS18, BIS23, KD1, WS17, NCIB8288, and DCP3 and Rhodopseudomonas palustris type strain NCIMB8252 from another study were also included (Oda et al., submitted). Strains BIS3, BIS6, BIS10, BIS11, BIS14, BIS17, BIS18, and BIS23 were previously isolated from the De Biesbosch sediment by an enrichment culture technique.
Amplification of 16S rRNA gene and sequencing.
Nearly complete 16S rRNA genes were amplified by using fD1 and rD1 as primers (50). PCR products were purified with a QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany). Sequencing was performed with an ABI PRISM BigDye terminator cycle sequencing Ready Reaction kit (PE Applied Biosystems) and an ABI PRISM 310 genetic analyzer (PE Applied Biosystems). Sequencing primer 536F (Escherichia coli positions 519 to 536) was used, and the 16S rRNA gene sequences (lengths, 584 to 714 bp) were compared to sequences in the GenBank database by using the Basic Local Alignment Search Tool (BLAST) (3). 16S rRNA gene sequences (630 bp) of the Rhodopseudomonas palustris isolates from this study and another study (Oda et al., submitted) were aligned by using the CLUSTALW program (44). Evolutionary distances were calculated by the correction method of Jukes and Cantor of the program TREECON for Windows, version 1.3b (45), and a phylogenetic tree was constructed by the neighbor-joining method (36). A bootstrap analysis was carried out to test the reliability of the tree (11). The GenBank accession numbers of 16S rRNA gene sequences of previously described strains of Rhodopseudomonas palustris are AF314062 (strain AP1), AF416653 (strain BIS3), AF416654 (strain BIS6), AF314064 (strain BIS10), AF416655 (strain BIS11), AF416656 (strain BIS14), AF416657 (strain BIS17), AF416658 (strain BIS18), AF416659 (strain BIS23), AF314063 (strain KD1), AF416660 (strain WS17), AF416661 (strain NCIB8288), AF416662 (type strain NCIMB8252), and AF416663 (strain DCP3).
Diversity indices (Shannon indices) of genotypes within the same species were calculated by using −Σpilnpi, where pi is the proportion of individual bacteria found in the ith genotype (42).
Hybridization experiments.
Approximately 10 μg of genomic DNA was digested with EcoRI, and the fragments were separated by electrophoresis on 1.2% agarose gels and Southern blotted onto positively charged nylon membranes (Roche Diagnostics Netherlands B.V., Almere, The Netherlands). All hybridizations were carried out as recommended by the manufacturer with a hybridization temperature of 65°C for high stringency and a hybridization temperature of 60°C for low stringency. PCR-amplified BamHI-XbaI fragments containing either the large subunit of the form I (cbbL) ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) gene (GenBank accession number AF355196) or the benzoate-coenzyme A (benzoate-CoA) ligase (badA) gene (GenBank accession number AF355195) from Rhodopseudomonas palustris strain DCP3 and a 0.95-kb AatII-EcoRI fragment of pRPII5 containing the form II (cbbM) RubisCO gene (GenBank accession number AF355197) from Rhodopseudomonas palustris strain DCP3 (Oda et al., submitted) were used to make randomly primed digoxigenin-labeled gene probes.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of genotypes used for phylogenetic tree construction have been deposited in the GenBank database under accession numbers AF417943 (genotype HaAB1), AF417944 (genotype HaAB2), AF417945 (genotype HaA5), AF417946 (genotype HaA7), AF417947 (genotype HaA8), AF417948 (genotype HaB6), AF417949 (genotype HaB7), AF417950 (genotype HaB8), AF417951 (genotype BiesA5), AF417952 (genotype BiesA18), AF417953 (genotype BiesA19), AF417954 (genotype BiesA20), AF417955 (genotype BiesA21), AF417956 (genotype BiesA22), AF417957 (genotype BiesB1), AF417958 (genotype BiesB5), AF417959 (genotype BiesB17), AF417960 (genotype BiesB18), AF417961 (genotype BiesB19), AF417962 (genotype BiesB20), and AF417963 (genotype BiesB21).
RESULTS
BOX-PCR genomic DNA fingerprint analyses.
Sediment suspensions of two samples (locations A and B) from two different sampling sites were spread on LCM medium plates containing benzoate as the primary carbon source. This approach, rather than selective enrichment cultures, was used in an effort to maximize the diversity of isolates obtained. After approximately 2 weeks of incubation under anoxic conditions in the light, red, pink, and brown colonies appeared on LCM medium plates, and a total of 128 isolates were obtained as pure cultures; 33 isolates were obtained from Haren A, 34 isolates were obtained from Haren B, 30 isolates were obtained from De Biesbosch A, and 31 isolates were obtained from De Biesbosch B. The cell suspensions from the Haren site were mostly red or red-brown, whereas approximately equal proportions of red, red-brown, and peach-brown cell suspensions were obtained with strains from the De Biesbosch site (Table 1).
TABLE 1.
Distribution of genotypes and species and hybridization analysis with the form I (cbbL) and form II (cbbM) RubisCO genes
| Sampling location | Genotypea | No. of isolates | Color of cell suspensionb | Cell shape | Species (% identity)c | RFLP patternd
|
|
|---|---|---|---|---|---|---|---|
| cbbL | cbbM | ||||||
| Haren A | HaAB1 | 19 | Red-brown | Rod | Rps. palustris (99) | IA | IIA |
| HaAB2 | 3 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| HaAB3e | 1 | Brown | Ovoid rod | Rmi. vannielii (99) | IB | — | |
| HaA4 | 4 | Red | Spiral | Rsp. rubrum (100) | — | IIB | |
| HaA5e | 2 | Red-brown | Rod | Rps. palustris (100) | U | IIA | |
| HaA6 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| HaA7 | 1 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| HaA8 | 1 | Red-brown | Rod | Rps. palustris (100) | IA | IIA | |
| HaA9 | 1 | Yellow-brown | Curved rod | Rvi. gelatinosus (100) | U | — | |
| Haren B | HaAB1 | 17 | Red-brown | Rod | Rps. palustris (100) | IA | IIA |
| HaAB2 | 8 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| HaAB3e | 3 | Brown | Ovoid rod | Rmi. vannielii (98) | IB | — | |
| HaB4e | 1 | Brown | Ovoid rod | Rmi. vannielii (99) | IB | — | |
| HaB5e | 1 | Brown | Ovoid rod | Rmi. vannielii (99) | U | — | |
| HaB6 | 1 | Red-brown | Rod | Rps. palustris (98) | IC | IIB | |
| HaB7 | 1 | Red-brown | Rod | Rps. palustris (99) | IC | IIA | |
| HaB8 | 1 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| HaB9 | 1 | Red | Spiral | Rsp. rubrum (100) | — | IIB | |
| De Biesbosch A | BiesA1 | 2 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC |
| BiesA2 | 2 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesA3 | 2 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IID | |
| BiesA4 | 2 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesA5 | 2 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| BiesA6e | 2 | Brown | Spiral | Rsp. fulvum (98) | — | U | |
| BiesA7 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesA8 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IID | |
| BiesA9 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesA10 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesA11 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesA12 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesA13 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesA14 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IID | |
| BiesA15 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesA16 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IID | |
| BiesA17 | 1 | Pink | Rod | Rpl. roseus (98) | U | — | |
| BiesA18 | 1 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| BiesA19 | 1 | Red-brown | Rod | Rps. palustris (98) | ID | IIE | |
| BiesA20 | 1 | Red-brown | Rod | Rps. palustris (98) | ID | IIE | |
| BiesA21 | 1 | Red-brown | Rod | Rps. palustris (100) | ID | U | |
| BiesA22e | 1 | Red-brown | Rod | Rps. palustris (98) | IA | IIE | |
| BiesA23 | 1 | Red | Spiral | Rsp. rubrum (100) | — | U | |
| BiesA24 | 1 | Yellow-brown | Curved rod | Rvi. gelatinosus (99) | U | Uf | |
| De Biesbosch B | BiesB1e | 7 | Red-brown | Rod | Rps. palustris (100) | U | IIA |
| BiesB2 | 2 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesB3 | 2 | Pink | Rod | Rpl. elegans (99) | U | Uf | |
| BiesB4e | 2 | Pink | Rod | Rpl. elegans (99) | U | — | |
| BiesB5e | 2 | Red-brown | Rod | Rps. palustris (98) | U | U | |
| BiesB6 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesB7 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesB8 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesB9 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesB10 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesB11 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | U | |
| BiesB12 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIC | |
| BiesB13 | 1 | Peach-brown | Curved rod | Rfx. fermentans (98) | — | IIB | |
| BiesB14e | 1 | Brown | Ovoid rod | Rmi. vannielii (98) | U | — | |
| BiesB15e | 1 | Pink | Rod | Rpl. elegans (99) | U | — | |
| BiesB16e | 1 | Pink | Rod | Rpl. roseus (99) | U | — | |
| BiesB17 | 1 | Red-brown | Rod | Rps. palustris (99) | IA | IIA | |
| BiesB18 | 1 | Red-brown | Rod | Rps. palustris (98) | U | IIF | |
| BiesB19 | 1 | Red-brown | Rod | Rps. palustris (98) | U | IIF | |
| BiesB20 | 1 | Red-brown | Rod | Rps. palustris (99) | IA | U | |
| BiesB21 | 1 | Red-brown | Rod | Rps. palustris (98) | U | IIF | |
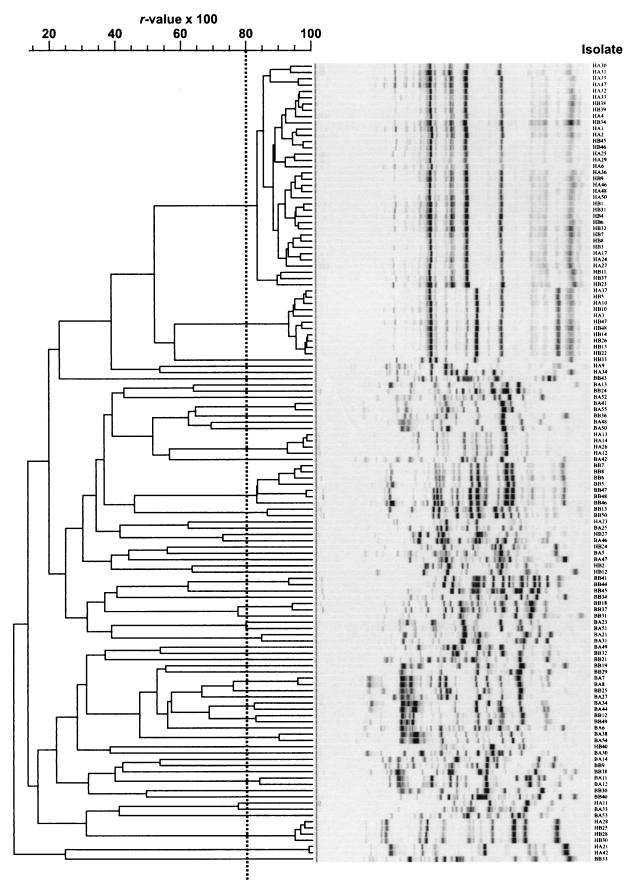
BOX-PCR genomic fingerprint patterns having r values of more than 0.8 (Fig. 1) were considered to be the same genotype. The designation HaAB indicates genotypes found in both Haren A and Haren B samples.
Cultures were grown in LCM liquid media containing 15 mM malate under anoxic conditions in the light.
Closest bacterial relative as determined by comparison of partial sequences of 16S rRNA genes (length, 584 to 714 bp) from a single representative of each genotype with known sequences in GenBank by using the Basic Local Alignment Search Tool (BLAST). Due to the difference in the natural habitat of the type strain of Rhodoferax antarcticus, which was isolated from an Antarctic microbial mat (26), our isolates were tentatively identified as Rhodoferax fermentans. Abbreviations: Rfx., Rhodoferax; Rmi., Rhodomicrobium; Rpl., Rhodoplanes; Rps., Rhodopseudomonas; Rsp., Rhodospirillum; Rvi., Rubrivivax.
A single representative of each genotype was probed. RFLP patterns were determined based on the sizes of the hybridized fragments (Fig. 3). RFLP patterns of the cbbL gene: IA, 0.85- and 0.49-kb fragments; IB, 7.5-kb fragment; IC, 1.3-kb fragment; ID, 5.5-kb fragments; U, either a unique pattern or a fragment of more than 23 kb long; —, no homology. RFLP patterns of the cbbM gene: IIA, 3.4-kb hybridized fragment; IIB, 6.6-kb hybridized fragment; IIC, 15.0-kb hybridized fragment; IID, 4.0-kb hybridized fragment; IIE, 1.7-kb hybridized fragment; IIF, 2.8-kb hybridized fragment; U, either a unique pattern or a hybridized fragment more than 23 kb long; —, no homology.
Benzoate-degrading genotype.
Signal detected only under low-stringency hybridization conditions.
Genetic differences among the isolates were assessed by using genomic DNA fingerprints obtained by rep-PCR genomic DNA fingerprinting performed with the BOX A1R primer (34). The reproducibility of BOX-PCR genomic fingerprinting was confirmed by repeatedly testing genomic DNA from several isolates. Cluster analysis of the resulting genomic fingerprints showed that independently obtained fingerprints of an isolate had r values of more than 0.9. To obtain a conservative estimate of the diversity of the 128 strains obtained in this study, we considered isolates with genotypes having r values greater than 0.8 (5) to be representatives of a single genotype (Fig. 1). Based on this criterion, a total of 60 unique genotypes were identified, including 9 genotypes from Haren A, 9 genotypes from Haren B, 24 genotypes from De Biesbosch A, and 21 genotypes from De Biesbosch B (Table 1). None of the genotypes found at the Haren site were found at the De Biesbosch site, indicating that the strains isolated appeared to be endemic to the sampling sites. Two genotypes (HaAB1 and HaAB2) were numerically dominant at the Haren site and accounted for 47 of the 67 isolates obtained. In contrast to the Haren site, a significant proportion of the isolates from the De Biesbosch site were unique genotypes, and none of the genotypes found at De Biesbosch A were found at De Biesbosch B.
FIG. 1.
Computer-assisted product-moment-unweighted pair group method using arithmetic averages cluster analysis of BOX-PCR genomic fingerprints of 128 isolates. BOX-PCR genomic fingerprint patterns having r values of more than 0.8 (dotted line) were considered to be the same genotype. Abbreviations: HA, Haren A; HB, Haren B: BA, De Biesbosch A; BB, De Biesbosch B.
Genotypic diversity within the species of purple nonsulfur bacteria.
The phylogenetic relatedness of isolates was assessed based on partial 16S rRNA gene sequences. The six isolates with the genotype HaAB1 had identical sequences (data not shown). Therefore, BOX-PCR genomic fingerprint patterns of the same genotype (based on rep-PCR fingerprints) were presumed to be representative of the same phylotype. Subsequently, the partial 16S rRNA gene sequence of a single representative of each genotype was determined and compared to sequences in the GenBank database.
The majority of the isolates from the Haren site were most closely related to Rhodopseudomonas palustris and showed more than 98% sequence identity. Based on the high degree of identity to the 16S rRNA gene sequences of Rhodopseudomonas palustris in the GenBank database, the colors of cell suspensions, and microscopic observation of cell morphology (Table 1), a total of 26 isolates from Haren A and 28 isolates from Haren B were presumptively identified as Rhodopseudomonas palustris. The remaining isolates (Table 1) were identified as organisms related to Rhodoferax antarcticus or Rhodoferax fermentans (based on 98% sequence identity of 16S rRNA genes), Rhodomicrobium vannielii (99% sequence identity), Rhodospirillum rubrum (100% sequence identity), and Rubrivivax gelatinosus (100% sequence identity).
In contrast to the Haren site, the majority of the isolates from the De Biesbosch site were identified as organisms related to Rhodoferax fermentans (18 and 10 isolates from De Biesbosch A and De Biesbosch B, respectively) and Rhodopseudomonas palustris (7 and 14 isolates from De Biesbosch A and De Biesbosch B, respectively). Strains of Rhodopseudomonas palustris and Rhodoferax fermentans comprised 59 and 23%, respectively, of the isolates in our collection. A small number of isolates were identified as organisms related to Rhodomicrobium vannielii, Rhodospirillum rubrum, and Rubrivivax gelatinosus. The low frequency with which Rhodomicrobium vannielii and Rhodospirillum rubrum strains were isolated suggests that these species may not be numerically abundant at our sampling sites. Species of Rhodoplanes elegans, Rhodoplanes roseus, and Rhodospirillum (Phaeospirillum) fulvum were isolated from the De Biesbosch site but were not isolated from the Haren site. The high level of genotypic diversity found among strains of Rhodopseudomonas palustris and Rhodoferax fermentans (Fig. 2) was reflected in the Shannon indices calculated from differences in genomic DNA fingerprints. Diversity indices within the species Rhodopseudomonas palustris showed that there were significant differences between the Haren and De Biesbosch sites (Table 2), indicating that there may be differences in either the tempo of diversification processes or the selective pressures experienced by Rhodopseudomonas palustris strains at the two sampling sites.
FIG. 2.
Cluster analysis of BOX-PCR genomic fingerprints showing the genotypic diversity within the Rhodopseudomonas palustris and Rhodoferax fermentans isolates from the Haren (A) and De Biesbosch (B) sites. When a given genotype was isolated from both Haren A and Haren B, the location from which the strain was isolated is indicated by either A (for Haren A) or B (for Haren B) in parentheses. Abbreviations: Rps., Rhodopseudomonas; Rfx., Rhodoferax.
TABLE 2.
Shannon indices calculated for genotype diversity among isolates identified as organisms belonging to the same species
| Speciesa | Shannon indices for the following sampling sitesb:
|
||||||
|---|---|---|---|---|---|---|---|
| Haren A | Haren B | Haren | De Biesbosch A | De Biesbosch B | De Biesbosch | Total | |
| Rps. palustris | 0.93 | 1.02 | 1.09 | 1.75 | 1.57 | 2.26 | 2.01 |
| Rfx. fermentans | NDc | —d | ND | 2.58 | 2.16 | 3.08 | 3.13 |
| Rmi. vannielii | ND | 0.95 | 0.87 | — | ND | ND | 1.15 |
| Rsp. rubrum | ND | ND | ND | ND | — | ND | 0.87 |
Abbreviations: Rps., Rhodopseudomonas; Rfx., Rhodoferax; Rmi., Rhodomicrobium; Rsp., Rhodospirillum.
The Shannon indices were calculated by using −Σpilnpi, where pi is the proportion of individual bacteria found in the ith genotype.
ND, not determined since only one genotype was isolated.
—, no isolates.
Southern hybridization of genomic DNA with the form I (cbbL) and form II (cbbM) RubisCO genes.
Only a limited number of species of purple nonsulfur bacteria have been reported to possess both the cbbL and cbbM genes (38, 49). Southern hybridization analyses were performed to investigate the distribution of these two genes among the isolates obtained in this study. Genomic DNA from a total of 63 isolates that represented each genotype was probed with both genes (Fig. 3). This analysis was intended not only to survey the distribution of the two genes among species of purple nonsulfur bacteria but also to identify RFLPs of the genes found in the same species. Only genotypes of Rhodopseudomonas palustris hybridized to both genes under high-stringency hybridization conditions (Table 1). Genotypes of Rhodomicrobium vannielii, Rubrivivax gelatinosus, Rhodoplanes elegans, and Rhodoplanes roseus hybridized to the cbbL gene, while genotypes of Rhodoferax fermentans, Rhodospirillum fulvum, and Rhodospirillum rubrum hybridized to the cbbM gene under high-stringency hybridization conditions. Some genotypes of Rubrivivax gelatinosus and Rhodoplanes elegans hybridized to the cbbM gene under low-stringency hybridization conditions.
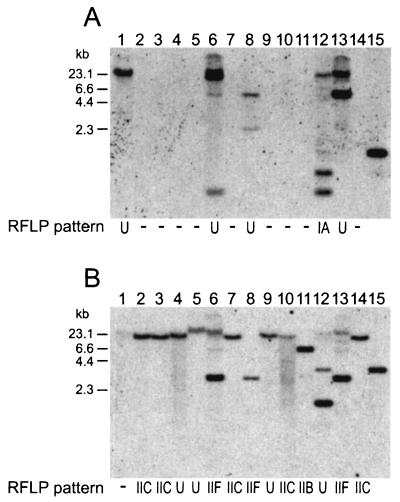
FIG. 3.
Distribution of the form I (cbbL) and form II (cbbM) RubisCO genes among the isolates of purple nonsulfur bacteria as determined by Southern hybridization analyses under high-stringency conditions. EcoRI-digested DNA from a single representative of each genotype and Rhodopseudomonas palustris strain DCP3 was probed with the cbbL (A) and cbbM (B) genes. Lane 1, genotype BiesB3; lane 2, BiesB6; lane 3, BiesB7; lane 4, BiesB8; lane 5, BiesB9; lane 6, BiesB18; lane 7, BiesB10; lane 8, BiesB19; lane 9, BiesB11; lane 10, BiesB12; lane 11, BiesB13; lane 12, BiesB20; lane 13, BiesB21; lane 14, BiesB2; lane 15, Rhodopseudomonas palustris strain DCP3. The RFLP patterns of the cbbL gene are designated as follows: IA, 0.85- and 0.49-kb hybridized fragments; U, either a unique pattern or a hybridized fragment more than 23 kb long; −, no homology. The RFLP patterns of the cbbM gene are designated as follows: IIB, 6.6-kb hybridized fragment; IIC, 15.0-kb hybridized fragment; IIF, 2.8-kb hybridized fragment; U, either a unique pattern or a hybridized fragment more than 23 kb long; −, no homology.
Based on the sizes of the fragments that hybridized with the cbbL and cbbM genes as probes, four and six distinct RFLP patterns were identified, respectively. The majority of the Rhodopseudomonas palustris genotypes exhibited pattern A with the cbbL and cbbM gene probes (patterns IA and IIA in Table 1, respectively), and genotypes of Rhodoferax fermentans exhibited pattern C with the cbbM gene probe (pattern IIC in Table 1).
Metabolism of benzoate.
To investigate the prevalence of the ability to degrade aromatic compounds, all isolates were tested for the ability to degrade benzoate under anoxic conditions in the light. A total of 25 of the 128 isolates were able to degrade benzoate, and strains able to degrade benzoate were most frequently isolated from De Biesbosch B (14 isolates). The small number of the isolates able to degrade benzoate (19.5%) was surprising since benzoate was the primary carbon source in the agar plates used for isolation of the strains. Apparently, yeast extract (0.01%) in the media, organic compounds from the sediment suspensions used as inocula, or benzoate metabolites served as growth substrates for most of the purple nonsulfur bacteria that grew. None of the isolates belonging to a single genotype differed with respect to the ability to degrade benzoate. Five species, Rhodopseudomonas palustris, Rhodomicrobium vannielii, Rhodospirillum fulvum, Rhodoplanes elegans, and Rhodoplanes roseus, were able to degrade benzoate, and Rhodopseudomonas palustris and Rhodomicrobium vannielii were the species that were isolated frequently. The benzoate-degrading isolates represented 12 genotypes (Table 3).
TABLE 3.
Anaerobic growth on benzoate, hybridization with the benzoate-CoA ligase (badA) gene, and utilization of 3-chlorobenzoate by the benzoate-degrading genotypes
| Genotype (no. of isolates)a | Speciesb | Growth rate on benzoate (h)c | Hybridization with badAd | Utilization of 3-chlorobenzoatee
|
|
|---|---|---|---|---|---|
| Without benzoate | With benzoate | ||||
| BiesB1 (7) | Rps. palustris | 31.7 (1.2) | + | − | + |
| BiesB5 (2) | Rps. palustris | 16.5 (0.5) | + | − | + |
| HaA5 (2) | Rps. palustris | 18.0 (0.5) | + | − | + |
| BiesA22 (1) | Rps. palustris | 12.0 (0.5) | + | − | +++ |
| HaAB3 (4) | Rmi. vannielii | 13.7 (0.8) | − | − | − |
| HaB4 (1) | Rmi. vannielii | 14.7 (0.6) | − | − | − |
| HaB5 (1) | Rmi. vannielii | 16.3 (0.8) | − | − | − |
| BiesB14 (1) | Rmi. vannielii | 13.7 (0.8) | − | − | − |
| BiesA6 (2) | Rsp. fulvum | 5.3 (0.3) | − | − | − |
| BiesB4 (2) | Rpl. elegans | 27.0 (1.0) | − | − | − |
| BiesB15 (1) | Rpl. elegans | 14.0 (0.5) | − | − | − |
| BiesB16 (1) | Rpl. roseus | 19.2 (1.0) | − | − | − |
A single representative of each genotype was tested.
Abbreviations: Rps., Rhodopseudomonas; Rmi., Rhodomicrobium; Rsp., Rhodospirillum; Rpl., Rhodoplanes.
Doubling time in media containing 2 mM benzoate. The data shown are averages of three measurements. The values in parentheses are standard deviations.
+, homology; −, no homology.
Utilization of the initial amount (1 mM) of 3-chlorobenzoate in the presence or absence of 1 mM benzoate after 3 weeks, indicated as follows: −, <5%; +, 5 to 25%; +++, >50%.
Characterization of the benzoate-degrading genotypes.
A more detailed analysis was performed with 12 benzoate-degrading isolates that represented all genotypes. The doubling times on benzoate ranged from 5.3 to 31.7 h, and Rhodospirillum fulvum grew most rapidly (Table 3). For the genotypes of Rhodopseudomonas palustris, the doubling times on benzoate ranged from 12.0 to 31.7 h. This was in contrast to the genotypes of Rhodomicrobium vannielii, all of which had similar growth rates. Southern hybridization experiments showed that only genotypes of Rhodopseudomonas palustris hybridized to the benzoate-CoA ligase (badA) gene, indicating that other benzoate-degrading species either possess a nonhomologous badA gene or metabolize benzoate by pathways that do not involve the conversion of benzoate to benzoyl-CoA by benzoate-CoA ligase (10, 16).
All 25 benzoate-degrading isolates were tested further for the ability to degrade 3-chlorobenzoate in the presence or absence of benzoate. None of the isolates was able to degrade 3-chlorobenzoate as the sole carbon source. Only isolates of Rhodopseudomonas palustris were able to degrade 3-chlorobenzoate if benzoate was present as a cosubstrate (Table 3). This indicates that the ability to degrade chlorinated benzoates was restricted to the species Rhodopseudomonas palustris.
Comparison of genotypic and phenotypic characteristics of strains of Rhodopseudomonas palustris.
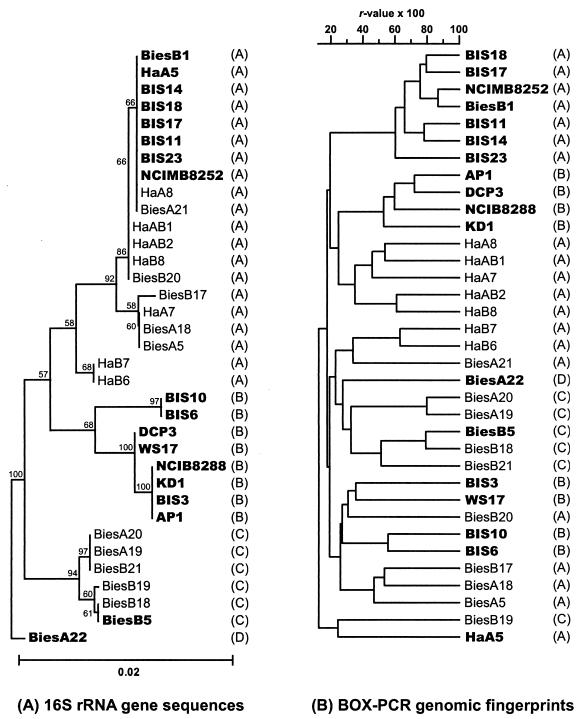
Interestingly, the majority (84.0%) of the isolates identified as Rhodopseudomonas palustris were unable to degrade benzoate (Table 1) in spite of the fact that degradation of benzoate has been recognized as a feature that is characteristic of the species (16, 20). To determine the phylogenetic relationships among the 21 different genotypes of Rhodopseudomonas palustris in this study, 13 strains of Rhodopseudomonas palustris used in another study (Oda et al., submitted), and Rhodopseudomonas palustris type strain NCIMB8252, a phylogenetic tree based on 16S rRNA gene sequences was constructed (Fig. 4A). Overall, the level of similarity of the sequences was more than 97.5%, and four distinct clusters (clusters A to D) were found. Cluster B contained only previously described benzoate-degrading strains, and cluster C contained only genotypes from the De Biesbosch site. Although genotypes and strains belonging to the same 16S rRNA cluster were distributed throughout the dendrogram generated by cluster analysis of BOX-PCR genomic fingerprints (Fig. 4B), there were certain trends: genotypes and strains belonging to the same 16S rRNA cluster were in the same branch of the genomic fingerprint dendrogram, and genotypes and strains isolated from the same sampling site (Ha genotypes versus Bies genotypes and BIS strains in Fig. 4B) were genetically more similar to one another than to genotypes and strains isolated from the other sampling site. These results indicate that certain strains were regionally endemic, and this may have resulted from divergent evolution of strains that are related by descent. It should also be noted that the strains described elsewhere, most which of were isolated by selective enrichment culture (Oda et al., submitted), clustered together, possibly indicating that enrichment bias influenced the outcome of experiments done to isolate these strains. The benzoate-degrading genotypes and strains were distributed throughout the genomic fingerprint dendrogram and were found in clusters that were distinct from the non-benzoate-degrading genotypes. The differences in genetic characteristics between benzoate-degrading and non-benzoate-degrading genotypes were also apparent in RFLPs of the form I and form II RubisCO genes. All benzoate-degrading genotypes isolated in this study had unique combinations of hybridization patterns with both genes, whereas many of the non-benzoate-degrading genotypes had the same RFLP patterns (Table 1). These results indicate that the benzoate-degrading strains were members of distinct lineages in Rhodopseudomonas palustris.
FIG. 4.
Phylogenetic and genetic relationships of the genotypes of Rhodopseudomonas palustris from this study, previously described strains of Rhodopseudomonas palustris (AP1, BIS3, BIS6, BIS10, BIS11, BIS14, BIS17, BIS18, BIS23, KD1, WS17, NCIB8288, and DCP3), and Rhodopseudomonas palustris type strain NCIMB8252 based on 16S rRNA gene sequences (630 bp) (A) and BOX-PCR genomic fingerprints (B). Cluster designations (clusters A to D) based on 16S rRNA gene sequences are shown in parentheses. Bootstrap values (based on 100 replicates) are given at branch points. Bar = 0.02 substitution per site. The benzoate-degrading genotypes and strains are indicated by boldface type.
DISCUSSION
Purple nonsulfur bacteria are likely to play an important role in nutritional cycles in natural environments (37), yet little is known about the extent of phylogenetic, genotypic, and phenotypic diversity within this group of bacteria. This might be due to the fact that most purple nonsulfur bacteria studied previously have been isolated by selective enrichment culturing (15, 20). The enrichment techniques may impose limits on studies of microbial diversity since most enrichment cultures favor fast-growing or numerically dominant organisms (9). In another study we observed that only strains of Rhodopseudomonas palustris were isolated from enrichment cultures for phototrophic bacteria that contained a mixture of 3-chlorobenzoate and benzoate regardless of the geographical and ecological origin of the inocula (Oda et al., submitted). In contrast, the procedure used for isolation of bacteria in this study involved plating sediment suspensions directly onto selective media. Based on 16S rRNA gene sequence homology, this approach yielded eight different species of purple nonsulfur bacteria and 60 distinct genotypes identified by BOX-PCR genomic fingerprinting (Table 1).
Among the isolates identified as organisms belonging to the species Rhodopseudomonas palustris, two genotypes, HaAB1 (36 of the 67 isolates) and HaAB2 (11 isolates), were repeatedly isolated (70.1%) from both the Haren samples. Since the patterns of amplified fragments generated by BOX-PCR genomic fingerprinting are complex, evolutionary convergence to the same genotype is highly unlikely. Thus, the repeated recovery of isolates with the same or nearly the same genotype suggests that these isolates are related by descent from a common ancestor. The selection and maintenance of these genotypes resemble a clonal population structure that is analogous to that observed for many pathogenic and commensal bacterial species found in homeostatic environments (1, 7, 27, 29, 51). In contrast, most of the isolates of Rhodopseudomonas palustris from the De Biesbosch site were genotypically distinct from one another. This is reflected in the cluster analysis of BOX-PCR genomic fingerprints that resulted in an extensively branched dendrogram (Fig. 2). Several studies (21, 51, 52) have contrasted clonal populations, in which there is limited genetic exchange and variation is produced largely by mutation, and freely recombining populations, in which diversity arises through parasexual mechanisms of gene transfer achieved through transduction, transformation, and conjugation. Typically, the population structure of clonal populations is characterized by limited but deeply branching lineages, whereas freely recombining populations are characterized by extensive but shallow branching patterns that reflect genetic differences that arise from genetic exchange. While BOX-PCR genomic DNA fingerprints do not provide information on amino acid sequences like multilocus enzyme electrophoresis analyses do, the branching patterns of dendrograms based on BOX-PCR data suggest that the genetic structures of Rhodopseudomonas palustris populations found at the Haren site and the De Biesbosch site were analogous to the genetic structures of clonal and freely recombining populations, respectively.
Previous studies have provided evidence for a positive relationship between genetic diversity and environmental heterogeneity (18, 28, 35). For example, data obtained by McArthur et al. (28) indicated that the degree of habitat variability correlated with the degree of genetic diversity of Burkholderia cepacia in soil samples. If genetic diversity increases with habitat variability, our observation of significant differences in genotypic diversity within the species Rhodopseudomonas palustris at the two sampling sites (Table 2) suggests that the De Biesbosch site exhibits greater habitat variability than the Haren site, and differences in the population structures (i.e., clonal and freely recombining) between the two sampling sites may reflect differences in local environmental conditions. However, this is speculative, and the ecological significance of the observed genotypic diversity among local populations of Rhodopseudomonas palustris remains unclear.
Aromatic compounds are some of the major sources of combined carbon available in natural environments (37). Photometabolism of benzoate by Rhodopseudomonas palustris, Rhodospirillum fulvum, and Rhodocyclus purpureus is well recognized, and several other species of purple nonsulfur bacteria have also been shown to grow at the expense of benzoate (16, 37); also, previous investigators have found that benzoate provides highly selective conditions in phototrophic enrichment cultures for the growth of Rhodopseudomonas palustris (20). This is the first reported isolation of Rhodoplanes elegans and Rhodoplanes roseus strains able to metabolize benzoate. Such strains may not have been isolated in previous studies in which selective enrichment cultures were used if Rhodopseudomonas palustris has a strong competitive advantage under the conditions used for enrichment over these species. Interestingly, the direct isolation procedures used in this study yielded a relatively low proportion of Rhodopseudomonas palustris isolates among the benzoate-degrading isolates. These observations suggest that Rhodopseudomonas palustris may not be the most numerically abundant benzoate-degrading species of purple nonsulfur bacteria in natural environments.
Both benzoate-degrading and non-benzoate-degrading strains of Rhodopseudomonas palustris were isolated. Furthermore, the growth rates on benzoate for benzoate-degrading genotypes of Rhodopseudomonas palustris varied from 12.0 to 31.7 h. This was in contrast to the isolates of Rhodomicrobium vannielii, all of which were able to degrade benzoate and had similar growth rates (Table 3). These results indicate that strains identified as Rhodopseudomonas palustris (or closely related organisms based on 16S rRNA gene sequences) are not only genetically more diverse but also phenotypically more diverse than hitherto assumed based on characterization of strains obtained by conventional enrichment techniques. Our phenotypic and genotypic characterization study of the 3-chlorobenzoate-degrading strains of Rhodopseudomonas palustris suggested that there may be a correlation between certain phenotypic properties (e.g., growth rate on benzoate) and variation in genome organization as determined by rep-PCR genomic DNA fingerprinting (Oda et al., submitted). The results of the combined genotypic and phenotypic analyses of the isolates identified as Rhodopseudomonas palustris in this study support this notion. The benzoate-degrading genotypes were separated from the non-benzoate-degrading genotypes in the dendrogram generated by cluster analysis of BOX-PCR genomic fingerprints (Fig. 4B). Moreover, the benzoate-degrading genotypes hybridized to the benzoate-CoA ligase gene (Table 3), whereas the non-benzoate-degrading genotypes did not hybridize to this gene (data not shown). It could be that all strains of Rhodopseudomonas palustris could degrade benzoate at one time and that this ability was lost from certain lineages. A second possibility is that some strains acquired the ability to degrade benzoate through lateral gene transfer. While the importance of such horizontal gene transfer in bacterial speciation is increasingly recognized (8, 30), it is impossible to distinguish between these two possibilities based on the data presented here. Nonetheless, distinct clades found within the species Rhodopseudomonas palustris may reflect an incipient speciation event that arises from adaptive evolution to local environmental conditions. Further investigation of the extent to which genetic diversity is reflected in phenotypic diversity in the strains of Rhodopseudomonas palustris may lead to a better understanding of the ecology and evolutionary biology of this species.
Acknowledgments
We are grateful to H. Bolhuis for valuable suggestions and discussions.
This work was supported by a grant from the Ubbo Emmius Foundation of the University of Groningen, Haren, The Netherlands.
REFERENCES
- 1.Aakre, R. K., A. Jenkins, B.-E. Kristiansen, and L. O. Frøholm. 1998. Clonal distribution of invasive Neisseria meningitidis isolates from the Norwegian county of Telemark, 1987 to 1995. J. Clin. Microbiol. 36:2623-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Arber, W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J.-C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruijn, F. J. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2128. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egland, P. G., J. Gibson, and C. S. Harwood. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 177:6545-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto, S., B. Marshall, and M. J. Blaser. 1994. PCR-based restriction fragment length polymorphism typing of Helicobacter pylori. J. Clin. Microbiol. 32:331-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulthorpe, R. R., A. N. Rhodes, and J. M. Tiedje. 1998. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl. Environ. Microbiol. 64:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerritse, J., B. J. van der Woude, and J. C. Gottschal. 1992. Specific removal of chlorine from the ortho-position of halogenated benzoic acids by reductive dechlorination in anaerobic enrichment cultures. FEMS Microbiol. Lett. 100:273-280. [DOI] [PubMed] [Google Scholar]
- 15.Gest, H., J. F. Favinger, and M. T. Madigan. 1985. Exploitation of N2-fixation capacity for enrichment of anoxygenic photosynthetic bacteria in ecological studies. FEMS Microbiol. Ecol. 31:317-322. [Google Scholar]
- 16.Gibson, J., and C. S. Harwood. 1995. Degradation of aromatic compounds by nonsulfur purple bacteria, p. 991-1003. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Gordon, D. M. 1997. The genetic structure of Escherichia coli populations in feral house mice. Microbiology 143:2039-2046. [DOI] [PubMed] [Google Scholar]
- 18.Hedrick, P. W., M. E. Ginevan, and E. P. Ewing. 1976. Genetic polymorphism in heterogeneous environments. Annu. Rev. Ecol. Syst. 7:1-32. [Google Scholar]
- 19.Hiraishi, A., and Y. Ueda. 1994. Rhodoplanes gen. nov., a new genus of phototrophic bacteria including Rhodopseudomonas rosea as Rhodoplanes roseus comb. nov. and Rhodoplanes elegans sp. nov. Int. J. Syst. Bacteriol. 44:665-673. [Google Scholar]
- 20.Imhoff, J. F., and H. G. Trüper. 1992. The genus Rhodospirillum and related genera, p. 2141-2155. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 21.Istock, C. A., K. E. Duncan, N. Ferguson, and X. Zhou. 1992. Sexuality in a natural population of bacteria—Bacillus subtilis challenges the clonal paradigm. Mol. Ecol. 1:95-103. [DOI] [PubMed] [Google Scholar]
- 22.Johnsen, K., S. Andersen, and C. S. Jacobsen. 1996. Phenotypic and genotypic characterization of phenanthrene-degrading fluorescent Pseudomonas biovars. Appl. Environ. Microbiol. 62:3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laguerre, G., P. van Berkum, N. Amarger, and D. Prévost. 1997. Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl. Environ. Microbiol. 63:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little, E. L., R. M. Bostock, and B. C. Kirkpatrick. 1998. Genetic characterization of Pseudomonas syringae pv. syringae strains from stone fruits in California. Appl. Environ. Microbiol. 64:3818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louws, F. J., D. W. Fulbright, C. T. Stephens, and F. J. de Bruijn. 1994. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madigan, M. T., D. O. Jung, C. R. Woese, and L. A. Achenbach. 2000. Rhodoferax antarcticus sp. nov., a moderately psychrophilic purple nonsulfur bacterium isolated from an Antarctic microbial mat. Arch. Microbiol. 173:269-277. [DOI] [PubMed] [Google Scholar]
- 27.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McArthur, J. V., D. A. Kovacic, and M. H. Smith. 1988. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc. Natl. Acad. Sci. USA 85:9621-9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musser, J. M., P. M. Schlievert, A. W. Chow, P. Ewan, B. N. Kreiswirth, V. T. Rosdahl, A. S. Naidu, W. Witte, and R. K. Selander. 1990. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc. Natl. Acad. Sci. USA 87:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 31.Pearson, K. 1926. On the coefficient of racial likeness. Biometrika 18:105-117. [Google Scholar]
- 32.Pooler, M. R., D. F. Ritchie, and J. S. Hartung. 1996. Genetic relationships among strains of Xanthomonas fragariae based on random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus PCR data and generation of multiplexed PCR primers useful for the identification of this phytopathogen. Appl. Environ. Microbiol. 62:3121-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rademaker, J. L. W., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. de Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. E vol. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 34.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1997. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 1-26. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, supplement 3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 35.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sasikala, C., and C. V. Ramana. 1998. Biodegradation and metabolism of unusual carbon compounds by anoxygenic phototrophic bacteria. Adv. Microb. Physiol. 39:339-377. [DOI] [PubMed] [Google Scholar]
- 38.Shively, J. M., W. Devore, L. Stratford, L. Porter, L. Medlin, and S. E. Stevens, Jr. 1986. Molecular evolution of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). FEMS Microbiol. Lett. 37:251-257. [Google Scholar]
- 39.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco, Calif.
- 40.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 42.Stiling, P. D. 1999. Ecology: theories and applications, 3rd ed. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 43.Suerbaum, S., J. Maynard Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Peer, Y., and R. de Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 46.van der Woude, B. J., M. de Boer, N. M. J. van der Put, F. M. van der Geld, R. A. Prins, and J. C. Gottschal. 1994. Anaerobic degradation of halogenated benzoic acids by photoheterotrophic bacteria. FEMS Microbiol. Lett. 119:199-208. [DOI] [PubMed] [Google Scholar]
- 47.van Rossum, D., F. P. Schuurmans, M. Gillis, A. Muyotcha, H. W. van Verseveld, A. H. Stouthamer, and F. C. Boogerd. 1995. Genetic and phenetic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl. Environ. Microbiol. 61:1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinuesa, P., J. L. W. Rademaker, F. J. de Bruijn, and D. Werner. 1998. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl. Environ. Microbiol. 64:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson, G. M. F., and F. R. Tabita. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol. Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]
- 50.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittam, T. S. 1992. Sex in the soil. Curr. Biol. 2:676-678. [DOI] [PubMed] [Google Scholar]
- 52.Wise, M. G., L. J. Shimkets, and J. V. McArthur. 1995. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl. Environ. Microbiol. 61:1791-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]