Abstract
The bacteriocin nisin is produced only by some strains of Lactococcus lactis, and to date production in other lactic acid bacteria has not been achieved. Enterococcus sp. strain N12β is a nisin-immune transconjugant obtained from a nisin-producing donor (L. lactis ATCC 11454) and a dairy recipient (Enterococcus sp. strain S12β), but it does not produce nisin. In this study, using PCR amplification, we confirmed that the whole nisin operon is likely present in Enterococcus sp. strain N12β. Northern hybridization of total RNA from strain N12β with a nisA probe and the results of reverse transcriptase PCR showed the lack of nisA transcription in this strain. However, nisA transcription was partially restored in strain N12β upon growth in the presence of exogenous nisin, and the nisA transcription signal was intensified after an increase in the external nisin level. Furthermore, bioassays showed that active nisin was produced in a dose-dependent fashion by strain N12β following induction by exogenous nisin. These results indicated that expression of the nisin genes in Enterococcus sp. strain N12β depended on autoinduction via signal transduction. However, the amount of external inducing signal required was significantly greater than the amount needed for autoinduction in L. lactis.
Nisin is a ribosomally synthesized bacteriocin which contains lanthionine residues and can be classified as a lantibiotic (19, 20). It is produced only by certain strains of Lactococcus lactis, which are extensively characterized bacteria used in the production of many fermented foods. It has a relatively wide antimicrobial spectrum and can inhibit the proliferation of most gram-positive bacteria (16). Nisin is heat stable and active at low pH, which makes it a good candidate for a natural food preservative. Indeed, it is used in this capacity in many different food products worldwide, in which it is particularly effective at preventing the development of clostridial spores, which is a concern in many processed foods. These foods include meats, salad dressings, canned vegetables, pasteurized liquid egg, etc. (6, 7, 8). In the United States, the Food and Drug Administration permits the use of pure nisin for the prevention of clostridial growth in processed cheeses. Currently this is the only approved use for a purified bacteriocin as a food ingredient in the United States. However, generally regarded as safe, nisin-producing lactococci are often used in fermented foods or cultured ingredients as natural source preservatives to increase the safety and shelf life of many food products. While many other lactic acid bacteria (LAB) used in the production of fermented food products also produce bacteriocins, the spectrum of antimicrobial capacity of these compounds is generally narrower than that of nisin. Widening the potential applications of nisin in foods by engineering other LAB used in food fermentations to produce nisin should be useful for the food industry.
The genetic nature and complex process of nisin biosynthesis inherently complicate expression of nisin in other LAB. Production of nisin is encoded by a cluster of genes proposed to be transcriptionally arranged as nisABTCIP, nisRK, and nisFEG, and this cluster is closely linked with sucrose utilization genes on a large, conjugative transposon (17, 23). This gene cluster encodes the nisin precursor protein (NisA), as well as proteins involved in posttranslation modifications, immunity for the producing cell, transcriptional regulation, transport, and processing of the prepeptide (11, 12, 13, 18, 21, 22, 32, 35, 36). The precursor is an inactive peptide that is chemically modified by the products of nisB and nisC (33). The chemical processes include dehydration of serine and threonine residues and formation of the thioether bridges as meso-lanthionines and β-methyllanthionines, which are characteristics of lantibiotics (31). The modified precursor peptide is transported by NisT and processed by a subtilisin-like protease, NisP, which cleaves the 23-amino-acid leader peptide to form an extracellular mature nisin peptide (21). The mature nisin peptide can then function as an autoinducer to regulate expression of the nisin genes through a two-component regulatory system, NisRK (23). The proposed model predicts that the extracellular mature nisin accumulates to a critical level and activates the sensor kinase, NisK, by autophosphorlation on a histine residue at the expense of ATP. It is proposed that the phosphoryl group is subsequently transferred to an asparate residue on the regulator protein, NisR, which can then activate the transcription of nisABTCIP and nisFEG. In addition to this nisin autoregulation, there are also other factors that influence the transcription of the nisin biosynthetic genes in a NisRK-independent fashion (4). To protect the producing cell, the membrane-associated NisI and NisFEG function together as immunity proteins (13, 22, 29, 32). However, the precise mechanism of immunity has not yet been elucidated.
Expression of nisin in a food grade manner in other LAB is essential to successful incorporation of these LAB in food systems. The conjugative genes present on the nisin transposons make it possible to transfer the nisin gene cluster to other bacteria in a nonrecombinant fashion. This has been achieved previously by Broadbent et al. (1), who used a dairy Enterococcus strain as a recipient for conjugative transfer of the nisin transposon Tn5307 from L. lactis ATCC 11454. While this transconjugant did exhibit immunity to nisin, it did not produce active nisin, as determined by standard bioassay procedures. As nisin gene expression is governed by the processes of transcription, translation, posttranslation modification, secretion, processing, and signal transduction, a block in any of these steps can sabotage nisin biosynthesis. In this study, we investigated this Enterococcus transconjugant to elucidate the cause of the nisin-production-negative phenotype and also to evaluate the feasibility of initiating nisin production by this dairy enterococcal bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. lactis subsp. lactis ATCC 11454, a nisin A-producing strain, was propagated in M17 broth (Difco, Detroit, Mich.) containing 0.5% glucose (M17G) at 30°C under stationary conditions. Enterococcus sp. strain S12β was isolated from a commercial cheese (1). Enterococcus sp. strain N12β, formerly called SI12β (1), is an S12β transconjugant containing transposon Tn5307 from L. lactis ATCC 11454. Enterococci were cultured under stationary conditions in M17G at 37°C. Micrococcus luteus ATCC 1040 was propagated aerobically in Luria-Bertani broth at 30°C.
Immunity assay.
To test for immunity to nisin, overnight cultures were inoculated (1%) into 10-ml portions of M17G containing 0 to 3,000 IU of nisin (Sigma, St. Louis, Mo.) per ml. The MIC of nisin was determined after incubation for 8, 12, and 30 h.
Nisin bioassay.
Nisin activity was determined by a rapid plate method as described previously (3). Five microliters of a sample to be tested was spot inoculated onto a 0.45-μm-pore-size nitrocellulose filter membrane (Millipore Corporation, Bedford, Mass.) prewetted by immersion in 0.01 M HCl-1% Tween 80. After diffusion for 14 h at 4°C, the filters were removed, and the plates were overlaid with soft agar seeded with M. luteus as an indicator. A standard curve of nisin inhibition zones versus units of commercial nisin was drawn, and from this curve the nisin concentrations in test samples were estimated.
Trypsin digestion.
Trypsin treatment of cell extracts was carried out essentially as described by Nelis et al. (26). The reaction mixtures contained NaOH (5.3 mM) and trypsin (1 mg/ml) in 0.05 M phosphate buffer (pH 8.0).
Nisin induction experiments.
Attempts were made to induce nisin production by Enterococcus sp. strain N12β by adding exogenous commercial nisin to growing cultures. In some cases (data not shown), nisin was added when cultures reached an optical density at 600 nm (OD600) of 0.4. The amount of nisin added was subtracted from batch nisin concentrations to calculate production by Enterococcus.
Kinetics of nisin production.
Overnight cultures of Enterococcus sp. strains N12β and S12β and L. lactis ATCC 11454 were inoculated (1%) into M17G containing 50 IU/ml of nisin. Samples were taken at zero time and after 2, 4, 6, 8, 10, 12, 24, and 48 h and frozen at −70°C. Samples were then tested for nisin by using the bioassay outlined above.
Reverse-phase high-pressure liquid chromatography (RP-HPLC).
Culture supernatants and cell extracts were tested for precursor and mature nisin essentially as described by Liu and Hansen (25). Assays were carried out by using a μRC C18 Sc2.1/10 column (Pharmacia Biotech, Piscataway, N.J.) and an AKTA HPLC pump and detection system (Pharmacia Biotech). Solvent A was 0.1% trifluoroacetic acid (Aldrich Chemical Co., Milwaukee, Wis.), and solvent B was 0.1% trifluoroacetic acid in acetonitrile. The detection UV wavelength was 220 nm. The gradient was 0 to 100% solvent B over 60 column volumes. Commercial nisin (Sigma) was used as a reference control, and Enterococcus sp. strain S12β was used as a negative control.
DNA manipulations.
A template for PCR was obtained by using crude culture lysates prepared as follows. One milliliter of cells was pelleted and then agitated at the maximum speed in a MiniBeater-8 (Biospec Products, Bartlesville, Okla.) with 0.5 volume of acid-washed glass beads (diameter, <106 μm; Sigma) for 30 s. Lysates were diluted 10−2 prior to use in PCR. PCR amplifications of nisin and thyA genes were performed by using the primers indicated in Table 1. All PCRs were performed with a Robocycle (Stratagene, La Jolla, Calif.). The reaction mixtures (final volume, 50 μl) contained 1 μl of template, 1 μl of each primer (30 μM), 1 μl of deoxynucleoside triphosphates (each at a concentration of 10 mM), and 0.5 μl of Taq DNA polymerase. The amplification conditions were as follows: one cycle of 92°C for 2 min; 30 cycles of 92°C for 30 s, 55°C for 45 s, and 72°C for 1 min; and one cycle of 72°C for 3 min. Sequencing reactions were performed with an ABI Prism dye terminator cycle sequencing kit by using AmpliTaq DNA polymerase FS, and the products were separated with an ABI 377 automatic sequencer (Applied Biosystems, Foster City, Calif.).
TABLE 1.
Primers used for amplification of nisin and thyA genes
| Gene | Primer | GenBank accession no.a | Positionb |
|---|---|---|---|
| nisA | 5′-GGATAGTATCCATGTCTG-3′ | M65089 | 250-269 |
| 5′-CAATGATTTCGTTCGAAG-3′ | M65089 | 516-498 | |
| nisB | 5′-CGCTTTGCTATGGAGACGAAT-3′ | L16226 | 1050-1070 |
| 5′-GAGCTCCTATGCCAAATGTA-3′ | L16226 | 1553-1534 | |
| nisT | 5′-GAAGAATACATGAAATGAGG-3′ | L16226 | 3419-3438 |
| 5′-TAACTTTCCAGCTGTCCC-3′ | L16226 | 3721-3704 | |
| nisC | 5′-CAGAGCAATATGAGGATAATG-3′ | L16226 | 5221-5241 |
| 5′-TTCTCATTTCCTCTTCCCTCC-3′ | L16226 | 6490-6469 | |
| nisI | 5′-ATTGTGGCCTTAATAGGG-3′ | L16226 | 6504-6521 |
| 5′-TAGCGACTTGTCAGAAGC-3′ | L16226 | 6785-6768 | |
| nisRK | 5′-TATAAAAGCGAGAGGAACG-3′ | X76884 | 3270-3284 |
| 5′-GTACATCCGACTTGACAT-3′ | X76884 | 3881-3864 | |
| nisF | 5′-CAGGTGCTACAAGATATCAG-3′ | U17255 | 189-208 |
| 5′-ACAACTCCGCAATACCATCAG-3′ | U17255 | 610-630 | |
| thyA | 5′-AACAGGTTTAGAAGTGG-3′ | AF028811 | 242-259 |
| 5′-GTTGTTCGATTTGGTAACGG-3′ | AF028811 | 660-641 |
GenBank sequence with which the primers were designed.
Positions in the GenBank sequence that correspond to the primer.
DNA fingerprinting.
Cultures were fingerprinted by using a triplicate arbitrary primed PCR (TAP-PCR) procedure described by Cusick and O'Sullivan (5).
RNA manipulations.
A modified RNA isolation procedure was used to isolate total RNA from cultures at OD600 of 0.8 to 0.9 (2). To remove any residual DNA, total RNA was treated with DNase I as recommended by the manufacturer (GIBCO BRL, Gaithersburg, Md.). The concentration of total RNA was determined with a DU-70 spectrophotometer (Beckman, Fullerton, Calif.) and was verified visually by gel electrophoresis. For RNA slot blot hybridizations, total RNA samples (0.5 μg) were transferred onto a Zeta-Probe membrane (Bio-Rad, Hercules, Calif.) by using a Bio-Dot slot blot apparatus (Bio-Rad). Probe labeling and hybridization detection kits were used according to the instructions of the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
The enzymes SuperScript II (Gibco BRL) and and Taq DNA polymerase (Promega, Madison, Wis.) were used for reverse transcriptase (RT) PCR, as described by the manufacturer. One microgram of total RNA was used as the template for RT-PCR. An identical PCR mixture with total RNA but without added RT was used as a negative control.
To quantify the amount of specific nisA mRNA transcripts, total RNA was diluted in series prior to hybridization with a nisA probe. The intensities of the hybridization signals were measured by using the densitometer capabilities of an IS-2000 digital imaging system (Alpha Innotech Corporation, San Leandro, Calif.).
RESULTS
Confirmation of the presence of the nisin gene operon in Enterococcus sp. strain N12β.
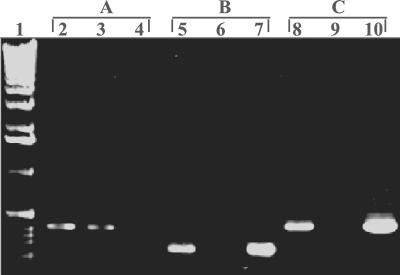
Broadbent et al. (1) previously confirmed the presence of nisA in Enterococcus sp. strain N12β by hybridization with a nisA probe. In this study, we further substantiated that the nisin gene cluster is present in this strain by amplifying each of the genes involved in nisin production (nisABTCIPRK) and also nisF, which is part of the downstream nisFEG operon involved in nisin immunity. Figure 1 shows the amplification results for nisA and nisF genes obtained by using PCR, as well as the results for amplification of the enterococcal thyA gene, which verified that the nisin genes amplified were indeed from Enterococcus. The data indicated that the nisin gene cluster is most likely fully intact in Enterococcus sp. strain N12β. To further verify the integrity of the structural gene for nisin, the nisA gene was sequenced in its entirety, and no mutations were observed.
FIG. 1.
Confirmation that the nisin gene cluster is present in Enterococcus sp. strain N12β. The primers used targeted the thyA gene from Enterococcus faecalis (A), the nisA gene (B), and the nisF gene (C). Lane 1, 1-kb DNA ladder (GIBCO BRL, Rockville, Md.); lanes 2, 5, and 8, Enterococcus sp. strain N12β; lanes 3, 6, and 9, Enterococcus sp. strain S12β; lanes 4, 7, and 10, L. lactis ATCC 11454.
Confirmation of the parentage of enterococcal strains N12β and S12β.
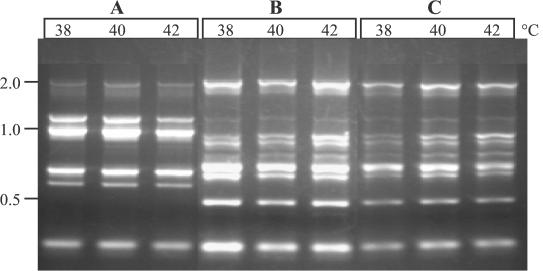
To confirm that strain S12β was the direct parent of N12β and that the conjugation event which was used in the construction of strain N12β (1) did not result in major DNA rearrangements, a sensitive DNA fingerprinting technique (TAP-PCR) was employed. This technique has previously been shown to differentiate organisms to the strain level in many instances (5). As Fig. 2 shows, the fingerprints of N12β and S12β appear to be identical and are distinct from the fingerprint of L. lactis ATCC 11454, which was the donor strain used for conjugation of Tn5307 into strain N12β.
FIG. 2.
TAP-PCR DNA fingerprints of L. lactis ATCC 11454 (A), Enterococcus sp. strain S12β (B), and Enterococcus sp. strain N12β (C). The three lanes for each strain show the results for triplicate PCRs conducted at the annealing temperatures indicated. Molecular sizes (in kilobases) are indicated on the left.
Evaluation of the contribution of enterococcal proteases to nisin stability.
One possible explanation for the failure to detect active nisin from the Enterococcus transconjugant, strain N12β, is that there were proteases that inactivated the peptide bacteriocin. To evaluate this possibility, 10-IU portions of nisin were added to 100-μl aliquots of overnight supernatants of strains N12β and S12β and incubated at 37°C. Bioassays performed at zero time and after incubation for 1, 5, and 12 h and overnight indicated there was no loss of nisin activity, suggesting that stability of the nisin peptide was not the reason for the inability to detect active nisin in strain N12β.
Investigation of posttranslational events for nisin production in strain N12β.
The fact that Enterococcus sp. strain N12β is immune to nisin (1) and the proposal that an immunity gene (nisI) is cotranscribed with the nisin structural, modification, transport, and processing genes (28) suggested that the nisin genes may be transcribed in this new host. If this were the case, then the block in production of active nisin by strain N12β may be due to a posttranslational event. To investigate this possibility, RP-HPLC and standard bioassay methods were used to detect nisin or its precursor in supernatants and cell extracts with and without trypsin treatment. Trypsin can activate nisin by cleaving the prepeptide from the inactive precursor (37). However, both bioassays and RP-HPLC did not indicate that there was any active nisin (data not shown). Furthermore, there were no apparent differences between the RP-HPLC profiles of both culture supernatants and cell extracts from strain N12β and the RP-HPLC profiles of the parent strain, S12β (data not shown). These data suggested that the lack of nisin production by N12β was probably not primarily due to posttranslational downstream events.
Quantification of nisin immunity in strain N12β.
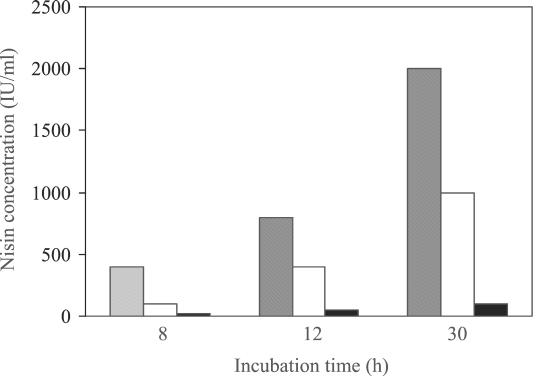
Nisin immunity is proposed to be conferred by both NisI and the NisFEG complex (29). While strain N12β was previously shown to be immune to nisin (1), the level of immunity was never compared to that of the original parent, L. lactis ATCC 11454. A nisin immunity assay for enterococcal strains N12β and S12β and the L. lactis ATCC 11454 nisin transposon Tn5307 donor was therefore performed. As Fig. 3 shows, the parent Enterococcus sp. strain S12β had a low natural resistance to nisin and could grow in the presence of up to ∼100 IU of nisin per ml. Strain N12β could grow in the presence of nisin at concentrations up to nearly 1,000 IU/ml, whereas L. lactis ATCC 11454 could grow in the presence of nisin at concentrations up to 2,000 IU/ml. This suggested that Enterococcus sp. strain N12β exhibited ∼50% of the immunity to nisin exhibited by the donor strain, L. lactis ATCC 11454. These data suggest that the nisABTCIP operon may not be efficiently expressed in strain N12β.
FIG. 3.
Histogram depicting the levels of nisin immunity exhibited by L. lactis ATCC 11454 (shaded bars), Enterococcus sp. strain N12β (open bars), and Enterococcus sp. strain S12β (solid bars), as measured by the lack of growth after 8, 12, and 30 h of incubation.
Investigation of nisA transcription in Enterococcus sp. strain N12β.
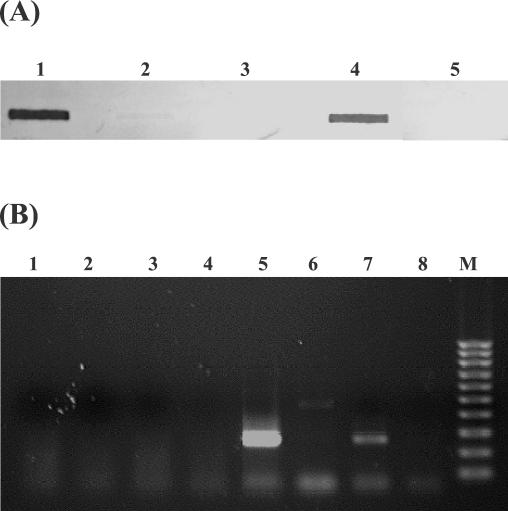
To investigate transcription of the nisA gene in strain N12β, slot blot Northern hybridizations with a nisA probe were conducted by using total RNA isolated from strains N12β and S12β and the positive control L. lactis ATCC 11454 (Fig. 4A). No nisA mRNA was detected in enterococcal strain S12β or N12β, while L. lactis ATCC 11454 exhibited a strong signal during growth in M17G. This result was substantiated by RT-PCR of total RNA (Fig. 4B). Lack of transcription from the nisA promoter was therefore the primary reason for the lack of nisin production by Enterococcus sp. strain N12β.
FIG. 4.
(A) Investigation of nisA transcription by RNA slot blot hybridization with a nisA probe. Each slot contained 5 μg of total RNA. Slot 1, L. lactis ATCC 11454; slot 2, Enterococcus sp. strain S12β; slot 3, Enterococcus sp. strain N12β; slot 4, Enterococcus sp. N12β grown in the presence of a subinhibitory nisin concentration; slot 5, Enterococcus sp. strain S12β grown in the presence of added nisin. (B) RT-PCR with nisA-specific primers of nisA transcription. Lanes 1 to 4, negative controls without added RT; lanes 1 and 5, L. lactis ATCC 11454; lanes 2 and 6, Enterococcus sp. strain N12β; lanes 3 and 7, Enterococcus sp. N12β grown in presence of exogenous nisin; lanes 4 and 8, Enterococcus sp. strain S12β; lane M, 100-bp DNA marker (Bio-Rad).
Transcription of nisA in strain N12β can be induced in the presence of external nisin.
Transcription of the nisA promoter in L. lactis is under positive control mediated by the two-component NisRK regulatory system and the concentration of exogenous nisin (23). To investigate if adding exogenous nisin to Enterococcus sp. strain N12β could stimulate transcription of the nisA promoter, a sublethal concentration of nisin (50 IU/ml) was added to log-phase cultures of strain N12β. After the cultures grew to an OD600 of 0.9, total RNA was isolated and investigated for the presence of nisA mRNA. Northern hybridization and RT-PCR both confirmed that exogenous nisin could stimulate transcription from the nisA promoter in strain N12β, albeit to a lower degree than in L. lactis ATCC 11454 (Fig. 4).
Induction of nisA transcription in Enterococcus sp. strain N12β results in production of active nisin.
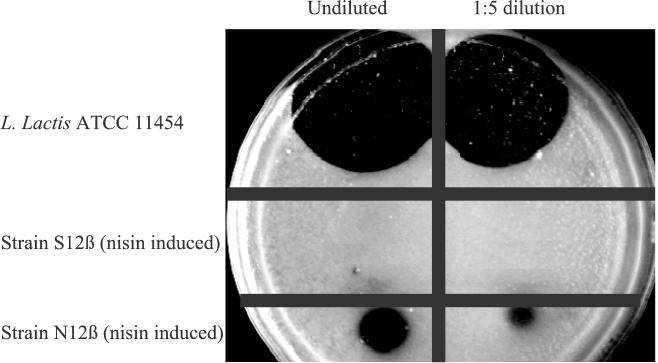
To ascertain whether N12β produced active nisin in the presence of added exogenous nisin, a quantitative bioassay was performed with both strains N12β and S12β following induction by added nisin. Strain S12β was used as a negative control because its genetic background was otherwise identical to that of strain N12β. As Fig. 5 shows, the supernatant from an induced culture of strain S12β did not inhibit the growth of the indicator organism M. luteus, while a clear inhibition zone was observed with supernatant from a culture of strain N12β. This indicated that active nisin was produced by strain N12β following induction by exogenous nisin, albeit at a significantly lower level than in L. lactis ATCC 11454.
FIG. 5.
Bioassay for the production of nisin by L. lactis ATCC 11454 and enterococcal strains S12β and N12β following growth in M17G containing 50 IU of added nisin per ml.
Investigation of the kinetics of induced nisin production in Enterococcus sp. strain N12β.
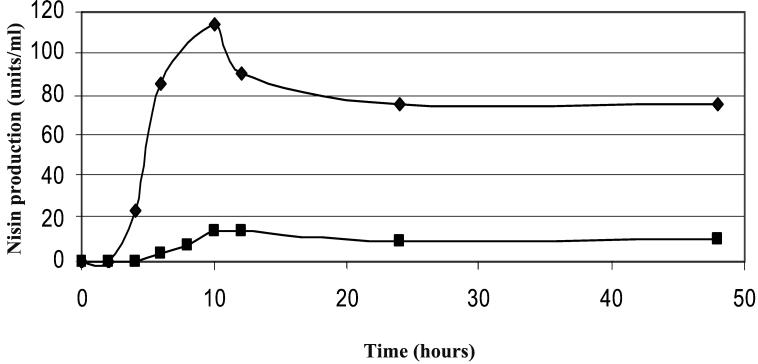
As nisin production in strain N12β was not fully restored following induction with exogenous nisin, the kinetics of nisin production were monitored for 48 h to determine if they followed a pattern similar to that in L. lactis. As the kinetics depicted in Fig. 6 show, the pattern of production detected in Enterococcus sp. strain N12β is very similar to that detected in L. lactis, although the amounts were smaller, and maximum production occurred after 10 h in both cases.
FIG. 6.
Kinetics of nisin production by L. lactis ATCC 11454 (♦) and Enterococcus sp. strain N12β (▪).
Dose-dependent relationship of nisA transcription and nisin production with addition of exogenous nisin.
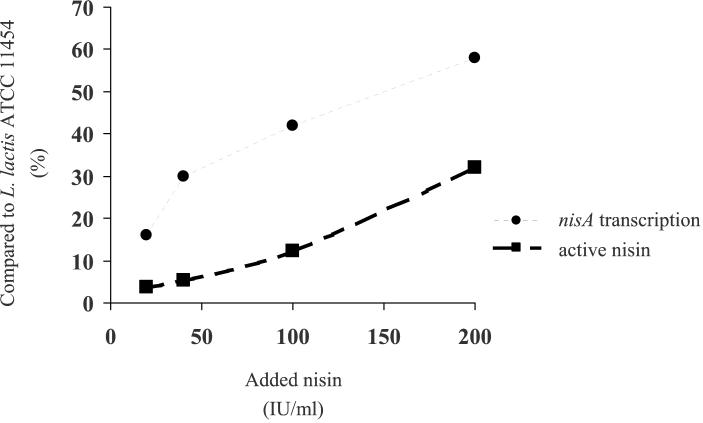
As the autoinduction of nisA transcription in L. lactis is approximately linearly correlated to the amount of external nisin (23), the effects of different amounts of added nisin on nisA transcription and nisin production by Enterococcus sp. strain N12β were investigated. Following induction of strain N12β with nisin concentrations ranging from 0 to 200 IU/ml, Northern hybridizations showed that there was an approximately linear increase in nisA transcription with increasing nisin concentrations (Fig. 7). However, when the concentration of nisA mRNA was compared with the concentration of nisA mRNA in L. lactis ATCC 11454, induction of strain N12β with 200 IU of nisin per ml was ≈60% (Fig. 8). Concomitant measurements of nisin production by strain N12β also showed that there was an approximately linear increase, but maximum induction with 200 IU of nisin per ml resulted in only ≈30% of the amount produced by L. lactis ATCC 11454 (Fig. 8). These studies indicate that the expression of active nisin from the Tn5307 transposon in Enterococcus strain N12β is similar to its expression in L. lactis ATCC 11454, except that significantly more external inducer (nisin) is required to activate the NisRK two-component induction system.
FIG. 7.
Slot blot Northern hybridization with a nisA probe of total RNA isolated from Enterococcus sp. strain N12β following induction by different amounts of exogenous nisin. Slot 1, no added nisin; slot 2, 20 IU of added nisin per ml; slot 3, 40 IU of added nisin per ml; slot 4, 100 IU of added nisin per ml; slot 5, 200 IU of added nisin per ml.
FIG. 8.
Effect of increasing concentrations of added nisin on induction of nisA transcription and production of active nisin by Enterococcus sp. strain N12β. The values are percentages of the maximum levels obtainable by L. lactis ATCC 11454.
DISCUSSION
Enterococcus sp. strain N12β was constructed by conjugation of the nisin-encoding transposon Tn5307 from L. lactis ATCC 11454 into Enterococcus sp. strain S12β (1). Although DNA hybridization revealed that strain N12β had in fact received the structural gene for nisin, nisA, and also demonstrated immunity to nisin, this strain could not produce active nisin. In the present study, strain N12β was investigated to determine why it did not produce nisin. To eliminate the possibility that there were deletions within the nisin gene cluster, PCR was used to confirm that genes extending throughout the nisin gene cluster were present in strain N12β, and sequence analysis of nisA confirmed that the structural gene was identical to the donor L. lactis ATCC 11454 structural gene.
The fact that strain N12β demonstrated immunity to nisin suggested that the nisin genes were likely expressed in this strain. Therefore, one possible explanation for the lack of active nisin was that the final step of precursor processing was not functioning, resulting in a buildup of modified but inactive precursor nisin. As trypsin has the same cleavage specificity as the nisin protease (37), it was quite feasible to investigate this possibility. However, trypsin treatment of both culture supernatants and cell extracts indicated that no modified precursor existed in strain N12β cultures. RP-HPLC profiles of both culture supernatants and cell extracts confirmed that no peaks corresponding to nisin were present. The results also indicated that strain N12β had a profile similar to the profile of its parent strain, S12β, and did not produce a detectable peak at the expected position for precursor nisin. These results therefore suggested that problems in expression of the nisin structural gene operon may occur.
To investigate this possibility, transcription of nisA was examined by using Northern hybridization and RT-PCR. Both of these techniques confirmed that transcription of nisA in strain N12β was blocked. This was surprising as one of the four immunity genes, nisI, is also in the same transcription operon as nisA. However, it has previously been determined that partial but significant nisin immunity could still occur in the absence of NisI (29). Quantification of the nisin immunity exhibited by strain N12β revealed that it was only ≈50% that of L. lactis ATCC 11454 (Fig. 3), suggesting that sufficient NisI may not be present. However, as the other three nisin immunity genes, nisFEG, are transcriptionally regulated like the nisA promoter (28), it seems unlikely that their transcription would not also be affected. A possible explanation for the immunity exhibited by strain N12β was that the addition of nisin to the culture during the test for immunity stimulated transcription of the nisin promoters via the two-component NisRK regulatory system. This is plausible given our current understanding of regulation of nisin expression in L. lactis (9, 28), and it suggests that addition of exogenous nisin to cultures of strain N12β may stimulate transcription of the nisin promoters and result in the production of active nisin.
Investigation of this possibility revealed that nisA transcription could be restored in strain N12β following nisin induction (Fig. 4), together with restoration of nisin production (Fig. 5). This confirmed that the NisRK signal transcription system was working in Enterococcus, albeit at a much lower efficiency than in L. lactis. Interesting, the kinetics of nisin production were very similar for Enterococcus and L. lactis (Fig. 6), suggesting that the expression mechanisms were similar in the two backgrounds, except that they were less efficient in Enterococcus. Induction of cultures with different amounts of nisin revealed an approximately linear relationship between the amount of added nisin and the levels of both nisin transcription and production (Fig. 8). However, addition of nisin at concentrations up to 200 IU/ml could not fully restore nisA transcription or nisin production in N12β. This indicated that a much higher concentration of the nisin signal was required to stimulate the NisRK signal transduction system in Enterococcus than in L. lactis. It is also noteworthy that an induced culture of strain N12β quickly lost the ability to produce nisin when it was subcultured at levels up to 20% in a medium without added nisin (data not shown), further substantiating the lower sensitivity of the NisRK two-component induction system in the Enterococcus background. This is in contrast to the results obtained with L. lactis ATCC 11454 cultures, which can be diluted 10−9 and still continue to produce nisin (Li and O'Sullivan, unpublished data). Production of bacteriocins that are regulated by two-component systems in other lactic acid bacteria can also be hampered by subculturing with diluted inocula. These include plantaricin production by Lactobacillus plantarum C11 (10), enterocin A and B production by Enterococcus faecium CTC492 (27), and carnobacteriocin production by Carnobacterium piscicola LV17 (30). However, in these cases the reason was limited amounts of the inducer. In the case of N12β, the reason appears to be an inefficient signal transfer from the external inducer, nisin, to the internal response regulator, NisR.
While the reason for the inefficient functioning of the NisRK two-component system in Enterococcus sp. strain N12β is presently unknown, there are a number of possible explanations. One possibility is that there are some other unknown factors needed for expression of the nisin genes in L. lactis that were not transferred into strain N12β from the donor, L. lactis ATCC 11454. Previous studies have linked the loss of nisin production to the loss of a plasmid (15, 24, 34). However, in these studies it was not determined if the nisin transposon remained in the plasmid-cured strains. Another possible reason for inefficient signal transduction is cross talk between two different two-component systems in the host. Different two-component regulatory systems that exhibit extensive similarity in protein structure can each affect the signal transduction of the other (14, 38). For example, the kinase sensor protein VanS of the vancomycin resistance regulon was shown to activate PhoB (regulator protein of phosphate synthesis) in Escherichia coli (14). It is therefore possible that there is some signal competition among different two-component systems in strain N12β, which impedes full expression of the nisin genes.
It was also notable that while induction of Enterococcus sp. strain N12β with increasing amounts of nisin gave increasing amounts of nisA transcription, up to a maximum of nearly 60% of the amount in L. lactis ATCC 11454, the amounts of active nisin detected by the bioassay were significantly lower (maximum, ≈30% of the amount in L. lactis ATCC 11454) (Fig. 8). Trypsin treatments of cell homogenates did not release any further nisin, indicating that unprocessed nisin was not trapped within the cells (data not shown). HPLC analysis of culture supernatants could not detect any nisin (or other) peaks, as the amount of nisin present was less than the sensitivity of the HPLC assay used (0.2 IU/μl) (data not shown). The smaller amount of active nisin compared to the transcription amount was probably not due to a partially modified nisin peptide, as prior studies have shown that a lack of modification by either of the two modification enzymes, NisB and NisC, does not result in a bioactive peptide. Specifically, Ra et al. (29) obtained an in-frame deletion in nisB which eliminated nisin production, but transcription of the nisin genes could be partially restored following external induction with nisin, analogous to the situation in Enterococcus sp. strain N12β in this study. However, active nisin could not be restored in the induced nisB mutant, confirming that a lack of NisB modification functions does not result in a bioactive peptide. Similarly a mutation in nisC eliminated transcription, but transcription could be partially restored following external induction with nisin. However, the mutation of the induced nisC mutant did not result in production of a bioactive peptide or buildup of an intracellular unprocessed peptide. The requirement for both NisB and NisC modifications to obtain bioactivity strongly suggests that the bioactivity observed with Enterococcus sp. strain N12β is due to mature nisin. The reduced production compared to the level of transcription may be due to translation differences and/or mRNA processing differences between the Enterococcus and Lactococcus backgrounds.
In conclusion, Enterococcus sp. strain N12β does not produce nisin because the efficiency of the NisRK signal transduction system is significantly lower than that in L. lactis, resulting in a lack of transcription of the nisin genes. Uncovering the reason for inefficient NisRK signal transduction in Enterococcus is necessary to enable efficient nisin production in this heterologous host.
Acknowledgments
We thank Jeff Broadbent for providing Enterococcus sp. strains N12β and S12β for this study.
This work was supported in part by Dairy Management Inc. and by the Minnesota Agricultural Experimental Station.
REFERENCES
- 1.Broadbent, J. R., W. E. Sandine, and J. K. Kondo. 1995. Characteristics of Tn5307 exchange and intergeneric transfer of genes associated with nisin production. Appl. Microbiol. Biotechnol. 44:139-146. [DOI] [PubMed] [Google Scholar]
- 2.Chandrapati, S. 2001. Elucidating of regulatory mechanisms effecting nisin biosynthesis in Lactococcus lactis ATCC 11454. Ph.D. thesis. University of Minnesota, St. Paul.
- 3.Chandrapati, S., and D. J. O'Sullivan. 1998. Procedure for quantifiable assessment of nutritional parameters influencing nisin production by Lactococcus lactis subsp. lactis. J. Biotechnol. 63:229-233. [DOI] [PubMed] [Google Scholar]
- 4.Chandrapati, S., and D. J. O'Sullivan. 1999. Nisin independent induction of the nisA promoter in Lactococcus lactis during growth in lactose or galactose. FEMS Microbiol. Lett. 170:191-198. [DOI] [PubMed] [Google Scholar]
- 5.Cusick, S. M., and D. J. O'Sullivan. 2000. Use of a single, triplicate arbitrary primed-PCR procedure for molecular fingerprinting of lactic acid bacteria. Appl. Environ. Microbiol. 66:2227-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delves-Broughton, J. 1990. Nisin and its use as a food preservative. Food Technol. 44:100-112. [Google Scholar]
- 7.Delves-Broughton, J., P. Blackburn, R. J. Evans, and J. Hugenholtz. 1996. Application of the bacteriocin nisin. Antonie Leeuwenhoek 69:193-202. [DOI] [PubMed] [Google Scholar]
- 8.Delves-Broughton, J., G. C. Williams, and S. Wilkinson. 1992. The use of nisin as a food preservative in pasteurized liquid whole egg. Lett. Appl. Microbiol. 15:133-136. [DOI] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1995. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Microbiol. 18:631-639. [DOI] [PubMed] [Google Scholar]
- 11.Dodd, H. M., N. Horn, and M. J. Gasson. 1990. Analysis of the genetic determinant for the production of the peptide antibiotic nisin. J. Gen. Microbiol. 136:555-566. [DOI] [PubMed] [Google Scholar]
- 12.Engelke, G., Z. Gutochowski-Eckel, M. Hammelman, and K. D. Entian. 1992. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl. Environ. Microbiol. 58:3730-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelke, G., Z. Gutochowski-Eckel, M. Hammelman, and K. D. Entian. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl. Environ. Microbiol. 60:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, S. L., W. Jiang, B. L. Wanner, and C. T. Walsh. 1995. Cross-talk between the histidine protein kinase VanS and the response regulator PhoB. J. Biol. Chem. 270:23143-23149. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, C. F., and B. S. Kunka. 1985. Transfer of sucrose-fermenting ability and nisin production phenotype among lactic streptococci. Appl. Environ. Microbiol. 49:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst, A. 1981. Nisin. Adv. Appl. Microbiol. 27:85-123. [Google Scholar]
- 17.Immonen, T., and P. E. J. Saris. 1998. Characterization of the nisFEG operon of the nisin Z producing Lactococcus lactis subsp. lactis N8 strain. DNA Sequence 9:263-274. [DOI] [PubMed] [Google Scholar]
- 18.Immonen, T., S. Ye, R. Ra, M. Qiao, L. Paulin, and P. E. J. Saris. 1995. The codon usage of the nisZ operon in Lactococcus lactis N8 suggests a non-lactococcal origin of the conjugative nisin-sucrose transposon. DNA Sequence 5:202-218. [DOI] [PubMed] [Google Scholar]
- 19.Ingram, L. 1969. Synthesis of the antibiotic nisin: formation of lanthionine and β-methyllanthionine. Biochim. Biophys. Acta 184:216-219. [DOI] [PubMed] [Google Scholar]
- 20.Ingram, L. 1970. A ribosome mechanism for synthesis of peptides related to nisin. Biochim. Biophys. Acta 224:263-265. [DOI] [PubMed] [Google Scholar]
- 21.Kuipers, O. P., M. M. Beerthuyzen, W. M. De Vos, and R. J. Siezen. 1993. Biosynthesis and secretion of a precursor of nisin Z by Lactococcus lactis directed by the leader peptide of the homologous lantibiotic subtilin from Bacillus subtilis. FEBS Lett. 330:23-27. [DOI] [PubMed] [Google Scholar]
- 22.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of the expression of the nisA and nisI genes for producer immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers, O. P., M. M. Beerthuyzen, P. G. G. A. De Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 24.LeBlanc, D. J., V. L. Crow, and L. N. Lee. 1980. Plasmid-mediated carbohydrate catabolic enzymes among strains of Streptococcus lactis, p. 31-41. In C. Stuttard and K. R. Rozee (ed.), Plasmid and transposons: environmental effects and maintenance mechanisms. Academic Press, New York, N.Y.
- 25.Liu, W., and J. N. Hansen. 1990. Some chemical and physical properties of nisin, a small-protein antibiotic produced by Lactococcus lactis. Appl. Environ. Microbiol. 56:2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelis, N. J., L. D. Vuyst, K. Goubert, B. V. Devreese, J. J. Van Beeumen, J. G. Raymackers, E. J. Vandamme, and A. P. De Leenheer. 1996. Detection of cleavage of a prenisin-mimicking decapeptide by Lactococcus lactis subsp. lactis endopeptidase activity. Enzyme Microbiol. Technol. 18:52-58. [DOI] [PubMed] [Google Scholar]
- 27.Nilsen, T., I. F. Nes, and H. Holo. 1998. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J. Bacteriol. 180:1848-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiao, M., S. Ye, O. Koponen, R. Ra, M. Usabiaga, T. Immonen, and P. E. J. Saris. 1996. Regulation of the nisin operons in Lactococcus lactis N8. J. Appl. Bacteriol. 80:626-634. [DOI] [PubMed] [Google Scholar]
- 29.Ra, R., M. M. Beerthuyzen, W. M. de Vos, P. E. Saris, and O. P. Kuipers. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145:1227-1233. [DOI] [PubMed] [Google Scholar]
- 30.Saucier, L., A. S. Paradkar, L. S. Frost, S. E. Jensen, and M. E. Stiles. 1997. Transcriptional analysis and regulation of carnobacteriocin production in Carnobacterium piscicola LV17. Gene 188:271-277. [DOI] [PubMed] [Google Scholar]
- 31.Sen, A. K., A. Narbad, N. Horn, H. M. Dodd, A. J. Parr, I. Colquhoun, and M. J. Gasson. 1999. Post-translational modification of nisin—the involvement of NisB in the dehydration process. Eur. J. Biochem. 261:524-532. [DOI] [PubMed] [Google Scholar]
- 32.Siegers, K., and K. D. Entian. 1995. Gene involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegers, K., S. Heinzmann, and K. D. Entian. 1996. Biosynthesis of lantibiotic nisin. Posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J. Biol. Chem. 271:12294-12301. [DOI] [PubMed] [Google Scholar]
- 34.Steel, J. L., and L. L. McKay. 1986. Partial characterization of the genetic basis for sucrose metabolism and nisin production in Streptococcus lactis. Appl. Environ. Microbiol. 51:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steen, M. J., Y. J. Chung, and J. N. Hansen. 1991. Characterization of the nisin gene as a part of a polycistronic operon in the chromosome of Lactococcus lactis ATCC 11454. Appl. Environ. Microbiol. 57:1181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandermeer, J. R., J. Polman, J., M. M. Beerthuyen, R. J. Siezen., O. P. Kuipers, and W. M. de Vos. 1993. Characterization of the nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandermeer, J. R., H. S. Rollema, R. J. Siezen, M. M. Beerthuyzen, O. P. Kuipers, and W. M. de Vos. 1994. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J. Biol. Chem. 269:3555-3562. [PubMed] [Google Scholar]
- 38.Wright, G. D., T. R. Holman, and C. T. Walsh. 1993. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32:5057-5063. [DOI] [PubMed] [Google Scholar]