Abstract
Glucose-dependent growth of the luxCDABE reporter bacterium Pseudomonas fluorescens HK44 was monitored noninvasively in quartz sand under unsaturated-flow conditions within a 45- by 56- by 1-cm two-dimensional light transmission chamber. The spatial and temporal development of growth were mapped daily over 7 days by quantifying salicylate-induced bioluminescence. A nonlinear model relating the rate of increase in light emission after salicylate exposure to microbial density successfully predicted growth over 4 orders of magnitude (r2 = 0.95). Total model-predicted growth agreed with growth calculated from the mass balance of the system by using previously established growth parameters of HK44 (predicted, 1.2 × 1012 cells; calculated, 1.7 × 1012 cells). Colonization expanded in all directions from the inoculation region, including upward migration against the liquid flow. Both the daily rate of expansion of the colonized zone and the population density of the first day's growth in each newly colonized region remained relatively constant throughout the experiment. Nonetheless, substantial growth continued to occur on subsequent days in the older regions of the colonized zone. The proportion of daily potential growth that remained within the chamber declined progressively between days 2 and 7 (from 97 to 13%). A densely populated, anoxic region developed in the interior of the colonized zone even though the sand was unsaturated and fresh growth medium continued to flow through the colonized zone. These data illustrate the potential of a light transmission chamber, bioluminescent bacteria, and sensitive digital camera technology to noninvasively study real-time hydrology-microbiology interactions associated with unsaturated flow in porous media.
Field scale bioremediation of the vadose zone continues to be an unpredictable process, in part because of a poor understanding of the factors that influence microbial growth under conditions of unsaturated flow in the subsurface. The U.S. Department of Energy has identified as a priority research goal for the next decade the need to understand better the linkage between microbial contaminant interactions and hydrologic processes in the subsurface (40). To address facets of this issue, we have developed a novel method to examine the interactions between microbial growth and solute transport under dynamic flowing conditions in unsaturated porous media. The method uses a light transmission chamber to quantify pore water content and hydraulic flow paths and bioluminescent bacteria to measure microbial activity and growth in response to solute movement.
The light transmission method for determining water content in thin slabs of porous media was introduced by Hoa (15) and further developed by Glass et al. (12) and Tidwell and Glass (37). Selker et al. (32) extended the method to the observation of solute distribution by application of dyed solutes. Niemet and Selker (22) made significant refinements in water content quantification in quartz sands by this method. The potential to use light transmission chambers and associated charge-coupled device (CCD) technology to examine microbial-hydrologic interactions has not been examined. In recent years, bacterial lux genes have been used to detect and to monitor the fate of microorganisms introduced into natural environments (8, 10, 11, 26, 33) and as an indicator of bacterial metabolic state (9, 18). In addition, fusions of lux genes to inducible catabolic genes have been used to detect bioavailable concentrations of organic contaminants and heavy metals in the environment (2, 4, 6, 13, 14, 17, 31, 34). A recent study describes the use of a fiber optic probe to monitor activity of the lux-marked bacterium Pseudomonas putida RB1353 in porous media under saturated flowing conditions (43). To our knowledge, there are no published studies in which lux gene-dependent bioluminescence has been used to noninvasively quantify bacterial population growth in porous media under unsaturated-flow conditions. Pseudomonas fluorescens HK44 carries a nahG-luxCDABE transcriptional fusion (17). Exposure of P. fluorescens HK44 to naphthalene or to its metabolite salicylate results in induction of the lux system and emission of light. Recent work in our laboratory demonstrated the ability to utilize the inducible bioluminescence response of HK44 to quantify its population density in saturated and unsaturated sand under static conditions (38).
The objectives of the work reported here were twofold. The first objective was to determine if light transmission chambers could be adapted for studying microbial growth under conditions of unsaturated flow. The second objective was to determine if the salicylate-inducible bioluminescence model could be used to quantify growth of HK44 under unsaturated-flow conditions in the light transmission chambers.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
P. fluorescens HK44 (supplied by Gary Sayler, University of Tennessee, Knoxville) hosts the plasmid pUTK21 containing the salicylate-inducible luxCDABE gene cassette and a tetracycline resistance marker (17). All media used in these experiments were amended with tetracycline (15 mg liter−1) to ensure maintenance of the plasmid. Stock liquid cultures were grown routinely in nitrate-free minimal mineral salts medium (MMS) supplemented with glucose (1 g liter−1). For chamber experiments the liquid medium was modified by reducing the glucose concentration to 0.25 g liter−1. Salicylate (0.1 g liter−1) was periodically added to induce bioluminescence. Although HK44 is capable of growth on salicylate, we have found that salicylate neither is consumed nor supports growth over 12 to 20 h after addition to glucose-grown cells (data not shown).
Porous medium.
The chamber was packed with an optically translucent 40/50-mesh size silica sand (Accusand; Unimin Corp., LeSueur, Minn.). The physical, chemical, and optical properties of this material have been described elsewhere (22, 28). The sand was prepared by soaking in a 5 M NaCl solution and thorough rinsing with distilled water to remove fine particulates and residual salt. The washed sand was autoclaved three times for 1 h each time and allowed to stand at room temperature for at least 24 h between each sterilization. After the third autoclaving, the sand was oven dried (50°C) to a constant weight.
Description of the light transmission chamber.
The chamber consists of two plate glass sheets, 51.0 cm wide by 61.0 cm high by 1.3 cm thick, separated by a 1.0-cm-thick U-shaped aluminum spacer and sealed to the spacer by fluorocarbon rubber O-ring stock (Viton; Dupont Dow Elastomers, Wilmington, Del.). The glass sheets can be drilled with various configurations of ports to allow inoculation or sampling of the chamber. For this work, the front sheet of chamber glass was drilled with a series of nine ports in an inverted T configuration (Fig. 1, first panel). The ports are sealed with high-temperature silicon sealer to allow injection or sampling via syringe. The lower side of the aluminum spacer contains a drain port and an integral manifold surmounted with a 10-μm-nominal-pore-size Twilled Dutch Weave stainless steel wire screen (Screen Technology Group, Inc., Washougal, Wash.). The bubbling pressure of the screen allows adjustment of the relative proportions of unsaturated and saturated sand within the chamber in response to the elevation of the open end of a tube attached to the drain port. All chamber materials were selected to be resistant to autoclaving. The chamber is clamped within an aluminum frame, which mounts to a fan-ventilated sheet metal housing containing a light bank of 10 fluorescent tubes (Philips F17T8/TL835, 61 cm). The light bank provides the source of illumination for creating light transmission images to determine water content of the sand (22). An illustration of the components of a typical light transmission chamber is provided by Niemet and Selker (22). The chamber-light box unit is attached to a sturdy mount that can be rotated to a horizontal position to facilitate sampling of the chamber contents at the conclusion of an experiment. The internal dimensions of the assembled chamber are 44.0 cm wide by 56.0 cm high by 1.0 cm thick. Water and feed solutions are applied to the surface of the chamber through an influent manifold composed of 11 23-guage syringe needle emitters spaced 4.1 cm apart. Each emitter is fed individually via a multichannel peristaltic pump (Cole-Parmer, Vernon Hills, Ill.), allowing precise control of the flow rate. Free exchange of air to the upper chamber is facilitated by the addition of several ports equipped with 0.2-μm-pore-size inline filters. The upper portion of the influent manifold housing contains a 15-W germicidal lamp (G15T8; General Electric, Cleveland, Ohio) that is used to irradiate the drip emitters and the surface of the sand with UV-C light. The lamp is operated periodically throughout the course of an experiment by use of a computer-controlled timer. Feed solutions are held in autoclaveable carboys (Nalgene, Rochester, N.Y.) with ventilated closures equipped with 0.2-μm-pore-size filters.
FIG. 1.
Temporal and spatial colonization of the light transmission chamber by P. fluorescens HK44 visualized by salicylate-inducible bioluminescence. The first panel shows a view of the chamber under room lighting. Drilled sampling port locations are marked by crosses. The superimposed rectangle defines the region of the chamber shown in each of the subsequent panels. The images have been artificially colorized to clearly show differences in emission intensity (arbitrary LU). The cross superimposed on each image indicates the location of the inoculation port, which is also shown as the uppermost port in the first panel. Digital image processing was carried out using Transform 3.4 (Fortner Software LLC), and processed images were prepared for publication using Photoshop 5.0 (Adobe Systems Inc., San Jose, Calif.).
Chamber preparation.
(i) Sterilization.
After assembly, the chamber was sterilized by autoclaving and allowed to dry for several days prior to being packed with sterile sand. Preliminary attempts to autoclave the chamber assembly after packing with sand were not successful, because differential expansion of the chamber materials during autoclaving caused the packed sand to slump. The influent manifold housing was washed with germicidal detergent, rinsed with hot water, and allowed to dry. The drip emitter fittings and tubing were autoclaved, and the manifold unit was assembled under a laminar flow hood. After assembly, all surfaces of the unit were sterilized by UV light. All other equipment was autoclaved prior to use.
(ii) Sand packing.
Chamber packing was carried out under a modified laminar flow hood, with its sides and top extended in order to accommodate the chamber and hopper assemblies. Sterile sand was added to the chamber from a funnel-hopper assembly until just above a fill line marked on the front glass sheet (48 cm above the surface of the effluent manifold screen). The fill was stopped shortly after the packing front had passed the fill line. The overburden was removed by vacuuming through a sterile tube. The upper interior of the chamber and the sand surface were then irradiated with UV light for 1 min. The porosity of the sand pack was 0.33 cm3 cm−3.
(iii) Influent flow rate calibration and initial chamber saturation.
After all tubing connections were made, the flow rates for the influent emitters were calibrated. The process was carried out under constant UV irradiation. The flow rate per individual dripper was 28.7 ± 0.4 ml h−1 (overall flow rate, 315.6 ± 4.6 ml h−1). After flow calibration, the influent manifold and UV unit were attached to the chamber, and the assembly was attached to the light box. The chamber was slowly purged through the drain port with 0.2-μm-pore-size-filtered CO2 gas to displace air from the sand and minimize the risk of trapping gas bubbles in the sand when the chamber was first filled with water (CO2 has greater water solubility than air). Sterile MMS was then slowly delivered through the drain port until ponding occurred at the sand surface. A siphon tube integrated into the influent manifold was opened, and several pore volumes of MMS were pumped through the system from the bottom up to remove CO2.
(iv) Inoculation.
P. fluorescens HK44 was grown overnight with shaking (150 rpm) at 27°C in MMS supplemented with glucose (1.0 g liter−1). The culture was centrifuged, washed with MMS basal medium, and resuspended to 5 × 108 CFU ml−1 in MMS supplemented with salicylate (0.1 g liter−1) to induce bioluminescence. After the culture was visibly bioluminescent, 5.0 ml was injected into the saturated sand through a port in the front glass sheet of the chamber at a depth of 15 cm below the sand surface. This resulted in an inoculated region with a circular surface area 4.4 cm in diameter and a bulk volume of approximately 15 cm3. After 1 h, flow through the influent drip emitters was started, and the effluent port was opened to allow the chamber to drain. MMS was applied through the outer 10 drip emitters, and MMS plus glucose (0.25 g liter−1) was applied through the center dripper only. The open end of the effluent drain tube was positioned at an elevation 1 cm above the lower edge of the sand pack. Under these conditions, the capillary fringe rose to the midpoint of the sand pack. The upper half of the sand pack remained unsaturated with a volumetric water content of about 0.12 cm3 of water per cm3 of total volume. The inoculated chamber was maintained at a temperature of 21°C (standard deviation, 1°C) during the course of the experiment. Because of the complexity of the system, the large number of manipulations required in its setup and during data collection, and the necessity of maintaining a fixed position of the chamber in relation to the light measuring system during the course of an experiment, only one chamber was operated per experiment. Nonetheless, the data were reproducible among experiments.
Data collection and analytical methods. (i) Light measuring system.
The light detection system consisted of a thermoelectrically cooled 14-bit digital CCD camera (ISI Systems, Santa Barbara, Calif.) with a Kodak KAF0400 (768 by 512 pixels) scientific-grade CCD array. The lens was a Nikon 35-mm, f-1.4-aperture lens (Nikon Corporation, Tokyo, Japan). Light transmission images were obtained using a 620-nm (red) center-wavelength 10-nm-bandpass filter (model 53930; Oriel Co., Stratford, Conn.) installed in the camera's internal filter wheel. Bioluminescence images were obtained using an optically clear glass filter to maintain a consistent optical path. The distance from the chamber to the lens was 3.86 m. With this system geometry, the entire chamber was included in each image, and each image pixel represented approximately 1 mm2 of chamber surface area. Each image provided more than 225,000 discrete measurements over the experimental domain. Digital image processing was performed using Transform 3.4 (Fortner Software LLC, Sterling, Va.) as described previously (22). The following measurements were made with this system.
(ii) Water content of porous medium.
Light transmission images were collected several times each day, and the water content was determined by the method of Niemet and Selker (22). Exposures of 0.8 s were made at an aperture setting of f-1.4 with the 620-nm-centered 10-nm-bandpass filter.
(iii) Quantification of the population with bioluminescence.
The bioluminescence response was measured once each day as follows. The outer 10 of the 11 influent drip emitters were switched to a solution of MMS and salicylate (0.1 g liter−1), and the central dripper was switched to a solution of MMS plus glucose (0.25 g liter−1) and salicylate (0.1 g liter−1). Salicylate was applied onto the chamber for 150 min, and then the feed systems were restored to the standard solutions. The bioluminescence response was recorded in a series of images (10-min exposure, f-1.4 aperture), with the clear filter in place, taken at 20-min intervals. Uesugi et al. (38) developed a model relating cell density to the time-dependent increase in light production after exposure to the inducer. For cells in the presence of sand, the model is expressed in the equation
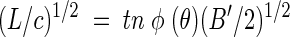 |
(1) |
where L is light units (LU) milliliter of pore liquid−1, c is the P. fluorescens HK44 cell density in CFU milliliter of pore liquid−1, t is the time in minutes since exposure to salicylate, and B′ is the rate of increase in cellular light production rate with units of LU (cell minute2)−1. The term nφ(θ) incorporates the attenuating effects of porosity, n, and the effect of variations in volumetric water content, φ(θ), on light detected from the sand. The φ(θ) relationship was determined empirically (38). The linear portion of a plot of (L/c)1/2 versus t has the following slope, m:
 |
(2) |
Solving equation 2 for B′ gives
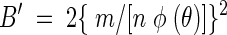 |
(3) |
When B′ is known, the following expressions equivalent to equations 1, 2, and 3 can be used to solve for cell density, c:
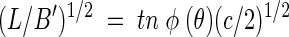 |
(4) |
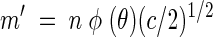 |
(5) |
 |
(6) |
where m′ is the slope of the linear portion of a plot of (L/B′)1/2 versus t.
Uesugi et al. (38) validated their model with nongrowing cells under nonflowing conditions. The induction time, t, for any given image was constant for the entire imaged area, and the volumetric water content, θ, used to obtain nφ(θ), was also constant. Under our experimental conditions, however, water content varied with position in the chamber, and time of first exposure to salicylate varied with both position and water content. The water content, and thus the nφ(θ) term, for any position in the chamber can be obtained from a light transmission image taken at about the same time as the emission images (i.e., just prior to the start of salicylate application). A reasonable value for the induction time, t, at a given position can also be obtained using the same light transmission image by assuming a uniform flow field across the width of the chamber and applying the following one-dimensional travel time equation:
 |
(7) |
where T is the travel time in minutes, z is the depth in centimeters, θ is the pixel row-averaged water content at that depth, and q is the areal flux rate in centimeters minute−1. In the work reported here, there was a 2-min delay (D) between the times that the salicylate pulse was started and that salicylate first reached the surface of the sand. This represented the time required for salicylate to travel through the influent lines to the drippers. The total travel time to move from the sand surface to the effluent manifold was 135 min. The induction time, t, for a given exposure becomes
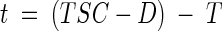 |
(8) |
where TSC is the elapsed time in minutes (at shutter closing) since the start of the pulse. Substituting equation 7 into equation 8 gives
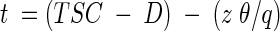 |
(9) |
The B′ values have dimensions of LU and thus are a function of camera sensitivity and position of the experimental system in relation to the camera. Therefore, we determined the value for B′ appropriate for our system by using the relationship described by equation 1 and experimentally measured values for population density determined by destructive sampling (see the next section) and the day 7 light emission data. Sixteen samples corresponding to regions of the colony visibly glowing on day 7 were selected for comparison. In these samples, volumetric water contents ranged from 0.11 to 0.33 cm3 cm−3, and measured cell densities ranged over 4 orders of magnitude. The results are presented in Table 1. The mean value for B′ was found to be 12.1 × 10−10 (95% confidence interval [CI], 8.4 × 10−10 to 15.8 × 10−10).
TABLE 1.
Determination of B′ valuea
| Measured population (1010 CFU ml−1) | Volumetric moisture (cm3 cm−3) | Slope (10−6) | r2 | B′ (10−10) |
|---|---|---|---|---|
| 0.00069 | 0.33 | 3.10 | 0.99 | 11.6 |
| 0.030 | 0.14 | 1.66 | 0.98 | 27.1 |
| 0.060 | 0.14 | 0.88 | 0.98 | 7.5 |
| 0.077 | 0.15 | 1.18 | 0.99 | 11.7 |
| 0.13 | 0.15 | 0.91 | 0.98 | 7.6 |
| 0.29 | 0.19 | 1.69 | 0.96 | 12.7 |
| 0.44 | 0.15 | 1.65 | 0.98 | 24.1 |
| 0.56 | 0.14 | 1.34 | 0.98 | 18.2 |
| 0.64 | 0.16 | 1.11 | 0.98 | 9.2 |
| 1.4 | 0.15 | 1.38 | 0.98 | 16.4 |
| 1.6 | 0.12 | 1.08 | 0.97 | 18.4 |
| 1.9 | 0.15 | 0.69 | 0.97 | 4.2 |
| 2.2 | 0.11 | 0.44 | 0.96 | 4.0 |
| 5.9 | 0.13 | 0.78 | 0.96 | 8.2 |
| 6.2 | 0.13 | 0.77 | 0.96 | 8.5 |
| 9.8 | 0.11 | 0.50 | 0.96 | 4.8 |
Samples (n = 16) were obtained from bioluminescing regions of the colonized zone. The slope and r2 values are for a plot of (L/c)1/2 against time (see text). Mean B′ value, 12.1 × 10−10 (95% CI, 8.4 × 10−10 to 15.8 × 10−10).
(iv) Biochemical analyses of chamber and effluent.
Effluent samples were analyzed for biomass protein and for residual glucose. The entire first 1,100 ml of chamber effluent was collected to ensure capture of the portion of the inoculum initially washed from the sand. Subsequently, 35-ml aliquots of chamber effluent were obtained at intervals throughout the course of the experiment. Cells were collected by centrifugation and washed with MMS. The first supernatant fraction and the cell pellets were stored at −20°C and analyzed later for residual glucose and total cell protein by using anthrone (5) and bicinchoninic acid (BCA), respectively. The Micro BCA protein assay kit (Pierce, Rockford, Ill.) was employed for protein measurements. Samples of pore liquid were obtained from each of the five lower sampling ports in the chamber front glass sheet at intervals throughout the experiment and analyzed for dissolved oxygen concentrations with a model 5300 Biological Oxygen Monitor (Yellow Springs Instrument Co., Yellow Springs, Ohio) equipped with a YSI model 5331 Clark-type oxygen microelectrode and a 1.8-ml water-jacketed sample chamber. At the end of the experiment, the chamber was rotated into a horizontal position, and samples (2.5 by 2.5 by 1.0 cm) of chamber contents were aseptically collected according to a predetermined grid pattern (100 sectors) and individually analyzed for viable cell count (by dilution plating on tryptic soy broth agar plates amended with tetracycline), total cell protein (using BCA), and gravimetric water content. A CCD image of the grid pattern provided a reference between light transmission-emission image data and sampling data.
RESULTS
Spatial development of colonization.
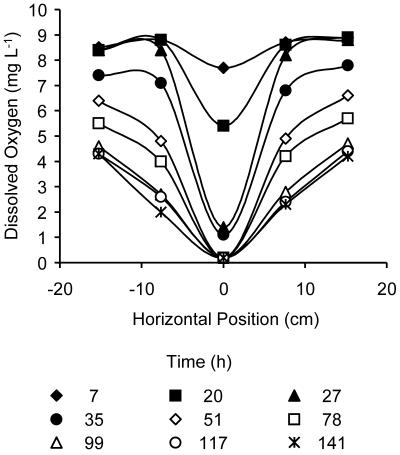
Figure 1 illustrates HK44 colonization of the light transmission chamber as visualized with images of the daily maximum bioluminescence for each day of the experiment. The image for day 1 was taken immediately after inoculation and shows the initial distribution of cells before flow was started. Microbial colonization spread from the 4.4-cm-diameter inoculated region on day 1 to cover a region of >17 cm in width by day 7. On day 2, aqueous-phase cells transported by the downward flow were clearly visible as streams of bioluminescence in the water-saturated region below the colonized zone. On subsequent days, light emission from the chamber was observed only in the unsaturated region. However, bioluminescence was observed in the effluent upon exposure to air as it exited the chamber. By day 4, the spatial pattern of bioluminescence developed a horseshoe-like appearance with a “dark” zone below the inoculation port that did not respond to the application of salicylate. This pattern prevailed for the remainder of the experiment. Measurement of dissolved O2 concentrations in pore water samples revealed that O2 was depleted most rapidly and extensively directly below the colonized region and had declined to <2 mg liter−1 by the second day of the experiment (Fig. 2). The zone of oxygen depletion expanded outward with time, paralleling the expansion of the colonized region. At the end of the experiment both viable plate counts and total cell protein analysis revealed that sufficient numbers (up to 7 × 109 CFU ml−1) of viable bacterial cells existed in the dark interior region of the colonized zone to produce a measurable bioluminescent response (data not shown). In addition, a dye tracer introduced through the central influent dripper demonstrated that salicylate solution passed through the entire colonized area (data not shown). The stability of the bioluminescent phenotype in the population recovered from the dark region was verified by observation of light emission after exposing plated colonies to naphthalene vapors. Presumably, anoxic conditions were established and maintained within the colonized zone despite the facts that the sand was unsaturated and that fresh air-saturated liquid medium was continuously supplied to the chamber.
FIG. 2.
Downstream dissolved oxygen dynamics in response to growth of P. fluorescens HK44 in the light transmission chamber. The horizontal axis indicates the position (distance to the left or right) of each sampling port with respect to the vertical center line of the chamber (defined as 0). The ports are marked as the horizontal row of crosses in the first panel in Fig. 1. The elapsed times after the start of the experiment are indicated.
Light response profiles.
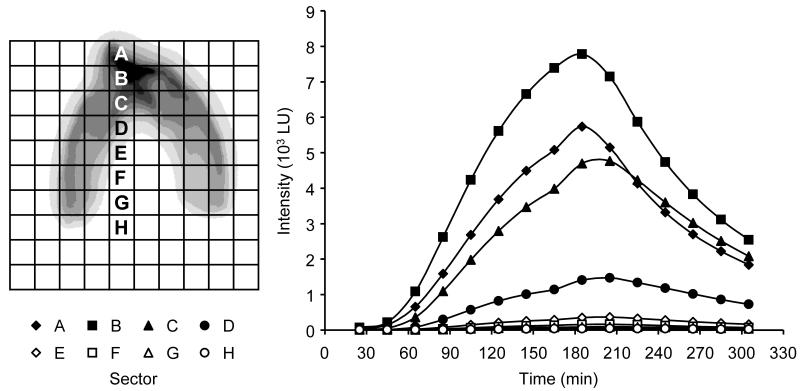
Figure 3 shows the day 7 bioluminescence response profiles for eight 2.5- by 2.5-cm sectors along a vertical transect through the colonized zone. LU are the mean values accumulated over each 10-min exposure for the 625 pixels within each of the sectors. Time zero indicates the start of the 150-min salicylate pulse. The elapsed time before emission detection, peak emission level, and time of peak emission all varied with depth through the colonized region. For example, light emission from the upper three sectors (A through C) exceeded 50 LU at 45 min after salicylate addition. The time increased by 20 min for each of the next four lower sectors (D through G). Emission for the lowest sector did not reach 50 LU until 205 min after the start of the salicylate pulse. Peak emission levels varied by over 2 orders of magnitude, with a high of 7,800 LU (sector B) and a low of 50 LU (sector H). Peak emission was recorded at 185 min for the upper two sectors (A and B) and at 205 min for the lower eight sectors (C through H). After salicylate was removed from the system, light emission declined in a logarithmic fashion. The half-life for light emission decay was 78.6 min (95% CI, 76.0 to 81.4 min) determined over all 57 sectors where the sector mean light emission reached >100 LU (10 LU min−1).
FIG. 3.
Day 7 P. fluorescens HK44 bioluminescence response profiles. Light emission values represent the mean cumulative counts recorded for each 2.5- by 2.5-cm region (A to H) shown in the left panel. Time zero is the start of the salicylate pulse (0.1 g liter−1).
Defining the extent of colonized area.
During the induction phase of bioluminescence, the intensity of light emitted from a given colonized point increases with time. Because the light originating at that point travels in all directions and is reflected and refracted at interfaces of air, water, and glass within the imaged area, the extent and intensity of an apparent halo of light about that point also increase (referred to here as the lantern effect). As a result, the apparent edge of the colonized area detected by the CCD shifts during the course of the bioluminescence response recorded on a given day. It is therefore necessary to delineate the borders of the colonized zone to avoid overestimating the extent of colonization. Figure 4A shows an emission profile for the fifth image collected during the salicylate pulse for a horizontal transect through the colonized zone at a depth of 15 cm (the inoculation depth) taken on day 3 of the experiment. The criteria for using the fifth image are that it falls within the middle of the increasing portion of the bioluminescence response curve (Fig. 3), all regions of the chamber have had sufficient time for induction, and salicylate is still present everywhere in the chamber. A similar profile was generated for each day of the experiment. The nearly vertical portion of each side of the curve was extrapolated to the baseline to define the edge of the colonized zone. The observed emission intensities at these points for the right and left sides were then averaged to obtain a threshold value for a given day. The threshold value was used to generate a contour line defining the perimeter of the colonized zone. Stacking of the daily perimeter threshold plots reveals the development of the colonized region with time (Fig. 4B). Moving from the middle outward, the contour lines represent the maximum extent of the colony on subsequent days. The outermost, dashed line represents the relative extent of the lantern effect observed on day 7. The number of pixels contained within each of the bioluminescent regions was used to calculate the changing area of the colony on each day (Table 2). The colonized area increased from 15.8 cm2 on day 1 to 224.4 cm2 on day 7. The average rate of daily increase in the colonized area was relatively constant (34.7 ± 7.2 cm2 day−1).
FIG. 4.
(A) Light emission profile of a horizontal transect through the P. fluorescens HK44-colonized zone at a depth of 15 cm (day 3). a, colonized region; b, light emission from the lantern effect. (B). Daily areal expansion of the colonized zone. Shaded regions (1 to 7) represent the newly colonized area on each day. The dashed line illustrates the apparent boundary of the colonized region on day 7 if uncorrected for the lantern effect.
TABLE 2.
Daily expansion of colonization and model-predicted populations in each of the regions (1 to 7) of the colonized zone defined in Fig. 4B
| Day | Colonized area
|
Predicted population (1010 CFU) in region:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (cm2) | Change (cm2 day−1)a | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1 | 15.8b | ||||||||
| 2 | 54.7 | 38.8 | 5.7 | 5.3 | |||||
| 3 | 83.7 | 29.0 | 13.1 | 12.9 | 3.6 | ||||
| 4 | 106.8 | 23.1 | 7.3 | 7.2 | 7.5 | 4.2 | |||
| 5 | 150.7 | 43.8 | 30.9c | 18.4c | 19.7c | 15.8c | 19.0 | ||
| 6 | 191.5 | 40.8 | 14.2 | 13.4 | 15.3 | 10.6 | 21.9c | 9.3 | |
| 7 | 224.4 | 32.9 | 7.0 | 7.8 | 9.3 | 5.5 | 15.3 | 11.1c | 4.1c |
Average change ± standard deviation, 34.7 ± 7.2 cm2 day−1.
Inoculum; the predicted population was not determined.
Maximum bioluminescence model-predicted population for the region. The sum of the maximum model-predicted values for all regions is 121.8.
Quantification of the HK44 population with bioluminescence.
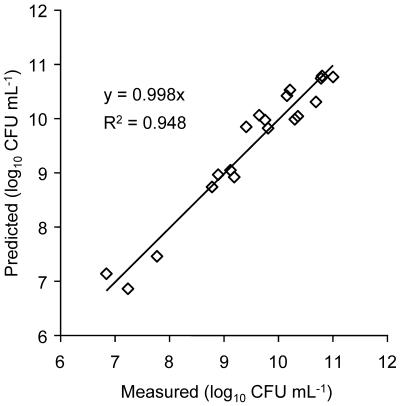
Figure 5 shows the population density (log CFU milliliter−1) predicted by the model for density-dependent bioluminescence using day 7 light emission data compared to population density (log CFU milliliter−1) measured on day 7 after destructive sampling. The comparison was made with 19 samples obtained from the visibly glowing outer regions of the colonized zone, where O2 was assumed to be nonlimiting. The slope of the fitted regression line was 0.998 (95% CI, 0.985 to 1.012), with an r2 value of 0.948. The average B′ value (12.1 × 10−10 [see Materials and Methods]) determined from the day 7 light emission data and population density measured after destructive sampling was used to apply the model to predict population densities for earlier days in the experiment. The bioluminescence data were transformed with the relationship (L/B′)1/2. The corresponding light transmission images for water content were transformed into a series of data sets representing the time, t, elapsed since the first exposure to salicylate for each position in the domain by use of equation 9. The same light transmission images were also transformed into data sets containing the water content-related correction associated with each position in the domain [nφ(θ)]. Next, the (L/B′)1/2 data sets and the time (t) data sets were used to construct a series of data sets containing the slope values m′ for B′ by linear regression. Finally, equation 6 was used to construct a data set containing the predicted population density, in CFU milliliter of pore liquid−1, for each position in the domain, using both the slope (m′) data sets and the water correction [nφ(θ)] data sets.
FIG. 5.
Model-predicted and measured population densities of P. fluorescens HK44 associated with 2.5- by 2.5-cm sand samples (n = 19) recovered from bioluminescing regions of the colonized zone.
Table 2 shows the predicted populations corresponding to the regions shown in Fig. 4B for each day of the experiment. With the exception of day 5, a relatively constant cell density (cells per square centimeter) developed in each new daily colonized region throughout the course of the experiment (1.5 × 109 ± 0.4 × 109 CFU cm−2, omitting day 5). There was also a trend for populations to continue to increase in regions first colonized on the previous day. Because the bioluminescence reaction requires oxygen, the subsequent decrease in predicted population for a given region might be a result of O2 depletion. Therefore, the maximum predicted population values for each region were summed to calculate the total number of cells produced during the experiment (1.2 × 1012 cells). Growth predicted by bioluminescence compared favorably with potential growth calculated by using glucose consumption, cell yield, and effluent biomass protein data (Table 3). Given the quantity of glucose consumed (861 mg), a yield coefficient of 0.54 mg of cells (dry weight) mg of glucose−1, protein representing 55% of cell dry weight, and an individual cell dry weight of 2.7 × 10−10 mg, the potential total cell production was 1.7 × 1012 cells. Overall, 64.8% of the calculated potential cell production was recovered in the effluent (301 mg of 465 mg of cells [dry weight]). Subtracting the effluent biomass from the potential production leaves 164 mg of cells associated with the sand at the end of the experiment, or the equivalent of 0.6 × 1012 cells.
TABLE 3.
Potential growth based on mass balance
| Day | Measured quantity
|
Inferred potential productiona
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucose consumedb (mg) | Effluent cellsc
|
Total
|
Sand associated
|
||||||
| mg | 1010 CFU | mg of biomass | 1010 CFU | Newd
|
Cumulative
|
||||
| mg of biomass | 1010 CFU | mg of biomass | 1010 CFU | ||||||
| 2 | 33.3 | 0.6 | 0.2 | 18.0 | 6.7 | 17.4 | 6.5 | 17.4 | 6.5 |
| 3 | 128.4 | 14.2 | 5.3 | 69.3 | 25.9 | 55.1 | 20.6 | 72.5 | 27.1 |
| 4 | 155.6 | 46.4 | 17.3 | 84.0 | 31.3 | 37.6 | 14.0 | 110.1 | 41.1 |
| 5 | 169.9 | 68.9 | 25.7 | 91.7 | 34.2 | 22.8 | 8.5 | 132.9 | 49.6 |
| 6 | 171.2 | 75.6 | 28.2 | 92.4 | 34.5 | 16.8 | 6.3 | 149.7 | 55.9 |
| 7 | 202.7 | 95.6 | 35.7 | 109.5 | 40.9 | 13.9 | 5.2 | 163.6 | 61.0 |
| Total | 861.1 | 301.3 | 112.3 | 464.9 | 173.5 | ||||
Based on glucose consumption and the following growth parameters: yield on glucose, 0.54; cell dry weight, 2.7 × 10−13 g; cell composition, 55% protein.
Glucose input minus glucose measured in effluent.
Based on measured effluent total cell protein.
Total daily potential growth minus daily measured effluent cells.
DISCUSSION
Previous work with the light transmission-flow cell chambers highlighted their ability to provide insight into hydrologic properties of porous media (22, 28, 29, 30, 32, 37). In this study we clearly show the potential to adapt these chambers for studying microbial-hydrologic interactions under unsaturated-flow conditions at a relatively small scale of resolution (<1 cm). The nonlinear model describing the relationship between the rate of increase in light production over time and population density of P. fluorescens HK44 (38) gave a good fit between predicted population density based on bioluminescence and measured population density of colonized regions over 4 orders of magnitude. The robustness of the model is remarkable considering that it was developed initially with homogeneous populations of culture-grown cells added to sand under nonflow conditions and not with a complex mixture of attached and unattached cells of different ages and physiologies that comprise a chamber-grown population. The induction phase of light production is the critical feature of the model which permits bioluminescence measurements to be made relatively quickly (in <3 h). Although other studies have reported a linear relationship between light emission and bacterial cell density, either when bioluminescence is measured at a fixed time point after induction or at peak bioluminescence (21, 26, 43), the necessity of waiting to capture the peak of bioluminescence may lead to complications if population changes occur more quickly than bioluminescence data can be accrued. There are limitations to using a lux-based reporter system for this kind of work. For example, oxygen is essential for bioluminescence, rapid growth rates might compromise the model, and the presence of catabolite-repressive substrates might prevent lux gene induction. The naphthalene pathway genes are not catabolite repressed (6, 27). We found that the magnitude of colonization in the interior of the colony was underpredicted. Taking into account the downstream dissolved oxygen data and the fact that bioluminescence was observed in the chamber effluent, we presume that this effect was largely due to O2 limitation. On the other hand, the observation of the development of the dark interior region of the colony points out the potential usefulness of bioluminescence to map the location and spatial and temporal development of aerobic and anaerobic zones in this dynamic experimental system. Establishment of a threshold to define the extent of the colonized zone, while necessary to avoid overprediction due to the lantern effect, is a compromise. There is no biological reason to assume that the spatial border of the colonized zone should be very sharp, and the true outer limit of the colonized zone containing a low density of cells might be underestimated. Addition of a second, constitutively expressed, light-based biomarker that functions independently of O2 availability would provide a considerable advantage over a solely lux-based system. Green fluorescent protein-lux-marked organisms (see, e.g., reference 39), for example, may permit independent observation of both microbial distribution and O2-dependent activity in the light transmission chambers.
Our experimental observations raise some interesting questions about the factors promoting spatially heterogeneous behavior in a bacterial population under unsaturated-flow conditions. For example, while both the rate of expansion of the colonized zone and the predicted population densities that developed daily in the newly colonized regions remained relatively constant throughout the experiment, populations also continued to increase in regions first colonized on the previous day, indicating that glucose and oxygen deliveries were maintained to some degree. It is not clear, therefore, to what extent flow and substrate movement were redirected in response to colonization. While redirection of flow around the colony might create substrate gradients that promote migration and growth away from the colonized zone, an attached biofilm-like behavior within the colonized zone is probably necessary to permit further increases in population density. In contrast to aquatic systems where biofilm structure has been well studied and modeled (7, 23, 24, 35), limited data are available on the structure of bacterial colonies formed under unsaturated conditions (3, 16). A number of studies in the literature indicate that under unsaturated conditions, bacteria may associate with air-water interfaces and are not necessarily attached to solid surfaces (see, e.g., references 25 and 41). Furthermore, it is well documented that physical heterogeneities in porous media locally modify flow, and these perturbations influence microbial growth and transport behavior (19, 20, 36). In our experiments, microbial growth itself created the heterogeneity in a previously homogeneous porous medium, thereby perturbing flow and promoting spatial heterogeneity in the microbial colony. Further studies are needed to determine the mechanisms underpinning the microbial-hydrologic interactions whereby unsaturated flow is modified to promote both distal growth and continuation of localized growth.
The proportion of daily potential cell production that remained in the chamber declined over time (day 2, 96.7%; day 7, 12.7%). The slope of a plot of log10 of that proportion against time was −0.19 (r2 = 0.98). It seems reasonable that as the total chamber population increases with time and occupies a greater volume of sand, the proportion of the constant daily flux of growth substrate flowing through both newly and previously colonized regions will decrease. As a consequence, the cell mass may become carbon limited and/or starved. It is unknown what daily proportion of effluent cells was composed of unattached, newly produced cells versus less metabolically active cells sloughed from older colonized regions. Others have reported that starvation conditions or the presence of surfactants may stimulate detachment of attached cells (see, e.g., references 1 and 25), that substrate loading and flow rate influence biofilm structure (23, 24), and that changes in O2 availability promote physiological heterogeneity in bacterial biofilms (42).
While the emphasis of this work was to highlight the use of digital camera technology in conjunction with light transmission chambers and bioluminescent bacteria for noninvasive measurements of hydrology and microbial growth, there is potential to gain more information about the changes in physiological activities and gene expression in the bacterial population in response to hydrologic changes. Strategic placement of sampling ports in the chamber will permit biopsy of grains of sand and small samples of pore water for this purpose.
Acknowledgments
This research was funded by grants from the National Science Foundation (no. 9630293) and the U.S. Department of Energy (no. DE-FG07-98ER14925) and by the Oregon Agricultural Experiment Station.
Footnotes
Technical Paper no. 11,840 of the Oregon Agricultural Experiment Station.
REFERENCES
- 1.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 2.Applegate, B. M., S. R. Kehrmeyer, and G. S. Sayler. 1998. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene sensing. Appl. Environ. Microbiol. 64:2730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auerbach, I. D., C. Sorensen, H. G. Hansma, and P. A. Holden. 2000. Physical morphology and surface properties of unsaturated Pseudomonas putida biofilms. J. Bacteriol. 182:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blouin, K., S. G. Walker, J. Smit, and R. F. B. Turner. 1996. Characterization of in vivo reporter systems for gene expression and biosensor applications based on luxAB luciferase genes. Appl. Environ. Microbiol. 62:2013-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brink, R. H., P. Dubach, and D. L. Lynch. 1960. Measurement of carbohydrates in soil hydrolyzates with anthrone. Soil Sci. 89:157-166. [Google Scholar]
- 6.Burlage, R. S., G. S. Sayler, and F. Larimer. 1990. Monitoring of naphthalene catabolism by bioluminescence with nah-lux transcriptional fusions. J. Bacteriol. 172:4749-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Z. Lewandowski, D. De Beer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176:2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.deWeger, L. A., P. Dunbar, W. Mahafee, B. Lugtenberg, and G. S. Sayler. 1991. Use of bioluminescence markers to detect Pseudomonas spp. in the rhizosphere. Appl. Environ. Microbiol. 57:3641-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan, S., L. A. Glover, K. Killham, and J. I. Prosser. 1994. Luminescence-based detection of activity of starved and viable but nonculturable bacteria. Appl. Environ. Microbiol. 60:1308-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flemming, C. A., K. T. Leung, H. Lee, and J. T. Trevors. 1994. Bioluminescent most-probable-number method to enumerate lux-marked Pseudomonas aeruginosa UG2Lr in soil. Appl. Environ. Microbiol. 60:3458-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemming, C. A., K. T. Leung, H. Lee, J. T. Trevors, and C. W. Greer. 1994. Survival of lux-lac-marked biosurfactant-producing Pseudomonas aeruginosa UG2L in soil monitored by nonselective plating and PCR. Appl. Environ. Microbiol. 60:1606-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, R. J., T. S. Steenhuis, and J.-Y. Parlange. 1989. Mechanism for finger persistence in homogenous unsaturated porous media: theory and verification. Soil Sci. 148:60-70. [Google Scholar]
- 13.Heitzer, A., K. Malachowsky, J. E. Thonnard, P. R. Bienkowski, D. C. White, and G. S. Sayler. 1994. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol. 60:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitzer, A., O. F. Webb, J. E. Thonnard, and G. S. Sayler. 1992. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol. 58:1839-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoa, N. T. 1981. A new method allowing the measurement of rapid variations on the water content in sandy porous media. Water Resour. Res. 17:41-48. [Google Scholar]
- 16.Holden, P. A., J. R. Hunt, and M. K. Firestone. 1997. Toluene diffusion and reaction in unsaturated Pseudomonas putida biofilms. Biotechnol. Bioeng. 56:656-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King, J. M. H., P. M. DiGrazia, B. M. Applegate, R. S. Burlage, J. Sanseverino, P. Dunbar, F. Larimer, and G. S. Sayler. 1990. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science 249:778-781. [DOI] [PubMed] [Google Scholar]
- 18.Meikle, A., K. Killham, J. I. Prosser, and L. A. Glover. 1992. Luminometric measurement of population activity of genetically modified Pseudomonas fluorescens in the soil. FEMS Microbiol. Lett. 99:217-220. [DOI] [PubMed] [Google Scholar]
- 19.Morley, L. M., G. M. Hornberger, A. L. Mills, and J. S. Herman. 1998. Effects of transverse mixing on transport of bacteria through heterogeneous porous media. Water Resour. Res. 34:1901-1908. [Google Scholar]
- 20.Murphy, E. M., T. R. Ginn, A. Chilakapati, C. T. Resch, J. L. Phillips, T. W. Wietsma, and C. M. Spadoni. 1997. The influence of physical heterogeneity on microbial degradation and distribution in porous media. Water Resour. Res. 33:1087-1103. [Google Scholar]
- 21.Neilson, J. W., S. A. Pierce, and R. M. Maier. 1999. Factors influencing expression of luxCDABE and nah genes in Pseudomonas putida RB1353(NAH7, pUTK9) in dynamic systems. Appl. Environ. Microbiol. 65:3473-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemet, M. R., and J. S. Selker. 2001. A new method for quantification of liquid saturation in 2-d translucent porous media systems using light transmission. Adv. Water Res. 24:651-666. [Google Scholar]
- 23.Peyton, M. B. 1996. Effects of shear stress and substrate loading rate on Pseudomonas aeruginosa biofilm thickness and density. Water Res. 30:29-36. [Google Scholar]
- 24.Picioreanu, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 1999. Discrete differential modeling of biofilm structure. Water Sci. Technol. 39:115-122. [Google Scholar]
- 25.Powelson, D. K., and A. L. Mills. 1998. Water saturation and surfactant effects on bacterial transport in sand columns. Soil Sci. 163:694-704. [Google Scholar]
- 26.Rattray, E. A. S., J. I. Prosser, K. Killham, and L. A. Glover. 1990. Luminescence-based nonextractive technique for in situ detection of Escherichia coli in soil. Appl. Environ. Microbiol. 56:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schell, M. A. 1990. Regulation of the naphthalene degradation genes of plasmid NAH7: example of a generalized positive control system in Pseudomonas and related bacteria, p. 165-176. In S. Silver, A. M. Chakrabarty, B. Iglewski, and S. Kaplan (ed.), Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. American Society for Microbiology, Washington, D.C.
- 28.Schroth, M. H., S. J. Ahearn, J. S. Selker, and J. D. Istok. 1996. Characterization of Miller-similar silica sands for laboratory hydrologic studies. Soil Sci. Soc. Am. J. 60:1331-1339. [Google Scholar]
- 29.Schroth, M. H., J. D. Istok, S. J. Ahern, and J. S. Selker. 1995. Geometry and position of light nonaqueous-phase liquid lenses in water-wetted porous media. J. Contam. Hydrol. 19:269-287. [Google Scholar]
- 30.Schroth, M. H., J. D. Istok, J. S. Selker, M. Oostrom, and M. D. White. 1998. Multifluid flow in bedded porous media: laboratory experiments and numerical simulations. Adv. Water Res. 22:169-183. [Google Scholar]
- 31.Selifonova, O. V., R. S. Burlage, and T. Barkay. 1993. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl. Environ. Microbiol. 59:3083-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selker, J. S., T. S. Steenhuis, and J.-Y. Parlange. 1992. Wetting front instability in homogeneous sandy soils under continuous infiltration. Soil Sci. Soc. Am. J. 56:1346-1350. [Google Scholar]
- 33.Shaw, J. J., F. Dane, D. Geiger, and J. W. Kloepper. 1992. Use of bioluminescence for detection of genetically engineered microorganisms released into the environment. Appl. Environ. Microbiol. 58:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sticher, P. M. C. M. Jaspers, K. Stemmler, H. Harms, A. J. B. Zehnder, and J. R. van der Meer. 1997. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol. 63:4053-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoodley, P., D. De Beer, and Z. Lewandowski. 1994. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 60:2711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szecsody, J. E., F. J. Brockman, B. D. Wood, G. P. Streile, and M. J. Truex. 1994. Transport and biodegradation of quinoline in horizontally stratified porous media. J. Contam. Hydrol. 15:277-304. [Google Scholar]
- 37.Tidwell, V. C., and R. J. Glass. 1994. X-ray and visible light transmission for laboratory measurement of two-dimensional saturation fields in thin-slab systems. Water Resour. Res. 30:2873-2882. [Google Scholar]
- 38.Uesugi, S. L., R. R. Yarwood, J. S. Selker, and P. J. Bottomley. 2001. A model that uses the induction phase of lux-gene dependent bioluminescence in Pseudomonas fluorescens HK44 to quantify cell density in translucent porous media. J. Microbiol. Methods 47:317-324. [DOI] [PubMed] [Google Scholar]
- 39.Unge, A., R. Tombolini, L. Molbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.U.S. Department of Energy. 2001. A national roadmap for vadose zone science & technology: understanding, monitoring, and predicting contaminant fate and transport in the unsaturated zone. DOE/ID-10871. Idaho National Engineering and Environmental Laboratory, Idaho Falls.
- 41.Wan, J., J. L. Wilson, and T. L. Keift. 1994. Influence of the gas-water interface on transport of microorganisms through unsaturated porous media. Appl. Environ. Microbiol. 60:509-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yolcubal, I., J. J. Pratt, S. A. Pierce, M. L. Brusseau, and R. M. Maier. 2000. Fiber optic detection of in situ lux reporter gene activity in porous media: system design and performance. Anal. Chem. Acta 422:121-130. [Google Scholar]