Abstract
The 2,4-dichlorophenoxyacetate (2,4-D)/α-ketoglutarate dioxygenase gene (tfdA) homolog designated tfdAα was cloned and characterized from 2,4-D-degrading bacterial strain RD5-C2. This Japanese upland soil isolate belongs to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in the α subdivision of the class Proteobacteria on the basis of its 16S ribosomal DNA sequence. Sequence analysis showed 56 to 60% identity of tfdAα to representative tfdA genes. A MalE-TfdAα fusion protein expressed in Escherichia coli exhibited about 10 times greater activity for phenoxyacetate than 2,4-D in an α-ketoglutarate- and Fe(II)-dependent reaction. The deduced amino acid sequence of TfdAα revealed a conserved His-X-Asp-X146-His-X14-Arg motif characteristic of the active site of group II α-ketoglutarate-dependent dioxygenases. The tfdAα genes were also detected in 2,4-D-degrading α-Proteobacteria previously isolated from pristine environments in Hawaii and in Saskatchewan, Canada (Y. Kamagata, R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje, Appl. Environ. Microbiol. 63:2266-2272, 1997). These findings indicate that the tfdA genes in β- and γ-Proteobacteria and the tfdAα genes in α-Proteobacteria arose by divergent evolution from a common ancestor.
The herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) has been used as a model compound to study evolution of bacterial catabolic genes for anthropogenic chemicals (10, 11, 16, 20, 21, 26, 35, 36, 37). A number of 2,4-D-degrading bacterial strains have been isolated and found to be distributed over many different phylogenetic groups (3, 5, 10, 12, 16, 20, 21, 25, 26, 35, 37). Most of the strains have been classified into two groups on the basis of the genetic and enzymatic properties of their 2,4-D-catabolizing traits. One group is composed of various genera in the β and γ subdivisions of the class Proteobacteria. They have 2,4-D-catabolizing genes that are homologous to tfd genes found in Ralstonia eutropha JMP134, a well-studied 2,4-D degrader (5, 26). The other group is composed of strains in the α subdivision of the class Proteobacteria. Their 2,4-D-catabolizing genes have shown less than 60% homology by hybridization to the tfd genes in the β- and γ-Proteobacteria (20, 21, 26, 37).
The tfdA gene encodes an α-ketoglutarate (α-KG)-dependent 2,4-D dioxygenase that converts 2,4-D into 2,4-dichlorophenol and glyoxylate while oxidizing α-KG to CO2 and succinate as an initial step of 2,4-D mineralization (8, 9, 31, 32). The tfdA sequences fall into three distinct classes for members of the β- and γ-Proteobacteria (26). The class I tfdA genes, including that of R. eutropha JMP134, are usually located on broad-host-range, self-transmissible plasmids (5, 19, 36). The class II tfdA gene, which is only found in the chromosome of Burkholderia sp. strain RASC, is 76% identical to the class I tfdA genes. The class III tfdA genes are 77 and 80% identical to the class I and class II tfdA genes, respectively. A tfdA phylogenetic tree with three distinct branches has been constructed, and interspecies gene transfer of the tfdA genes among the β- and γ-Proteobacteria was suggested by comparing the phylogenetic trees of the tfdA and 16S ribosomal (rDNA) genes (26).
An alignment of the tfdA genes was used to design PCR primer sets for amplifying tfdA-like genes from other sources (31, 37). In contrast to the 2,4-D-degrading β- and γ-Proteobacteria, none of 2,4-D-degraders belonging to the α-Proteobacteria has been found to yield tfdA-like PCR products (21, 26, 37). No hybridization was observed with tfdA even under low-stringency conditions, and α-KG-dependent 2,4-D dioxygenase activity was not detected. Therefore, the 2,4-D genes in this particular phylogenetic group were considered to be functionally and evolutionally different from the tfdA genes in the former phylogenetic groups (21, 26, 37).
In order to understand the origin and evolutionary relationship of the 2,4-D catabolic genes, it is important to characterize the 2,4-D genes in the α-Proteobacteria. Recently we isolated the 2,4-D-degrading bacterial strain RD5-C2 from Japanese upland soil (18). RD5-C2 is a member of the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster (14, 29) in the α-Proteobacteria, like the 2,4-D degraders previously isolated from the pristine environments in Hawaii and in Saskatchewan, Canada, and Chile (21). In this paper we report that RD5-C2 and other degraders isolated from the pristine environments possess tfdA-like genes that encode a protein belonging to the α-KG-dependent dioxygenase family (8, 9, 31, 32). Significantly, we show that the recombinant RD5-C2 protein exhibits greater activity for nonchlorinated phenoxyacetate over 2,4-D in contrast to the more uniform activities observed for the canonical TfdA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
RD5-C2 and the other 2,4-D-degrading strains isolated from the pristine environments (21), HW13, HWK12, and BTH, were cultured on 2,4-D-containing basal medium (18) or HM medium (27). Escherichia coli HB101 and E. coli DH5α were used for the cloning and expression experiments, respectively. Luria-Bertani (LB) medium was used to grow E. coli. The tfdA-like gene from RD5-C2 (hereafter referred to as tfdAα) was cloned into pUC118 and then subcloned into pUC119. The plasmid vector pMAL-cRI (New England BioLabs) was used to construct the MalE-TfdAα fusion protein. The tfdA-like gene was fused to the malE gene, which encodes maltose-binding protein, and was expressed by using the tac promoter and translation initiation signals of maltose-binding protein.
Cloning and sequencing of tfdAα.
To obtain whole DNA from the isolates, the cultures were grown on HM medium, washed with 1% NaCl, suspended in distilled water, frozen at −20°C, thawed at room temperature, and treated with proteinase K (10 μl of a 1-mg/ml concentration per 40 μl of sample) (Takara) in 40 mM Tris buffer solution (50 μl) containing 1% Tween 20, 0.5% Nonidet P-40 (Sigma), and 1 mM EDTA (pH 8.0). The mixture was incubated at 60°C for 20 min to digest proteins and then at 100°C for 5 min to inactivate the enzyme. After centrifugation (8,060 × g, 10 min) the supernatant was collected for gene amplification by PCR (15). A partial tfdAα fragment was amplified by using modified primers designed for the conserved regions of the tfdA genes from R. eutropha JMP134 and Burkholderia sp. strain RASC (3, 37): 5′-AC(C/G)GAGTTCTG(C/T)GA(C/T) ATG-3′ and 5′-(A/G)ACGCAGCG(G/A)TT(G/A)TCCCA-3′. The amplified tfdAα gene fragments were directly ligated into pT7Blue T-Vector (Novagen) and were transformed into E. coli HB101-competent cells according to the supplier's instructions. The inserted regions were amplified by PCR by using M13 primers M4 and RV (Takara) and were directly sequenced with an ABI 377 DNA sequencing system (Perkin-Elmer Japan) with the same primers.
The amplified tfdAα gene fragment was labeled with a DIG DNA Labeling Kit (Roche Molecular Biochemicals) and was shown to hybridize with a single ca. 4.0-kbp fragment of whole DNA SacI digest (data not shown). DNA fragments of around 4.0 kbp were extracted from the gel, ligated into pUC118 to produce pTFD1-1, and transformed into E. coli DH5α-competent cells to create a genomic library. The DNA fragments were then screened by colony hybridization with the tfdAα gene fragment as a probe.
Plasmid DNA was purified from five positive clones, and the SacI fragment was further digested with several restriction enzymes. The SalI fragment was subcloned into pUC119, and the DNA sequence of the inserted fragment was determined as described previously. On the basis of the gene sequence, the following new primer set was constructed to amplify tfdAα from HW13, HWK12, and BTH: 5′-AC(C/G)GAGTTC(G/T)(C/G)CGACATGCG-3′ and 5′-GCGGTTGTCCCACATCAC-3′. The tfdAα genes were amplified, and the full sequences for HW13 and HWK12 and the partial sequence for BTH were determined by using a PCR in vitro cloning kit (Takara). All procedures described here were conducted according to the supplier's instructions.
Analysis of tfdAα sequence.
The tfdAα sequences were compared with other available sequences by using the FASTA (28) and BLAST (2) algorithms. Sequences were aligned manually with the CLUSTALW program (34), and the sites where nucleotides were not resolved for all sequences were excluded from the alignments. Phylogenetic analysis was performed with three types of algorithms. CLUSTALW version 1.8 (34) was used as the software package for the neighbor-joining (NJ) analysis (30), and the bootstrap analysis based on 1,000 replicates was used to place confidence estimates on the tree. A maximum-likelihood (ML) analysis was carried out with the program package MOLPHY version 2.3b3 (1). An ML distance matrix was calculated by using NucML, and the NJ topology was reconstructed by using NJdist in MOLPHY as the starting tree for the ML method. An ML tree was obtained by using NucML with an R (local rearrangement search) option based on the HKY model (13). Local bootstrap probabilities were estimated by the resampling of estimated log-likelihood method (13, 24). A maximum-parsimony tree reconstruction was performed by using PAUP* software version 4.0b10 (33). A heuristic search was used with a random stepwise addition sequence of 100 replicates, tree-bisection-reconnection branch swapping, and the multrees option. A further analysis was run with 100 bootstrap replicates, each consisting of 30 additional random replicates. The Kishino-Hasegawa test (23) was carried out to survey the best topology having the highest log-likelihood value among the most parsimonious trees. A consensus tree was obtained by using the strict method.
Construction of malE-tfdAα fusion plasmid.
From pTFD1-1, plasmid pNK2 was constructed by subcloning the appropriate SalI fragment into pUC119. tfdAα of RD5-C2 was amplified by PCR by using the primers 5′-GCTCTAGAATGACGGTCCTGATCCGGCAG-3′ and 5′-CCCAAGCTTTGTTATTGTGCCCGCTACTCC-3′ to give XbaI and HindIII ends, respectively, and was inserted into the XbaI and HindIII cloning sites of the pMAL-cRI vector. The plasmid was transformed into E. coli DH5α-competent cells according to the supplier's instructions.
Preparation of cell extracts and enzyme assay.
Unless stated otherwise, the experiments were done at 4°C. E. coli DH5α (pUC119-tfdAα) was grown overnight at 30°C in LB medium supplemented with 100 μg of ampicillin/ml. E. coli DH5α (pMAL-cRI-tfdAα) was grown to mid-log phase at 30°C in LB medium supplemented with 2 g of glucose/liter and 100 μg of ampicillin/ml, followed by induction with isopropyl-[β]-d-thiogalactopyranoside at 0.3 mM for 2 h. Cells were harvested by centrifugation (6,000 × g, 15 min) and suspended in a solution containing 10 mM Tris, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM leupeptin (pH 7.6). The suspended cells were passed through a French press at 120 MPa, and cell extracts were obtained by centrifugation (100,000 × g, 45 min). The cell extracts were examined by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were assayed for TfdA enzyme activity. Gels were stained with Coomassie brilliant blue, and phosphorylase b (Mr, 97,400), bovine serum albumin (Mr, 66,200), ovalbumin (Mr, 45,000), carbonic anhydrase (Mr, 31,000), soybean trypsin inhibitor (Mr, 21,500), and lysozyme (Mr, 14,400) (Bio-Rad) were used as standards. The MalE-TfdAα fusion protein was further purified by using an amylose resin column (New England BioLabs) according to the supplier's instructions. The fusion protein eluate was dialyzed against 10 mM Tris-0.1 mM EDTA (pH 7.4) prior to the enzyme assay. Protein concentration was determined by using a protein assay kit (Bio-Rad) with bovine serum albumin as the standard.
The enzyme activity was measured by the modified 4-aminoantipyrine method (8, 22). The typical assay mixture contained 1 mM α-KG, 50 μM ascorbate, 50 μM (NH4)2Fe(SO4)2, and 1 mM 2,4-D in 10 mM imidazole buffer (pH 6.75). The enzyme reaction was conducted at 30°C and was quenched by the addition of EDTA to give a final concentration of 5 mM. The concentration of released phenol derivative was measured as follows. One hundred microliters of a pH 10 buffer solution, 10 μl of 2% 4-aminoantipyrine, and 10 μl of 8% potassium ferricyanide were added to 1 ml of the reaction mixture. The absorbance at 510 nm was measured after 20 min. The assay was simultaneously conducted with purified R. eutropha JMP134 TfdA (9) for reference.
For the other phenoxyacetate derivatives, the corresponding phenol compounds were used as the standard in the assay. For the other α-ketoacids, phenoxyacetate was used as the substrate because this exhibited the highest activity among the phenoxyacetate derivatives used.
Nucleotide sequence accession number.
The nucleotide sequences determined in this study have been deposited in the DDBJ/EMBL/GenBank database, and the accession numbers for tfdAα of RD5-C2, HWK12, HW13, and BTH are AB074490, AB074491, AB074492, and AB074493, respectively.
RESULTS
tfdAa gene and TfdAα amino acid sequences of RD5-C2.
The tfdAα sequence was found to most closely match that of tfdA of R. eutropha JMP134 and showed 56 to 60% identity to representative tfdA genes of JMP134, RASC, and Achromobacter xylosoxidans EST4002 (E. Vedler, V. Koiv, and A. Heinaru, Analysis of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pEST4011 of Achromobacter xylosoxidans subsp. denitrificans strain EST4002; direct submission to GenBank [accession no. U32188], 1995), which were reported to have the class I, II, and III tfdA genes, respectively (26). The deduced amino acid sequence of RD5-C2 TfdAα was aligned with those of representative TfdA proteins and taurine/α-KG dioxygenase (TauD) from E. coli, which all belong to group II of the α-KG-dependent dioxygenase family (7, 8, 9, 17, 32) (Fig. 1). The data indicated 80 to 93% identity of amino acids among the three classes of TfdA, whereas TfdAα showed lower identity (45 to 48%) to all classes of TfdA, as expected from the gene sequence analysis. TfdAα also showed lower identity to TauD (27%) than the other TfdA proteins (28%). Despite the divergence of TfdAα compared to the other sequences, all of these proteins possessed the two His and one Asp considered to be iron-binding ligands and an Arg suspected of forming a salt bridge with the C-5 carboxylate of α-KG (17). The four predicted active-site residues in TfdAα are His115, Asp117, His264, and Arg279.
FIG. 1.
Multiple alignment of the deduced amino acid sequences of RD5-C2 TfdAα and other α-KG-dependent dioxygenases. JMP134, RASC, and EST4002 are representative strains having class I, II, and III tfdA genes, respectively. TauD is taurine/α-KG dioxygenase of E. coli. Conserved residues are boxed.
TfdA activity of the MalE-TfdAα fusion protein.
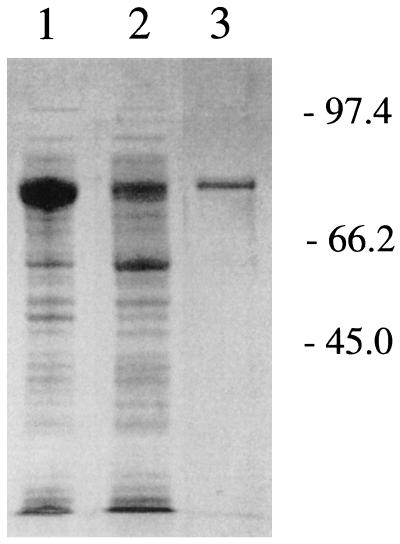
When tfdAα was overexpressed in E. coli by using a pUC119 derivative (pNK2), TfdAα protein was produced, as shown by a predominant band exhibiting an Mr of 36,000 following denaturing gel electrophoresis. However, most of the recombinant wild-type protein was insoluble and possessed no activity even when prepared from cells grown at 30 or 25°C (9) or when examined with cells cultured in 1 M sorbitol and 2.5 mM glycyl betaine (4). In contrast, the MalE-TfdAα fusion protein (Mr, 76,000) exhibited increased solubility, allowing its partial purification by amylose resin chromatography (Fig. 2). As shown in Table 1, the specific activity of purified MalE-TfdAα protein was greatly reduced in comparison to that of R. eutropha TfdA. Significantly, however, the MalE-TfdAα protein exhibited higher activity for phenoxyacetate derivatives that were less chlorinated than 2,4-D. In contrast, TfdA showed similar activity towards these phenoxyacetates, as reported previously (8). Besides the difference in specific activities between the substrates, MalE-TfdAα showed the same traits as TfdA (8) in terms of dependency on α-KG and ferrous iron (Table 2) and in exhibiting the highest activity for α-KG among the α-ketoacids (Table 3).
FIG. 2.
Purification of MalE-TfdAα fusion protein from RD5-C2 (pMAL-cRI-tfdAα). Lane 1, whole-cell disruptant of RD5-C2 (pMAL-cRI-tfdAα); lane 2, supernatant of the disrupted cells; lane 3, purified MalE-TfdAα fusion protein eluted from an amylose resin column.
TABLE 1.
Specific activity of the MalE-TfdAα fusion proteina
| Substrate | Sp act
|
|
|---|---|---|
| MalE-TfdAα (pmol/min · μg of protein) | TfdA (nmol/min · μg of protein) | |
| 2,4-D | 2.6 | 7.9 |
| 4-Chlorophenoxyacetate | 6.5 | 8.6 |
| Phenoxyacetate | 21 | 8.7 |
Experiments were performed at 30°C in 10 mM imidazole buffer (pH 6.75) containing 1 mM substrate, 1 mM α-KG, 50 μM ferrous iron, and 50 μM ascorbate.
TABLE 2.
Requirement of α-KG and ferrous iron for activity of the MalE-TfdAα fusion protein
| Conditions | Sp act (pmol/min · μg of protein) |
|---|---|
| Controla | 25.1 |
| −α-KGb | 3.8 |
| −Ascorbateb | 21.3 |
| −Ferrous ironb | 3.7 |
| −Enzymeb | 2.9 |
| −Phenoxyacetateb | 2.1 |
Experiments were performed at 30°C in 10 mM imidazole buffer (pH 6.75) containing 1 mM phenoxyacetate, 1 mM α-KG, 50 μM ferrous iron, and 50 μM ascorbate.
Each component was removed from the reaction mixture.
TABLE 3.
α-Ketoacid substrate specificity of the MalE-TfdAα fusion protein
| Substratea | Sp act (pmol/min · μg of protein) |
|---|---|
| α-KG | 27.5 |
| α-Ketoadipate | 6.3 |
| α-Ketovalerate | 3.8 |
| α-Ketobutyrate | 2.5 |
| α-Ketoisovalerate | 1.3 |
| α-Ketocaproate | 0.6 |
Experiments were performed at 30°C in 10 mM imidazole buffer (pH 6.75) containing 1 mM phenoxyacetate, 1 mM α-ketoacid, 50 μM ferrous iron, and 50 μM ascorbate.
Phylogenetic analysis of tfdAα genes from strain RD5-C2 and other 2,4-D-degrading α-Proteobacteria.
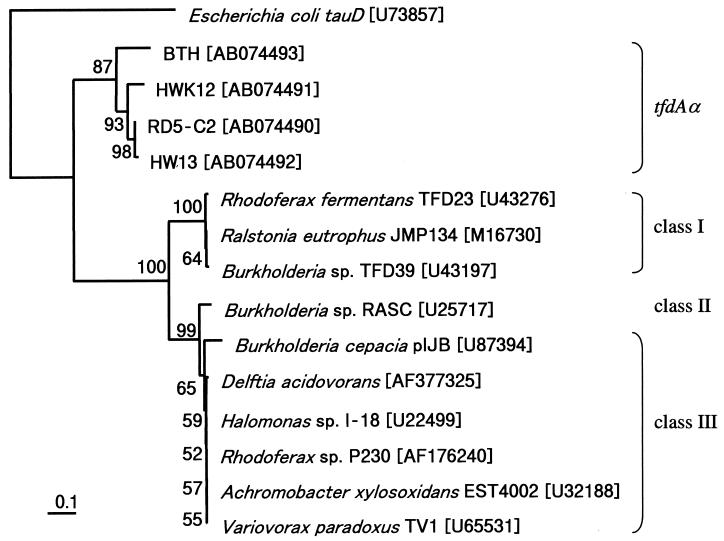
Three 2,4-D-degrading strains belonging to α-Proteobacteria, previously isolated from pristine environments (21), were tested for the presence of a tfdAα-type gene by PCR amplification. The resulting tfdAα genes were sequenced and, along with RD5-C2 tfdAα, were subjected to phylogenetic analysis (Fig. 3). Since only four full-length open reading frames for tfdA are available in the DDBJ/EMBL/GenBank database, partial sequences (307 bp) of tfdA and tfdAα were used for the analysis. The tfdAα genes of α-Proteobacteria comprise a distinct cluster compared to the three reported clusters associated with the representative tfdA genes. Gene sequence identity among the tfdAα representatives was 81 to 99%. Based on the full-length sequence comparisons, a similar phylogenetic tree was obtained. In addition, all three algorithms presented similar topologies in both the full-length and partial sequences (data not shown).
FIG. 3.
Phylogenetic position of tfdAα among other genes of the α-KG-dependent dioxygenase family. The tfdA genes of JMP134, TFD39, and TFD23 are found in class I, the tfdA gene of RASC is in class II, and the other previously reported tfdA genes are class III representatives. E. coli tauD encodes taurine/α-KG dioxygenase. Designations in parentheses are DDBJ/EMBL/GenBank accession numbers. The phylogenetic tree was constructed on the basis of common partial sequences (307 bp) by using the NJ method. Bootstrap values above 50% are shown at nodes. The scale bar indicates substitutions per site.
DISCUSSION
Previous studies of α-proteobacterial 2,4-D degraders indicated that the genes responsible for 2,4-D degradation were different from the canonical tfd genes and that these bacteria lack a TfdA-like enzyme, suggesting a different origin of the biodegradative pathway compared to the β- and γ-Proteobacteria (21, 26, 37). Our studies refute this interpretation; rather, strain RD5-C2 and other 2,4-D degraders within the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster of α-Proteobacteria do harbor a tfdA-like gene (tdfAα), and the RD5-C2 TfdAα protein exhibits TfdA-like activity. This is the first unequivocal evidence that tfdA-type genes and TfdA-like enzymes exist among α-, β-, and γ-proteobacterial 2,4-D degraders. Because the FASTA homology search with the DDBJ/EMBL/GenBank database indicated no tfdAα-like genes among microorganisms other than those presented in Fig. 3, it is difficult to envisage the original function of these genes. The low sequence identity (56 to 60%) of tfdAα genes with representative tfdA genes explains why they did not hybridize in prior studies, even under low-stringency conditions (21, 26, 37).
All α-Proteobacteria strains used in this study were isolated from pristine environments and Japanese arable soil that had had no history of chemical treatment, suggesting that tfdAα genes are distributed among soil strains independent of 2,4-D exposure. These tfdAα genes are likely to have some original function(s), and some strains with the appropriate tfdAα genes incidentally degrade 2,4-D.
On the basis of the 30% amino acid identity between TauD from E. coli and TfdA from R. eutropha JMP134, Eichhorn et al. (7) suggested that the two dioxygenases might have arisen by convergent evolution rather than from a common ancestor. In contrast, the tfdA and tfdAα genes are likely to have arisen by divergent evolution from a common ancestor, because they exhibit higher similarities and are separately distributed in β- and γ-Proteobacteria and α-Proteobacteria, respectively. Horizontal gene transfer is a well-known phenomenon, and interspecies gene transfer of tfdA genes was demonstrated by phylogenetic comparison of tfdA and 16S rDNA genes (26). On the other hand, phylogenetic trees of tfdAα and 16S rDNA genes are congruent (18), indicating evolution of the tfdAα genes without horizontal gene transfer occurring. On the basis of the findings that microorganisms isolated from pristine environments harbor tfdAα and that TfdAα has higher activity for nonchlorinated phenoxyacetate than 2,4-D, the tfdAα genes could be the origin of the tfdA genes. Horizontal transfer by transposons and/or plasmids and further adaptations to the anthropogenic environments would give rise to the well-studied tfdA representatives. Plasmid pJP4, carrying tfdA, was shown to be transferred to strains of α-, β- and γ-Proteobacteria; however, the 2,4-D-degrading phenotype could not be expressed in α-Proteobacteria such as Rhizobium sp. and Agrobacterium tumefaciens (5). Distribution of tfdA-like genes among non-2,4-D-degrading soil bacteria in many different phylogenetic groups was reported, and nucleotide sequence analyses of four isolates showed the highest homology with canonical tfdA (16). Sequence analysis of the tfdA genes from the other strains could reveal the evolutionary relationship between the tfdA and tfdAα genes.
TfdAα was experimentally found to be an α-KG-dependent dioxygenase, as previously observed for TfdA. The amino acid sequence of TfdAα clearly belongs to group II of the α-KG-dependent dioxygenase family, since it contains multiple conserved residues that are thought to function in catalysis (8, 17). The substrate for ancestral TfdA is unknown, but naturally occurring aryl-ether compounds released during degradation of lignin are a reasonable possibility (16). In addition, it is possible that the TfdA ancestor functioned in the metabolism of a cinnamic acid (6).
In the present study we showed that 2,4-D-degrading α-Proteobacteria possess tfdAα genes that arose from a common ancestor to the canonical tfdA genes, and it is considered that ancestral tfdAα genes may serve as the origin of the tfdA genes on the basis of the following reasons. First, the 2,4-D-degrading α-Proteobacteria were isolated from pristine environments, widely distributed in Hawaii, and in Saskatchewan, Canada, and Chile and Japan, that had not been exposed to 2,4-D. Second, TfdAα is an α-KG- and Fe(II)-dependent dioxygenase that exhibits higher specific activity for nonchlorinated substrates. Thus, the chemistry of the TfdAα enzymes could be retained while modifying the substrate binding site to allow enhanced reactivity toward highly chlorinated 2,4-D to give rise to the TfdA activities. Further studies are needed to clarify the evolutionary relationships between tfdAα and the canonical tfdA genes and to understand the adaptation process of bacteria to chlorinated compounds.
REFERENCES
- 1.Adachi, J., and M. Hasegawa. 1996. Computer science monographs, no. 28, MOLPHY: version 2.3. In M. Ishiguro, G. Kitagawa, Y. Ogata, H. Takagi, Y. Tamura, and T. Tsuchiya (ed.), Programs for molecular phylogenetics based on maximum likelihood. Institute of Statistical Mathematics, Tokyo, Japan.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amy, P. S., J. W. Schulke, L. M. Frazier, and R. J. Seidler. 1985. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 49:1237-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell, J. R., and R. Horgan. 1991. A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett. 295:10-12. [DOI] [PubMed] [Google Scholar]
- 5.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunning Hotopp, J. C., and R. P. Hausinger. 2001. Alternative substrates for 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J. Mol. Catal. 15:155-162. [Google Scholar]
- 7.Eichhorn, E., J. R. van der Ploeg, M. A. Kertesz, and T. Leisinger. 1997. Characterization of α-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem. 272:23031-23036. [DOI] [PubMed] [Google Scholar]
- 8.Fukumori, F., and R. P. Hausinger. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311-24317. [PubMed] [Google Scholar]
- 9.Fukumori, F., and R. P. Hausinger. 1993. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulthorpe, R. R., C. McGowan, O. V. Maltseva, W. H. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulthorpe, R. R., A. N. Rhodes, and J. M. Tiedje. 1996. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl. Environ. Microbiol. 62:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulthorpe, R. R., and R. C. Wyndham. 1992. Involvement of a chlorobenzoate catabolic transposon, Tn5271, in community adaptation to chlorobiphenyl, chloroaniline, and 2,4-dichlorophenoxyacetic acid in a freshwater ecosystem. Appl. Environ. Microbiol. 58:314-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 14.Hattori, T., H. Mitsui, H. Haga, N. Wakao, s. Shikano, K. Gorlach, Y. Kasahara, A. el-Beltagy, and R. Hattori. 1997. Advances in soil microbial ecology and the biodiversity. Antonie Van Leeuwenhoek 72:21-28. [DOI] [PubMed] [Google Scholar]
- 15.Hiraishi, A. 1992. Direct automated sequencing of 16S rRNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:21-213. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, D. A., D. H. Buckley, C. H. Nakatsu, T. M. Schmidt, and R. P. Hausinger. 1997. Distribution of the tfdA gene in soil bacteria that do not degrade 2,4-dichlorophenoxyacetic acid, 2,4-D. Microb. Ecol. 34:90-96. [DOI] [PubMed] [Google Scholar]
- 17.Hogan, D. A., S. R. Smith, E. A. Saari, J. McCracken, and R. P. Hausinger. 2000. Site-directed mutagenesis of 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase. Identification of residues involved in metallocenter formation and substrate binding. J. Biol. Chem. 275:12400-12409. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, K., R. Kanda, Y. Momoda, Y. Sumita, Y. Kamagata, K. Suyama, and H. Yamamoto. 2000. Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microb. Environ. 15:113-117. [Google Scholar]
- 19.Ka, J. O., and J. M. Tiedje. 1994. Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergenetic transfer. J. Bacteriol. 176:5284-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King, R. J., K. A. Short, and R. J. Seidler. 1991. Assay for detection and enumeration of genetically engineered microorganisms which is based on the activity of a deregulated dichlorophenoxyacetate monooxygenase. Appl. Environ. Microbiol. 57:1790-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 24.Kishino, H., T. Miyata, and M. Hasegawa. 1990. Maximum likelihood inference of protein phylogeny, and the origin of chloroplasts. J. Mol. Evol. 31:151-160. [Google Scholar]
- 25.Maltseva, O., C. McGowan, R. Fulthorpe, and P. Oriel. 1996. Degradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology 142:1115-1122. [DOI] [PubMed] [Google Scholar]
- 26.McGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieuwkoop, A. J., Z. Banfalvi, N. Deshmane, D. Gerhold, M. G. Schell, K. M. Sirotkin, and G. Stacey. 1987. A locus encoding host range is linked to the common nodulation genes of Bradyrhizobium japonicum. J. Bacteriol. 169:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito, A., H. Mitsui, R. Hattori, K. Manamisawa, and T. Hattori. 1998. Slow-growing and oligotrophic bacteria phylogenetically close to Bradyrhizobium japonicum. FEMS Microb. Ecol. 25:277-286. [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Streber, W. R., K. N. Timmis, and M. H. Zenk. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suwa, Y., A. D. Wright, F. Fukumori, K. A. Nummy, R. P. Hausinger, W. E. Holben, and L. J. Forney. 1996. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl. Environ. Microbiol. 62:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2002. PAUP* phylogenetic analysis using parsimony and other programs, version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonso, N. L., V. G. Matheson, and W. E. Holben. 1995. Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb. Ecol. 30:3-24. [DOI] [PubMed] [Google Scholar]
- 36.Top, E. M., W. E. Holben, and L. J. Forney. 1995. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl. Environ. Microbiol. 61:1691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vallaeys, T., R. R. Fulthorpe, A. M. Wright, and G. Soulas. 1996. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microb. Ecol. 20:163-172. [Google Scholar]