Abstract
The human gastrointestinal (GI) tract harbors a complex community of bacterial cells in the mucosa, lumen, and feces. Since most attention has been focused on bacteria present in feces, knowledge about the mucosa-associated bacterial communities in different parts of the colon is limited. In this study, the bacterial communities in feces and biopsy samples from the ascending, transverse, and descending colons of 10 individuals were analyzed by using a 16S rRNA approach. Flow cytometric analysis indicated that 105 to 106 bacteria were present in the biopsy samples. To visualize the diversity of the predominant and the Lactobacillus group community, denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA gene amplicons was performed. DGGE analysis and similarity index comparisons demonstrated that the predominant mucosa-associated bacterial community was host specific and uniformly distributed along the colon but significantly different from the fecal community (P < 0.01). The Lactobacillus group-specific profiles were less complex than the profiles reflecting the predominant community. For 6 of the 10 individuals the community of Lactobacillus-like bacteria in the biopsy samples was similar to that in the feces. Amplicons having 99% sequence similarity to the 16S ribosomal DNA of Lactobacillus gasseri were detected in the biopsy samples of nine individuals. No significant differences were observed between healthy and diseased individuals. The observed host-specific DGGE profiles of the mucosa-associated bacterial community in the colon support the hypothesis that host-related factors are involved in the determination of the GI tract microbial community.
The human gastrointestinal (GI) tract harbors a diverse community of obligate and facultative anaerobic bacteria. These bacteria have an important metabolic and protective function in the GI tract (23). The complex interactions between the host and the bacterial community are of considerable importance but are just starting to be understood (3, 9, 10). Most of the knowledge about bacterial diversity in the human GI tract has been obtained by selective cultivation of microbes from fecal samples. Recently, culture-independent approaches using the sequence variability of the 16S rRNA genes have shown that most of the predominant bacteria in human fecal samples have not been obtained in culture yet, which indicates that our knowledge of these predominant members is very limited (22, 26, 28). In addition, denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) analyses of fecal 16S ribosomal DNA (rDNA) and rRNA amplicons have shown to be powerful approaches in determining and monitoring the bacterial community in feces (28, 29). Such studies revealed that the predominant bacterial community in mammalian feces is stable in time, host specific, affected by ageing, and not altered after consumption of certain probiotic strains (11, 21, 24, 25, 28, 29). Furthermore, DGGE has been used to compare bacterial communities in fecal samples from infants with and without necrotizing enterocolitis, although no differences associated with this disease were observed (17).
Our present knowledge of the bacterial diversity associated with the human GI tract is based mainly on analysis of fecal samples, and in a few cases samples that originated from different parts of the intestine have been characterized. Most of these analyses with contents from sudden-death victims (15) or with biopsy samples from living individuals involved a culturing approach and focused on the attachment of certain probiotic strains (1, 2, 12), the presence of sulfate reducers (5, 27), and/or on bacterial population levels in diseased persons (6). Since biopsy samples are very small in size and therefore more easily exposed to oxygen during sampling, the number of viable strict anaerobes might be reduced easily. Not surprisingly, relatively high levels of facultative anaerobes were reported to be present in intestinal biopsy samples. So far, there have been no studies in which the bacterial composition of biopsy samples has been analyzed at the species level. Molecular approaches based on the sequence variability of 16S rRNA genes could be instrumental in analyzing the composition of bacterial communities in intestinal biopsy samples. Recently, such an approach was used to study the bacterial diversity within the human subgingival crevice (13). In another recent study, temporal TGGE analysis of 16S rDNA fragments was successfully used to compare the bacterial compositions in gastric biopsy samples and showed that Helicobacter is detectable in samples from healthy individuals and those suffering from gastritis (18). In addition, Marteau and colleagues reported significant differences in community structure between samples from feces and contents from the cecum by using culturing techniques and dot blot hybridization (16).
The aim of our research was to determine whether the bacterial composition in colonic biopsy samples was significantly different from that in fecal samples and to investigate whether differences in composition could be detected at different locations in the colon. We used DGGE approaches to characterize the 16S rDNA sequence variability of the predominant bacterial composition and that of the Lactobacillus-like species with general and specific PCR primers (8, 28). We focused on lactobacilli as a subgroup because of their potential probiotic effects in the human GI tract. The compositional variability in feces and colonic biopsy samples from the ascending, transverse, and descending colons of 10 volunteers was studied.
MATERIALS AND METHODS
Experimental approach.
To describe the bacterial diversity in fecal and biopsy samples, a 16S rRNA approach was used. DNA was isolated from these samples derived from the same individual, and the V6-to-V8 regions were PCR amplified with general primers and analyzed by DGGE. After scanning of the gels, similarity indices of DGGE profiles were compared and statistically analyzed. In addition, a specific PCR was performed to amplify the V2-to-V4 region of the Lactobacillus group that subsequently was separated by DGGE. To quantify the number of bacteria per biopsy sample, a flow cytometric approach was used.
Volunteers.
Fecal and biopsy samples as fresh as possible were collected from 10 adult human volunteers. The 10 volunteers donating biopsy and fecal samples were patients undergoing routine diagnostic colonoscopies. The procedure normally includes biopsies, and so the study did not cause any extra risk, pain, or discomfort to the participants. Informed consent was obtained from each volunteer before the sampling. The group consisted of five men and five women (Table 1). With the exception of various GI symptoms (including pains, bloating, and, in patients with ulcerative colitis, bouts of diarrhea) for which they underwent the examination, the volunteers considered themselves healthy. They did not follow any special dietary regimen, and none had recently received any antibiotic treatment.
TABLE 1.
Characteristics of the volunteers in this study
| Volunteer ID | Age (yr) | Sexa | Diagnosed illnessb | Bacterial cell counts inc:
|
SId
|
|||
|---|---|---|---|---|---|---|---|---|
| A | T | D | F-D | A-D | ||||
| 1 | 59 | F | None | ND | 2.0 × 105 | 1.4 × 105 | 83.1 | 97.6 |
| 2 | 70 | M | None | ND | ND | ND | 91.3 | 98.1 |
| 3 | 79 | M | P | ND | ND | ND | 67.1 | 95.9 |
| 4 | 36 | F | UC* | 8.6 × 104 | 1.1 × 105 | 1.1 × 105 | 81.9 | 56.5 |
| 5 | 43 | M | UC | 6.4 × 105 | 6.4 × 105 | 1.3 × 106 | 22.9 | 91.2 |
| 6 | 63 | M | None | ND | ND | ND | 25.8 | 98.0 |
| 7 | 45 | F | None | 5.7 × 105 | 3.8 × 106 | 1.2 × 106 | 24.7 | ND |
| 8 | 30 | F | UC* | 6.5 × 105 | 6.9 × 106 | 2.5 × 106 | 40.7 | 95.8 |
| 9 | 43 | M | None | 1.7 × 105 | 1.2 × 105 | 2.0 × 105 | 13.6 | 81.6 |
| 10 | 51 | F | None | 2.9 × 105 | 9.0 × 105 | 7.7 × 105 | 30.0 | 82.0 |
F, female; M, male.
P, polyposis; UC, ulcerative colitis; UC*, remission of UC.
A, T, and D, biopsy samples from ascending, transverse, and descending parts of the colon, respectively; ND, not determined.
SI, similarity index; -, comparison; F, fecal sample.
Colonoscopy, fecal sample collection, and treatment of samples.
The colonic evacuation before the colonoscopy was performed by using a laxative (Colonsteri; Orion Oy, Espoo, Finland) according to the instructions of the manufacturer. The instrument used for the actual colonoscopy and biopsies was Pentax EC-3801 L. Biopsy samples (∼0.5 mg) were obtained from the ascending (A), transverse (T), and descending (D) parts of the colon (two parallels per location). One of the parallel samples was stored in 0.05 M potassium phosphate buffer (pH 7.0), and the other was stored in phosphate-buffered saline (PBS) (containing, per liter, 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 [pH 7.2]) with 4% paraformaldehyde. To minimize contamination during sampling, the colonoscope jaws were carefully washed in tap water after each biopsy was performed. Fecal samples were obtained before the colonic evacuation. They were stored in the home freezers of the volunteers and collected immediately prior to the colonoscopy. Both fecal and biopsy samples were subsequently deeply frozen at −70°C, shipped in dry ice, and if appropriate, stored at −70°C. Samples were thawed in ice-water prior to further analysis.
Bacterial counts in biopsy samples.
The paraformaldehyde-fixed biopsy samples were washed twice with PBS and resuspended in 50% ethanol-PBS. After incubation for at least 1 h at −20°C, the biopsy samples were sonicated in an ultrasonic water bath for 2 min to separate the bacterial cells from the biopsy material. This treatment has shown to be optimal to separate viable cells from each other without damaging them (19). After centrifugation at 700 × g for 1 min to remove host cells and debris, the supernatant was centrifuged at 9,000 × g for 5 min to pellet the bacteria. The bacteria were resuspended in 490 μl of PBS (pH 8.4) and incubated with 5 μl of propidium iodide (PI) (1 mg/ml) at 37°C for 20 min so that the total number of cells could be determined. Before flow cytometric counts, 5 μl of 0.7-μm yellow-green (YG) beads with known concentration (Polysciences, Inc) was added according to the manufacturer's instructions in order to determine cell numbers. Samples were analyzed by a FACScalibur flow cytometer (Becton Dickinson). Illumination of the samples was done with an argon ion laser (488 nm), and fluorescence of the YG beads and PI were collected in the FL1 (515 to 545 nm) and FL3 (>600-nm long pass) detectors, respectively. The system threshold was set on forward scatter signals, and all bacterial analyses were performed at the low rate settings (12 μl/min). Collection and analysis of the data were performed as reported previously (30).
DNA isolation, PCR, and DGGE analysis.
Before DNA isolation, fecal samples were resuspended in 0.05 M potassium phosphate. DNA was isolated from the fecal and unfixed biopsy samples by using the bead beating method as described previously (30). In short, samples were incubated at 55°C for 1 h after addition of 50 μl of 10% sodium dodecyl sulfate and 10 μl of proteinase K (20 mg/ml), followed by addition of 150 μl of phenol (pH 7.5) and mechanical disruption at 5,000 rpm for 3 min. Phenol-chloroform extractions and one chloroform extraction were performed to remove impurities. Before ethanol precipitation at −20°C was performed, 1 μl of glycogen solution (20 mg/ml) was added. After washing of the pellets, DNA was resuspended in 100 μl of Tris-EDTA buffer.
DNA isolated from biopsy and fecal samples (<10 ng) was subsequently used as a template to amplify the V6-to-V8 regions of 16S rDNA with primers F-0968-GC and R-1401 (20). The amplification (35 cycles) and the analysis of 5 μl of amplicons on ethidium-stained 1.2% agarose gels were performed as described previously (28). DGGE analysis of the amplicons was performed on 8% polyacrylamide gels containing a urea-formamide gradient from 38 to 48% (a 100% urea-formamide solution consists of 7 M urea and 40% [vol/vol]) formamide). Electrophoresis and staining of the gels were performed as reported previously (29). Stained gels were scanned at 400 dots per inch and analyzed with the software of Molecular Analyst 1.12 (Bio-Rad). The similarities between the DGGE profiles were determined by calculating similarity indices of the densitometric curves of the profiles compared by using the Pearson product-moment correlation (7, 29). Unweighted pair group method using arithmetic averages (UPGMA), Ward's, and neighbor-joining algorithms were performed, and corresponding dendrograms showing the relationships between the DGGE profiles were constructed. Scanning and analysis of the gels were performed three times.
Amplification of 16S rDNA fragments from the Lactobacillus group population was performed by a nested-PCR approach. First, the complete 16S rDNA was amplified with the canonical primers Bact-0011f and Bact-1492r (14). After purification with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany), the Lactobacillus group-specific PCR was performed with primers Bact-0124-GCf and Lab-0677r followed by DGGE analysis on 8% polyacrylamide gels containing a urea-formamide gradient from 30 to 60% (8). For cloning and sequence analysis, the Lactobacillus group amplicons were purified, cloned, and sequenced as described previously (8).
Statistical analysis.
Paired and Student's t tests were used for statistical analysis of comparisons between the cell numbers and between similarity indices from the scanned DGGE profiles, respectively.
Nucleotide sequence accession numbers.
Sequences determined in this study were deposited in the GenBank database under accession numbers AY027791 and AY027792.
RESULTS
Colonoscopic examination of the volunteers.
Three of the 10 individuals (numbers 4, 5, and 8 [Table 1]) had previously diagnosed ulcerative colitis. The disease was in remission both clinically and histologically in individuals 4 and 8, while individual 5 was having a relapse at the time of the study. Polyposis was diagnosed for individual 3 (Table 1). These four individuals are subsequently indicated as individuals having a diagnosed illness. No intestinal disease could be detected in the remaining six individuals.
Bacterial numbers in the biopsy samples.
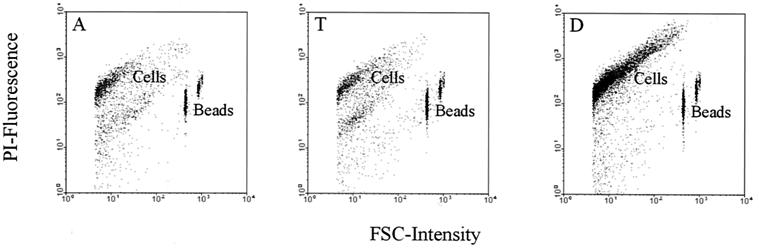
The bacteria in biopsy samples of approximately 0.5 mg were counted by a flow cytometric approach in order to quantify them in a culture-independent way. Since the bacteria were released from the biopsy material by a mild treatment and since it is difficult to determine how many cells were still attached after sonication, the total count of bacteria was determined as the minimal number per biopsy sample. PI-stained bacterial cells could be accurately counted when beads with known concentration were added, as illustrated in Fig. 1. The different biopsy samples revealed bacterial quantities that varied between 8.6 × 104 and 6.9 × 106 cells depending on the location in the colon and the individual (Table 1), with a mean count of 1.1 × 106 bacteria per sample. The detection limit for accurate counting was found to be 3.7 × 104 (± standard deviation [SD] of 1.4 × 104) cells per sample. The numbers of bacteria in specimens from the ascending colon seem to be slightly lower than from the other locations, although no significant differences in bacterial numbers at these locations (the lowest P2-tail was 0.075) were found by paired t test analyses.
FIG. 1.
Flow cytometric dot blots showing the discrimination between the PI-stained cells and the YG beads. The different biopsy locations for one individual with colitis ulcerosa (individual 5) are illustrated. A, T, and D, ascending, transverse, and descending parts of the colon, respectively.
Spatial distribution of the predominant bacterial community.
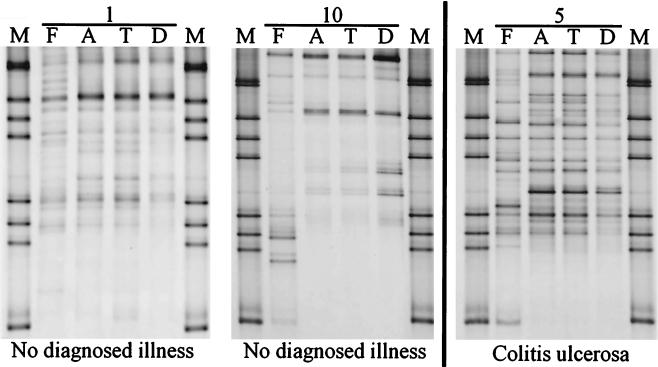
Following DNA isolation from the fecal and biopsy samples of the 10 individuals (Table 1), PCR was performed to amplify the V6-to-V8 regions of 16S rDNA. Amplicons were detected in all samples with the exception of the biopsy samples from the ascending and transverse colon of individual 7. DGGE analysis of the fecal and biopsy samples showed an enormous difference in the diversity of the amplicons in the profiles from the different individuals (as illustrated in Fig. 2). Dilution of biopsy specimen DNA (10 times) did not result in a change in the profile, indicating that the number of cells per biopsy sample was sufficient to obtain reliable and reproducible DGGE profiles. Remarkably, the predominant community in biopsy samples from all locations in the colon gave very similar profiles in each individual, despite the difference in diversity and diagnosed illness of the individuals (Fig. 2). In contrast, the fecal profiles were in most cases different from those obtained with the biopsy samples, indicating that it is very unlikely that fecal contamination took place during the colonoscopy. Since the biopsy samples were taken after evacuation of the colon it is very plausible that the bacteria detected in these specimens are mucosa-associated and therefore in close contact with the host cells.
FIG. 2.
Silver-stained DGGE gel showing profiles which represent the predominant community of feces (F) and biopsy samples (A, T, D) of individuals 1, 5, and 10. M represents the marker for DGGE analysis. The physiological conditions of the individuals are indicated.
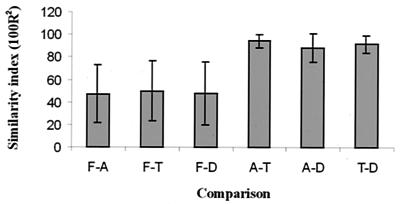
To determine whether communities from feces and biopsy samples were significantly different in single individuals, similarity indices of the DGGE profiles were calculated. It was observed that within comparisons between a fecal sample and one of the biopsy samples the individual variation was relatively high compared to the comparisons between different biopsy samples from the same individual. For example, the similarity indices for comparisons between feces and descending colon biopsy specimens varied from 13.6 to 91.3, while similarity indices between 81.6 and 98.1 were found when descending colon biopsy specimens were compared to ascending colon specimens (Table 1). Overall, indices for the similarity indices of comparisons between all biopsy samples from the same individual were very high (91.6 ± 9.6 [SD]), close to those calculated for the reproducibility of the procedures (93.4 ± 3.6). To rule out that the diagnosed illness of some of the individuals had an influence on the observed findings, the mean and standard variation of each similarity index within the healthy individuals and those with diagnosed illness were compared separately. Student's t test revealed that there was no significant difference between the similarity indices of both groups for each comparison, since the lowest P2-tail observed was 0.065 (7 df) for the similarity indices for comparison between ascending and transverse colonic biopsy samples.
The similarity indices between a fecal sample and one biopsy sample were compared with those between the remaining biopsy samples in order to obtain independent comparisons for statistical analysis (Fig. 3). All combinations of comparisons showed that the bacterial composition in fecal samples was significantly different from that in the biopsy samples. The highest P2-tail was 0.0012 (16 df) for comparison between the similarity indices of feces and transverse colon biopsy samples with ascending and descending colon biopsy samples.
FIG. 3.
Comparison between fecal and biopsy samples. F, fecal samples; A, T, and D, biopsy samples from the ascending, transverse, and descending parts of the colon, respectively; -, comparison. The means and SD of similarity indices are indicated.
Spatial distribution of the Lactobacillus community.
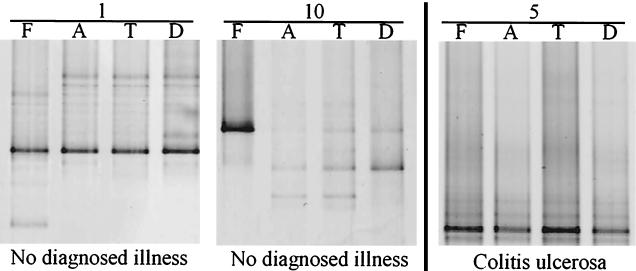
A nested-PCR approach was used to specifically amplify the V2-to-V4 regions of the 16S rDNA of the Lactobacillus group community, since no amplicons were retrieved by a direct specific-PCR approach. In contrast to the DGGE profiles of the predominant bacterial community, the Lactobacillus group-specific profiles were lower in diversity, as illustrated in Fig. 4. Because of this low diversity, similarity indices for the DGGE profiles cannot be determined. In contrast to the predominant bacterial community, the Lactobacillus communities in fecal and biopsy samples were very similar in 6 of the 10 individuals. In these individuals, only one amplicon was dominating (see, for example, the data for individuals 1 and 5 in Fig. 4). In the other individuals one of the fecal amplicons was the only predominant one in the biopsy samples or vice versa (such as for individual 10 in Fig. 4). Furthermore, for 3 of the 10 individuals some minor differences in the Lactobacillus group compositions between the biopsy samples were found. These small differences could not be explained by the physiological condition, age, or gender of the host since they were found in individuals 2 (healthy), 8 (remission of ulcerative colitis), and 10 (healthy).
FIG. 4.
Silver-stained DGGE gel showing profiles which represent the Lactobacillus group community of fecal (F) and biopsy (A, T, D) samples of individuals 1, 5, and 10. The physiological conditions of the individuals are indicated.
Comparison between healthy individuals and individuals with diagnosed illness.
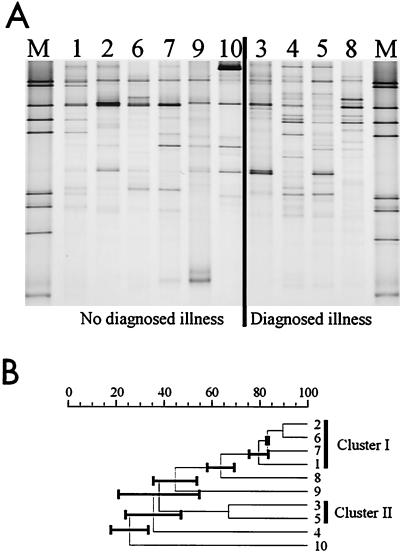
DGGE profiles of biopsy samples from the descending colons of individuals with and without a diagnosed illness were compared to see if the presence or absence of specific bacteria could be correlated to the illness. The descending colon was chosen, since all diagnosed illnesses were observed at least in this part. The profiles of the predominant bacterial community appeared to be unique for each individual, and no specific amplicon could be assigned to the presence or absence of a colonic illness (Fig. 5A). To analyze the predominant communities, similarity indices of the comparisons between the DGGE profiles were calculated. Repetitive comparisons between the UPGMA, Ward, and neighbor-joining algorithms were performed, and dendrograms were constructed. Only two clusters were found in all dendrograms, while the position of the others branches in the dendrogram changed depending on the clustering method. The large error bars of the nodes in the UPGMA tree (Fig. 5B) could be seen as an indication that the corresponding branches of these nodes may vary between the different algorithms. One of the repetitive clusters consisted of four healthy individuals (i.e., individuals 1, 2, 6, and 7), and the other consisted of the two individuals diagnosed with an active form of a GI tract disorder (i.e., individuals 3 and 5). This preliminary observation suggests that there might be differences in the predominant bacterial composition between healthy and diseased individuals, although the group of individuals in this study is too small to allow a definite conclusion.
FIG. 5.
(A) Silver-stained DGGE gel showing profiles which represent the predominant communities of the descending colon biopsy samples from individuals 1 to 10. M represents the marker for DGGE analysis. The physiological conditions of the individuals are indicated. (B) UPGMA dendrogram illustrating the correlation between the different DGGE profiles of panel A. Cluster I and II represent the repetitive clusters obtained using different algorithms. The black bars represent the error bars.
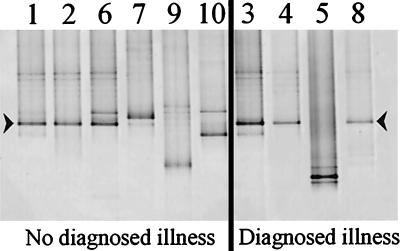
For the Lactobacillus group community also no specific differences could be found between the individuals with and those without diagnosed illness (Fig. 6). A striking observation was the presence of an amplicon with identical DGGE band positions for 9 of the 10 individuals. Since it appeared to be a Lactobacillus gasseri-like species (see below), we tested whether its predominance might be a result of preferential amplification. Our nested-PCR approach did not show any preference in favor of L. gasseri when mixtures of its DNA with that from Lactobacillus acidophilus and Lactobacillus paracasei were used as template DNA for PCR. Cloning and sequence analysis of the amplicons from the DGGE profiles of individual 1 (healthy) and individual 3 (with polyposis) showed that both sequences had 99% similarity with L. gasseri. Alignment of the two sequences showed that they differ by only one base (adenine in one and thymine in the other). This indicates that L. gasseri is likely to be a predominant Lactobacillus species in the biopsy samples.
FIG. 6.
Silver-stained DGGE gel showing profiles which represent the Lactobacillus group communities of the descending colon biopsy samples from individuals 1 to 10. The physiological conditions of the individuals are indicated. The arrowheads indicate the amplicons which have been identified by cloning and sequencing.
DISCUSSION
In this study we have used a culture-independent approach based on the 16S rDNA sequence variability to analyze bacterial communities in different parts of the colon. Fecal and biopsy samples were taken from people with and without a diagnosed illness. Since the colon was evacuated before biopsies were performed, it is very likely that the bacteria in the biopsy samples are mucosa associated. The minimum number of cells per biopsy sample as measured by flow cytometry is comparable to numbers found by cultivation of bacteria from biopsy samples which were obtained by a similar procedure (1). Considerable variation was found in the bacterial numbers from different biopsy samples. Factors that may cause this variation include the evacuation and sampling procedures, the sonication procedure, and individual differences. On the other hand, this variation in bacterial number may explain why no PCR product could be obtained from two biopsy samples.
DGGE analysis of 16S rDNA amplicons was used to determine, compare, and visualize the compositions of the predominant bacterial and of the Lactobacillus group communities. The DGGE profiles reflecting the predominant bacterial communities in biopsy samples from different locations in the GI tract were highly similar to each other, while they differed significantly from those of fecal samples (Fig. 3). Therefore, it seems that the mucosa-associated bacteria are equally distributed along the complete colon and that different populations are dominating in the mucosa and the feces. Recently, differences in the structures of communities in feces and cecal contents, observed through a dot blot hybridization and culturing approach, have been reported (16). Culture-dependent studies of contents from different parts of the colon (including the ascending, transverse, and descending parts) of sudden-death victims have revealed that the conditions, for example, pH and concentration of fermentation products, in these parts differ considerably from one another (15). This suggests that the uniform distribution of the attaching bacterial composition along the colon is very likely due to host-bacterium interactions at the mucosa. Several studies have already suggested that the bacterial community in the GI tract has a strong effect on the host and that signaling between host and bacterium is very important (3, 9, 10). In a recent study, a significant positive relationship between the genetic relatedness of the hosts and the similarity between their bacterial communities was found (29). However, it is not clear yet what the nature of these host-related factors is.
With a nested-PCR approach using group-specific primers (8), the Lactobacillus group-specific composition was analyzed. In contrast to those of the predominant community, the profiles of biopsy and fecal samples were quite similar for 6 of the 10 individuals. Furthermore, for 3 of the 10 individuals some minor differences between the biopsy samples were found in the Lactobacillus group composition. This suggests that the changing conditions in the GI tract influence the presence or absence of certain species belonging to the Lactobacillus group. Another explanation could be the detection limit of these bacteria in the biopsy specimens. Since we are focusing on a subpopulation in a community which contains approximately 106 bacteria, a small difference in the number of organisms of a certain species might have a large impact on its detection. Remarkably, one amplicon with the highest sequence similarity (99%) to L. gasseri was found in descending colon biopsy samples of 9 of the 10 individuals. Moreover, it was the most predominant one in most biopsy specimens. Since the 16S rDNA of this species was not preferentially amplified by the nested-PCR approach, L. gasseri may be regarded as a general mucosa-associated bacterium in humans.
Because colonic illnesses were observed during colonoscopy, samples from healthy individuals and those with a diagnosed illness were compared to each other. Since the illnesses are found especially in the descending colon, we compared the samples from these regions and compared them with those from healthy individuals. No significant difference could be detected with respect to the number of bacteria per biopsy specimen, the composition of the predominant bacterial community, and that of the Lactobacillus group community. This is supported by the observation that the profiles reflecting the predominant community were highly similar along the complete colon for both groups. Lactobacilli were detected in both feces and biopsy samples from all individuals, and no differences in the Lactobacillus group populations between healthy and diseased tissues were found.
The molecular approach used in this study can be influenced by preferential amplification and difference in DNA isolation efficiency of different species. However, DNA from biopsy samples could be diluted 10 times without changing the profiles. Furthermore, for two individuals the similarity between fecal and biopsy samples was very high, while the number of bacteria in the fecal samples was at least 103 times higher than that in the biopsy samples. These data indicate that it is very unlikely that procedures such as sampling, storage, and transport or preferential lysis of specific groups of bacteria have a major impact on our observations.
In conclusion, using a culture-independent approach we were able to clearly demonstrate that mucosa-associated bacterial communities in the colon are significantly different in composition from those in feces. A strikingly high similarity between bacterial communities from different locations in the colon was observed. This observation suggests strongly that host-related factors are important in the colonic ecosystem, which is in line with previous observations (3, 9, 10, 29). A culture-independent approach was also used to characterized subpopulations of the Lactobacillus genus. Similar approaches using group-specific primers can also be applied to study (sub)populations of other bacteria such as those implicated in the initiation and maintenance of ulcerative colitis (4, 5, 27). Hence, systematic culture-independent approaches could be instrumental in determining the roles of various GI tract subpopulations in the pathogenesis of colonic diseases.
Acknowledgments
This work was partly supported by the Wageningen Centre for Food Sciences.
We thank G. H. J. Heilig, P. Verbaarschot, M.-L. Kekäläinen, and M. Rekola for technical assistance and J. A. G. M. de Visser for advice on statistical analysis. In addition, we thank all volunteers for providing fecal and biopsy samples.
REFERENCES
- 1.Alander, M., R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1997. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett. Appl. Microbiol. 24:361-364. [DOI] [PubMed] [Google Scholar]
- 2.Alander, M., R. Satokari, R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bry, L., P. G. Falk, T. Midtvedt, and J. I. Gordon. 1996. A model of host-microbial interactions in an open mammalian ecosystem. Science 273:1381-1383. [DOI] [PubMed] [Google Scholar]
- 4.Campieri, M., and P. Gionchetti. 2001. Bacteria as the cause of ulcerative colitis. Gut 48:132-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson, G. R., J. H. Cummings, and G. T. Macfarlane. 1991. Growth and activities of sulphate-reducing bacteria in the gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol. Ecol. 86:103-112. [Google Scholar]
- 6.Gillian Hartley, M., M. J. Hudson, E. T. Swarbrick, M. J. Hill, A. E. Gent, M. D. Hellier, and R. H. Grace. 1992. The rectal mucosa-associated microflora in patients with ulcerative colitis. J. Med. Microbiol. 36:96-103. [DOI] [PubMed] [Google Scholar]
- 7.Häne, B. G., K. Jäger, and H. Drexler. 1993. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 14:967-972. [DOI] [PubMed] [Google Scholar]
- 8.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed]
- 9.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2000. Molecular analysis of host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson, M.-L., G. Molin, B. Jeppsson, S. Nobaek, S. Ahrné, and S. Bengmark. 1993. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl. Environ. Microbiol. 59:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In. E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons, Chichester, United Kingdom.
- 15.Macfarlane, G. T., G. R. Gibson, and J. H. Cummings. 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57-64. [DOI] [PubMed] [Google Scholar]
- 16.Marteau, P., P. Pochart, J. Doré, C. Béra-Maillet, A. Bernallier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millar, M. R., C. J. Linton, A. Cade, D. Glancy, M. Hall, and H. Jalal. 1996. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J. Clin. Microbiol. 34:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monstein, H.-J., A. Tiveljung, C. H. Kraft, K. Borch, and J. Jonasson. 2000. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J. Med. Microbiol. 49:817-822. [DOI] [PubMed] [Google Scholar]
- 19.Nebe-von-Caron, G., P. Stephens, C. H. Hewitt, J. R. Powel, and R. A. Badley. 2000. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 42:97-114. [DOI] [PubMed] [Google Scholar]
- 20.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannock, G. W. 1995. Normal microflora. An introduction to microbes inhabiting the human body. Chapman and Hall, London, United Kingdom.
- 24.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughan, E. E., G. H. J. Heilig, E. G. Zoetendal, R. Satokari, J. K. Collins, A. D. L. Akkermans, and W. M. de Vos. 1999. Molecular approaches to study probiotic bacteria. Trends Food Sci. Tech. 10:400-404. [Google Scholar]
- 26.Wilson, K. H., and R. H. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinkevich, V., and I. B. Beech. 2000. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol. Ecol. 34:147-155. [DOI] [PubMed] [Google Scholar]
- 28.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoetendal, E. G., A. D. L. Akkermans, W. M. Akkermans van-Vliet, J. A. G. M. de Visser, and W. M. de Vos. 2001. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb. Ecol. Health Dis. 13:129-134. [Google Scholar]
- 30.Zoetendal, E. G., K. Ben-Amor, A. D. L. Akkermans, T. Abee, and W. M. de Vos. 2001. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst. Appl. Microbiol. 24:405-410. [DOI] [PubMed] [Google Scholar]