Abstract
Nineteen numerically dominant heterotrophic bacteria from a freshwater biofilm were identified by 16S ribosomal DNA gene sequencing, and their coaggregation partnerships were determined. Phylogenetic trees showed that both distantly related and closely related strains coaggregated at intergeneric, intrageneric, and intraspecies levels. One strain, Blastomonas natatoria 2.1, coaggregated with all 18 other strains and may function as a bridging organism in biofilm development.
Coaggregation between bacteria occurs when two or more genetically distinct strains interact by specific cell-cell recognition (12). The phenomenon was first recognized between different oral plaque-forming bacteria, where both intergeneric and intrageneric coaggregation occurs (11). Coaggregation also occurs between bacteria isolated from a freshwater biofilm (3, 19), and it has been suggested that coaggregation may also mediate in the sequential integration of species of bacteria into freshwater biofilms (8, 20). Recently, Rickard et al. (19) used partial 16S rRNA gene sequencing to identify four coaggregating strains of Blastomonas natatoria and one strain of Micrococcus luteus from an established freshwater biofilm community (3). Six coaggregation partnerships between these five strains were found and shown to be mediated by growth-phase-dependent lectin-saccharide interactions (19). These five coaggregating strains of B. natatoria and M. luteus were part of a larger community of 19 coaggregating strains that were all isolated simultaneously from a biofilm formed on glass in a chemostat (3). The identities of the other members of the consortium are unknown, and the extent of intergeneric and intrageneric and intraspecies coaggregation between all members of the community has not been analyzed previously. Since coaggregation may be an adhesion mechanism involved with integrating and establishing bacteria in the biofilm community, it is important to know the extent of this specific adhesion mechanism in the freshwater biofilm. It is also relevant to know how closely related coaggregating strains are, since this has implications for understanding the biodiversity of the biofilm community. Therefore, the work reported here had three main objectives: (i) identification by 16S rRNA gene sequencing of all strains in the freshwater biofilm community; (ii) construction of phylogenetic trees by the computation of evolutionary distance matrices and maximum-likelihood rooted dendrograms; and (iii) analysis of intergeneric and intrageneric and intraspecies (interstrain) coaggregation partnerships between members of the biofilm community deduced using the phylogenetic trees.
All strains used in this study were isolated from a 14-day-old biofilm on a glass coupon in a two-stage chemostat kept at 4°C, which was initially inoculated with water from a borehole water source (Porton borehole, Salisbury, United Kingdom) (3). The ionic composition of the water and temperature within the chemostat were very similar to the conditions found in the source borehole water (4). Strains from the biofilm were initially grown on R2A agar (17) and were inoculated separately into conical flasks (250 ml) containing 100 ml of R2A broth and shaken at 200 rpm at 25°C in a G20 orbital shaker (New Brunswick Scientific, New Brunswick, N.J.).
The strains were identified by the method of Rickard et al. (19). Approximately 650 bases of the 16S rRNA gene were sequenced. Amplification of 16S ribosomal DNA was performed by removing a single bacterial colony from R2A agar plates to provide template DNA. Partial 16S rRNA gene sequences corresponding to the Escherichia coli 16S ribosomal DNA nucleotide positions 8 to 806 were amplified and sequenced using the universal primers 8FPL (22) and 806R (23). Partial 16S rRNA gene sequences of each of the strains were initially compared to those in the databases by using the FASTA3 program, and unambiguous positions of representative sequences were then aligned by using CLUSTALX version 1.64b (21). Maximum-likelihood analysis was conducted using DNAML (6), and the trees were viewed using TREEVIEW (15).
Table 1 shows the percent nucleotide sequence identity of all 19 biofilm strains to the closest sequence in the EMBL database. The five strains identified by Rickard et al. (19) have been included for completeness. Eleven of the fourteen strains identified in this study could be assigned to a genus, but no species identification was possible, since the closest strains in the database were not assigned species (Table 1). The biochemical and morphological characteristics of each strain supported the assigned identities (data not presented). Only strain 2.17 could not be identified to the genus level, since it had a very low percent sequence identity (87.6%) with the closest matching 16S rRNA gene sequence in the database, which belonged to Salinicoccus roseus (EMBL accession no. SR16SRRN1).
TABLE 1.
Identification of the aquatic biofilm strains by alignment with the sequences of organisms in the EMBL database
| Strain no. | Database accession no. | Highest % identity to sequence in database | Proposed identitya |
|---|---|---|---|
| 2.2 | AJ299221 | 99.35 | Afipia sp. 2.2 |
| 2.1 | AJ250434 | 99.71 | B. natatoria 2.1* |
| 2.3 | AJ250435 | 98.39 | B. natatoria 2.3* |
| 2.4 | AJ299222 | 99.71 | B. natatoria 2.4 |
| 2.6 | AJ250436 | 99.85 | B. natatoria 2.6* |
| 2.8 | AJ250437 | 99.85 | B. natatoria 2.8* |
| 2.7 | AJ299223 | 98.68 | Methylobacterium sp. 2.7 |
| 2.9 | AJ299224 | 98.05 | Methylobacterium sp. 2.9 |
| 2.13 | AJ250438 | 98.88 | M. luteus 2.13* |
| 2.14 | AJ299232 | 97.54 | Nocardioides sp. 2.14 |
| 2.20 | AJ299233 | 97.71 | Nocardioides sp. 2.20 |
| 2.21 | AJ299231 | 99.84 | P. marcusii 2.21 |
| 2.19 | AJ299230 | 99.07 | Pseudomonas sp. 2.19 |
| 2.17 | AJ299234 | 87.59 | Unknown |
| 2.10 | AJ299225 | 99.51 | Sphingomonas sp. 2.10 |
| 2.11 | AJ299226 | 94.77 | Sphingomonas sp. 2.11 |
| 2.12 | AJ299227 | 96.76 | Sphingomonas sp. 2.12 |
| 2.15 | AJ299228 | 96.94 | Sphingomonas sp. 2.15 |
| 2.18 | AJ299229 | 98.39 | Sphingomonas sp. 2.18 |
Strains with closest sequence identity were named. *, strains previously sequenced and named by Rickard et al. (19).
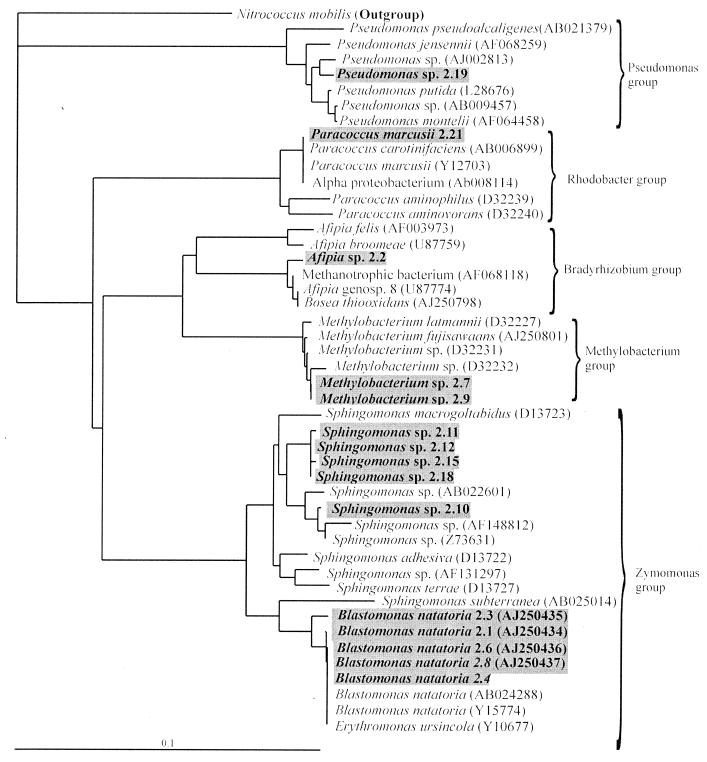
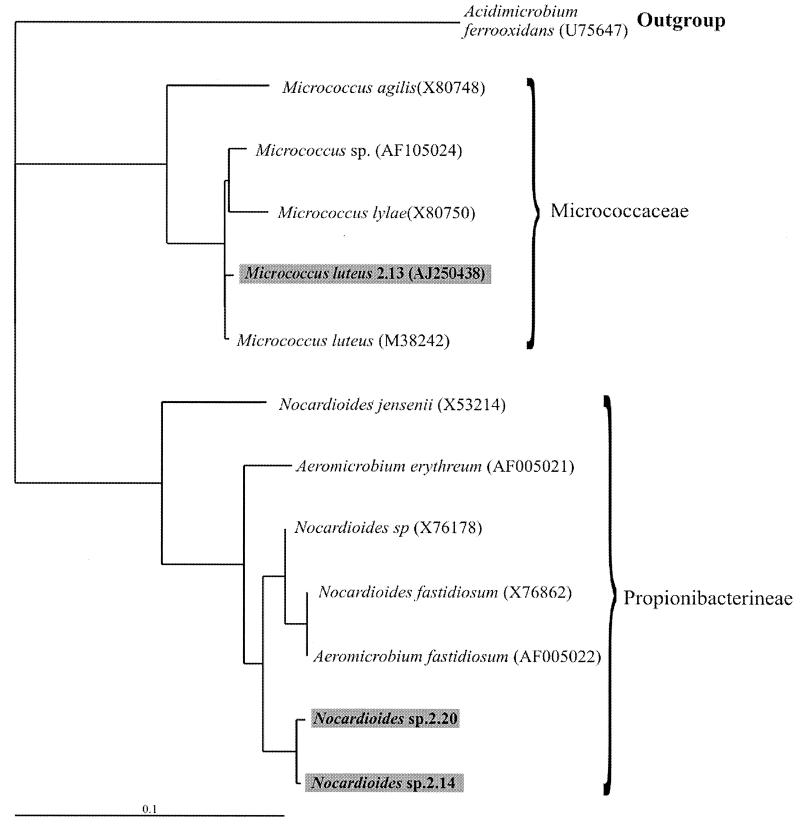
Maximum-likelihood phylogenetic trees inferred from the 16S rRNA sequences of all the 19 coaggregating biofilm strains showed that they are distributed over a wide range of bacterial taxonomic groups (Fig. 1 and 2). The 6 gram-negative genera are distributed between four groups of the alpha-Proteobacteria subclass of the Proteobacteria: the Zymomonas, Methylobacterium, Bradyrhizobium, and Rhodobacter groups, as well as a single group of the gamma-Proteobacteria subclass, the Pseudomonas group (Fig. 1). The two gram-positive genera, Micrococcus and Nocardiodes, were members of the high-G+C groups Micrococcaceae and Propionibacterineae (Fig. 2). Strain 2.17, which could not be identified, is not on the gram-positive phylogenetic tree but is a member of the low-G+C group occupied by S. roseus.
FIG. 1.
Maximum-likelihood phylogenetic tree constructed using partial 16S rRNA gene sequences of closely related speciated and nonspeciated strains belonging to the alpha- and gamma-Proteobacteria. The tree is rooted using Nitrococcus mobilis (L35510) as the outgroup. Nucleotide sequence database accession codes are shown in the brackets. The length of each branch is proportional to the estimated number of substitutions per position. Sequenced strains are highlighted in boxes; most-likely identities were assigned.
FIG. 2.
. Maximum-likelihood phylogenetic tree constructed using partial 16S rRNA gene sequences of closely related speciated and nonspeciated strains belonging to the high-G+C gram-positive bacteria. The tree is rooted using Acidimicrobium ferrooxidans (U75647) as the outgroup. Nucleotide sequence database accession codes are shown in the brackets. The length of each branch is proportional to the estimated number of substitutions per position. Sequenced strains are highlighted in boxes; most-likely identities were assigned.
Most of the identified strains in this study were from the alpha-Proteobacteria (74%), and no beta-Proteobacteria were found. R2A agar has been shown to support the growth of alpha and gamma Proteobacteria rather than the less easily cultivable beta-Proteobacteria (9, 10). This could explain why the majority of these freshwater biofilm strains that were originally isolated by Buswell et al. (3) were from the alpha-Proteobacteria. However, the objective of this study was to establish the extent of coaggregation between cultivable heterotrophic bacteria, and enough strains have been isolated and identified to reveal that coaggregation is a common property of strains in this ecosystem. A considerable taxonomic diversity of heterotrophs in the biofilm was found, and all except strain 2.17 are very closely related to organisms that have previously been isolated from freshwater or soil environments.
The phylogenetic trees revealed that the biofilm contained clusters of very closely related strains or clones. These clonal groups included five B. natatoria strains, four Sphingomonas strains, two Methylobacterium strains (Fig. 1), and two Nocardioides strains (Fig. 2). Each clonal group contained strains with different coaggregation partners (see Fig. 5). It has been proposed that biofilms can act as reservoirs of clones of the same species and that this may enhance the survival of the species during environmental fluctuations (2, 16). It is not yet clear why closely related clones with such distinct coaggregation patterns can coexist within a freshwater biofilm.
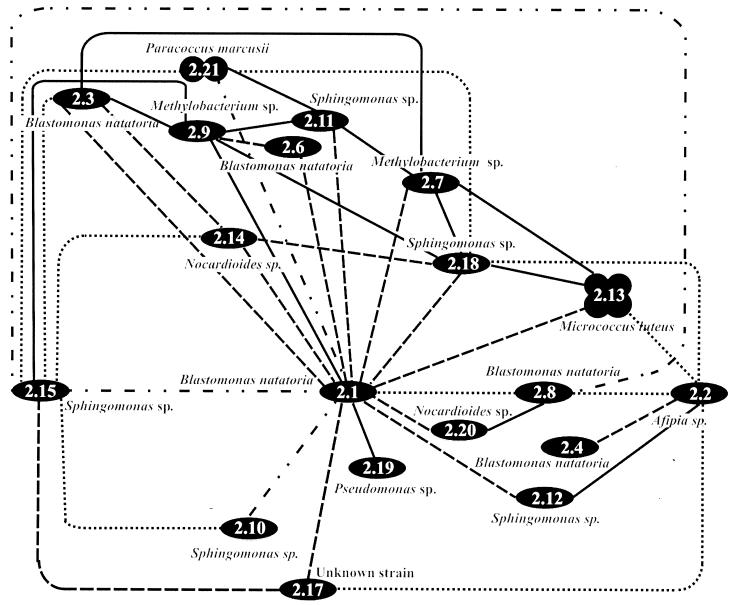
FIG. 5.
A diagrammatic representation of the specificity of coaggregation of all 19 identified strains when grown separately in batch culture and harvested after 36, 72, and 144 h. Optimal expression of coaggregation at either 36 (—), 72 (- - -), or 144 h is shown for each pair. Expression of coaggregation that was constant between pairs at 36, 72, and 144 h is also shown (-..-..-). Visual coaggregation scores below a score of 2 are not included.
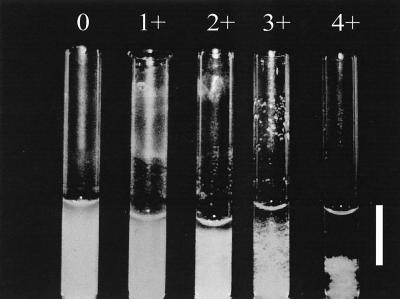
In order to detect all the coaggregation partnerships between strains, all 171 possible pairwise combinations of strains were tested using the visual coaggregation assay originally developed by Cisar et al. (5) and adapted by Rickard et al. (20). Because the ability to coaggregate has been found to be optimum for only a relatively short period in stationary phase and not to occur in exponential phase for B. natatoria 2.1 and M. luteus 2.13 (19), strains were harvested at three different times during stationary phase. The harvest times were the following: early stationary phase (36 h), mid-stationary phase (72 h), and later stationary phase (144 h) in batch culture (growth kinetics data not shown). In this way the optimum time in stationary phase at which coaggregation occurred could be determined. Cells were harvested and washed three times in sterile deionized water. Using a spectrophotometer (Cecil instruments, Cambridge, United Kingdom), the washed cells were then resuspended in deionized water to an optical density at 650 nm (OD650) of 1.0 and concentrated to give a calculated final OD650 of 1.5. For coaggregation, pairs of strains were mixed at an OD650 of 1.5 in equal volumes (200 μl) at room temperature in 6- by 50-mm silica Durham tubes (Scientific Lab Supplies, Nottingham, United Kingdom). Mixtures were vortexed for 10 s, and the tubes were rolled gently for 30 s. The degree of coaggregation between each pair was scored as follows: 0, no coaggregates in suspension; 1+, small uniform aggregates in a turbid suspension; 2+, easily visible coaggregates in a turbid suspension; 3+, clearly visible coaggregates which settle, leaving a clear supernatant; 4+, large flocs of coaggregates that settle almost instantaneously, leaving a clear supernatant. Control tubes of each isolate on its own were also included to assess autoaggregation. Where present, the autoaggregation score was deducted from the coaggregation score.
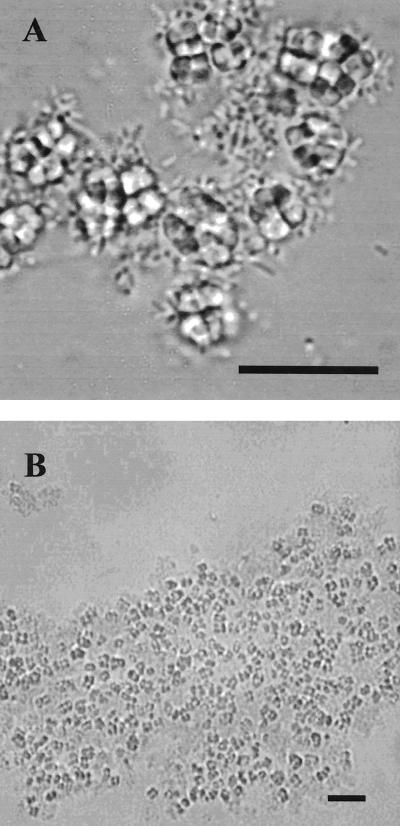
Maximum visual coaggregation scores for different pairs ranged from 1+ to 4+ (Fig. 3), and 82 pairs out of the total of 171 pairs (48%) gave a positive score. Scores were always reproducible after growth in batch culture for a set period of time, and three batches of all cultures were tested separately to confirm the reproducibility of coaggregation. For all pairs, a microscopic examination of the coaggregating pairs showed flocs of coaggregating cells and the size of the coaggregates increased with increasing visual coaggregation scores (Fig. 4).
FIG. 3.
Visual coaggregation scores for different pairs of strains. Cells were grown to stationary phase and harvested after 36, 71, or 144 h. Sphingomonas sp. strain 2.10 and Afipia sp. strain 2.2 (36 h) showed a score of 0 (no coaggregation), P. marcusii 2.21 and B. natatoria 2.3 (72 h) showed a score of 1+, Sphingomonas sp. strain 2.18 and Afipia sp. strain 2.2 (144 h) showed a score of 2+, B. natatoria 2.1 and Nocardioides sp. strain 2.20 (144 h) showed a score of 3+, and B. natatoria 2.1 with M. luteus 2.13 (72 h) showed a score of 4+. Bar, 1 cm.
FIG. 4.
Light micrographs showing rod-shaped cells of B. natatoria 2.1 coaggregating to tetrads of coccus-shaped cells of M. luteus 2.13. Bar, 10 μm.
In order to represent the complexities of the many coaggregation partnerships, a matrix of all the pairings giving a score of 2+ or greater was constructed (Fig. 5). The growth time in batch culture (36, 72, or 144 h) at which optimum coaggregation scores occurred is indicated on the matrix. For 76 of the 82 coaggregating pairs (93%) the ability to coaggregate was maximum at only one of the three harvest times in stationary phase. After 36 h of growth in batch culture, 38 pairs (22%) coaggregated, after 72 h, 62 pairs (36.2%) coaggregated, and after 144 h of growth, 36 pairs (21%) coaggregated. The majority of the possible coaggregation partnerships occurred after incubation in batch culture for 72 h, although 7% of the possible 171 coaggregation partnerships showed a consistent score which was not influenced by the harvest time (Fig. 5). In freshwater the bacteria in biofilms are probably in stationary phase for the majority of the time (7) and may therefore have evolved to express the ability to coaggregate while in this starved physiological state.
As shown in the coaggregation matrix (Fig. 5), B. natatoria 2.1 was able to coaggregate with all other strains tested. Other highly interactive strains included Sphingomonas sp. strain 2.15, with 15 partner strains, and Afipia sp. strain 2.2, with 13 partner strains. In contrast, Pseudomonas sp. strain 2.19 and B. natatoria 2.4 had the lowest number of partnerships, each coaggregating with two and four partners, respectively. Each strain possessed different coaggregation abilities, with respect to numbers and identity of partners as well as the size of the flocs formed.
When considering the phylogenetic relationships and coaggregation partnerships, coaggregation at an intergeneric level was extremely common and every strain tested coaggregated intergenerically with at least one other strain from a different genus at a level of 2+ or greater (Fig. 5). For example, strain B. natatoria 2.1 coaggregated with strains from seven other genera—Sphingomonas, Nocardioides, Paracoccus, Methylobacterium, Micrococcus, Pseudomonas, and strain 2.17. Coaggregation at the interstrain (intraspecies) level was common, occurring between closely related strains of Sphingomonas and Blastomonas clustering in the same phylogenetic groups. Coaggregation between B. natatoria 2.1 and three other strains of B. natatoria (2.3, 2.8, and 2.6) has been reported previously (19); however, this study shows that B. natatoria 2.4 is a very poor coaggregator and it cannot coaggregate at the interstrain level with the other Blastomonas strains. Intergeneric coaggregation is common between oral bacteria (13), but intraspecies (interstrain) coaggregation has not yet been reported between plaque bacteria. Thus, intraspecies coaggregation may well be a characteristic that is unique to freshwater biofilm bacteria.
While intergeneric and interstrain coaggregations are common between these aquatic biofilm community members, there is only one example of intrageneric coaggregation. Sphingomonas 2.10 and Sphingomonas 2.15 coaggregate with each other after 144 h (Fig. 5) with a score of 4+, and the evidence indicates that they are likely to be from different, as yet unnamed species. First, the phylogenetic tree (Fig. 1) shows a cluster of very closely related Sphingomonas strains (2.11, 2.12, 2.15, and 2.18) with strain 2.10 in a separate subgroup. The group of four strains all have >99.1% sequence homology to each other and none were 100% identical. However, strain 2.10 had only from 97.7 to 98.4% sequence homology with the four closely related strains. Secondly, typing of the Sphingomonas strains by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels of whole-cell proteins shows 20 visible protein bands for strain 2.10, while for the strains in the group of four, only five identical bands were visible (18). In addition, all the Sphingomonas strains possessed distinct colony and cell morphologies. Taken together, the evidence indicates that 2.10 is likely to be a different species from the other four Sphingomonas strains. Therefore, we propose that intrageneric coaggregation can occur in both aquatic and oral biofilms.
In dental plaque, Fusobacterium nucleatum can coaggregate with all other oral bacteria so far tested (1, 14) and has therefore been described as a “promiscuous” coaggregator which is proposed to have a very significant role as a bridging organism linking primary and secondary colonizers that cannot coaggregate with each other (11). Since B. natatoria 2.1 coaggregated with all 18 other biofilm strains, it may also be described as promiscuous. B. natatoria 2.1 may have a role equivalent to that of F. nucleatum in the development of this freshwater biofilm and may have the same role in other biofilms. It is not yet known whether colonization during freshwater biofilm formation involves a succession of primary and secondary colonizers, as occurs in the development of dental plaque. However, it is possible that highly coaggregating genera such as Blastomonas, Afipia, and Sphingomonas spp. could be quantitatively important members of freshwater biofilm communities and that their ability to adhere to other community members in a biofilm would give them a colonization advantage.
In conclusion, this study has revealed that intergeneric and intraspecies coaggregation between freshwater bacteria are common phenomena that occur between strains from a laboratory model aquatic biofilm. In addition, expression of coaggregation is dependent on cells being in the optimum physiological state for coaggregation, which usually occurs in stationary phase. It is therefore possible that since cells grow very slowly in nutrient-limited biofilms, a natural freshwater biofilm would provide suitable conditions for expression of coaggregation.
Nucleotide sequence accession numbers.
Partial 16SrRNA sequences for M. luteus 2.13 and the four B. natatoria strains (2.1, 2.3, 2.6, and 2.8) have been assigned accession numbers previously (19). The sequences of all the remaining identified strains were also deposited in the EMBL database. The EBML accession numbers of the sequences were the following: for Afipia sp. strain 2.2, accession no. AJ299221; for B. natatoria 2.1, 2.3, 2.4, 2.6, and 2.8, accession numbers AJ250434, AJ250435, AJ299222, AJ250436, and AJ250437, respectively; for Methylobacterium sp. strains 2.7 and 2.9, accession numbers AJ299223 and AJ299224, respectively; for M. luteus 2.13, accession number AJ250438; for Nocardioides sp. strains 2.14 and 2.17, accession numbers AJ299232 and AJ299233, respectively; for Paracoccus marcusii 2.21, accession no. AJ299231; for Pseudomonas sp. strain 2.19, accession no. AJ299230; for Sphingomonas sp. strains 2.10, 2.11, 2.12, 2.15, and 2.18, accession numbers AJ299225, AJ299226, AJ299227, AJ299228, and AJ299229, respectively; for unknown bacterial heterotroph 2.17, accession no. AJ299234.
REFERENCES
- 1.Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13:51-54. [DOI] [PubMed] [Google Scholar]
- 2.Bowden, G. H. W. 1999. Oral biofilm an archive of past events? p. 211-235. In H. N. Newman and M. Wilson (ed.), Dental plaque revisited: oral biofilms in health and disease. Bioline press, Cardiff, United Kingdom.
- 3.Buswell, C. M., Y. M. Herlihy, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1997. Coaggregation amongst aquatic biofilm bacteria. J. Appl. Microbiol. 83:477-484. [Google Scholar]
- 4.Buswell, C. M., Y. M. Herlihy, C. W. Keevil, P. D. Marsh, and S. A. Leach. 1999. Carbon load in aquatic ecosystems affects the diversity and biomass of water consortia and the persistence of the pathogen Campylobacter jejuni within them. J. Appl. Microbiol. Symp. Suppl. 85:161S-167S. [DOI] [PubMed] [Google Scholar]
- 5.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomycetes viscosus or Actinomycetes naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1989. PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 7.Halda-Alija, L., and T. C. Johnston. 1999. Diversity of culturable heterotrophic aerobic bacteria in pristine stream bed sediments. Can. J. Microbiol. 45:879-884. [PubMed] [Google Scholar]
- 8.Handley, P. S., A. H. Rickard, S. A. Leach, C. M. Buswell, and N. J. High. 2001. Coaggregation—is it a universal phenomenon? p. 1-10. In P. Gilbert, D. Allison, M. Brading, J. Verran, and J. Walker (ed.), Biofilm community interactions: chance or necessity? Bioline press, Cardiff, United Kingdom.
- 9.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 63:4164-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kämpfer, P., R. Erhart, C. Beimfohr, J. Böhringer, M. Wagner, and R. Amann. 1996. Characterisation of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32:101-121. [DOI] [PubMed] [Google Scholar]
- 11.Kolenbrander, P. E., R. N. Andersen, D. L. Clemens, C. J. Whittaker, and C. M. Klier. 1999. Potential role of functionally similar coaggregation mediators in bacterial succession. In H. N. Newman and M. Wilson (ed.), Dental plaque revisited: oral biofilms in health and disease, p. 171-186. Bioline press, Cardiff, United Kingdom.
- 12.Kolenbrander, P. E. 1997. Oral microbiology and coaggregation. In J. A. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms, p. 245-269. Oxford University Press, Oxford, United Kingdom.
- 13.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Appl. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E., K. D. Parrish, R. N. Andersen, and E. P. Greenberg. 1995. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect. Immun. 63:4584-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 16.Rainey, P. B., E. R. Moxon, and I. P. Thompson. 1993. Intraclonal polymorphism in bacteria, p. 263-300. In J. Gwynfryn Jones (ed.), Advances in microbial ecology. Plenum Press, New York, N.Y.
- 17.Reasoner, D. J., and E. E. Geldrich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickard, A. H. 2001. Coaggregation between aquatic biofilm bacteria. Ph.D thesis. University of Manchester, Manchester, United Kingdom.
- 19.Rickard, A. H., S. A. Leach, C. M. Buswell, N. J. High, and P. S Handley. 2000. Coaggregation between aquatic bacteria is mediated by specific-growth-phase-dependent lectin-saccharide interactions. Appl. Environ. Microbiol. 66:431-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickard, A. H., J. Thomas, S. A. Leach, C. M. Buswell, N. J. High, and P. S. Handley. 1999. Coaggregation amongst aquatic and oral bacteria is mediated by lectin-saccharide interactions, p. 343-354. In J. Wimpenny, P. Gilbert, J. Walker, M. Brading, and R. Bayston (ed.), Biofilms: the good, the bad and the ugly. Bioline press, Cardiff, United Kingdom.
- 21.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisberg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S Ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson, K. E., Blitchington, R. B., and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]