Abstract
To study the effect of plant species on the abundance and diversity of bacterial antagonists, the abundance, the phenotypic diversity, and the genotypic diversity of rhizobacteria isolated from potato, oilseed rape, and strawberry and from bulk soil which showed antagonistic activity towards the soilborne pathogen Verticillium dahliae Kleb. were analyzed. Rhizosphere and soil samples were taken five times over two growing seasons in 1998 and 1999 from a randomized field trial. Bacterial isolates were obtained after plating on R2A (Difco, Detroit, Mich.) or enrichment in microtiter plates containing high-molecular-weight substrates followed by plating on R2A. A total of 5,854 bacteria isolated from the rhizosphere of strawberry, potato, or oilseed rape or bulk soil from fallow were screened by dual testing for in vitro antagonism towards Verticillium. The proportion of isolates with antagonistic activity was highest for strawberry rhizosphere (9.5%), followed by oilseed rape (6.3%), potato (3.7%), and soil (3.3%). The 331 Verticillium antagonists were identified by their fatty acid methyl ester profiles. They were characterized by testing their in vitro antagonism against other pathogenic fungi; their glucanolytic, chitinolytic, and proteolytic activities; and their BOX-PCR fingerprints. The abundance and composition of Verticillium antagonists was plant species dependent. A rather high proportion of antagonists from the strawberry rhizosphere was identified as Pseudomonas putida B (69%), while antagonists belonging to the Enterobacteriaceae (Serratia spp., Pantoea agglomerans) were mainly isolated from the rhizosphere of oilseed rape. For P. putida A and B plant-specific genotypes were observed, suggesting that these bacteria were specifically enriched in each rhizosphere.
The study of root-associated bacteria and their antagonistic potential is important not only for understanding their ecological role in the rhizosphere and the interaction with plants but also for any biotechnological application, e.g., biological control of soilborne plant pathogens. Verticillium wilt caused by the soilborne fungus Verticillium dahliae Kleb. is an important disease responsible for dramatic yield losses in many crops (46). Since microsclerotia of V. dahliae that develop in the senescing tissues of the dead plant may persist in soil for several years in the absence of a susceptible host, chemical control is nearly impossible (25). In the coming years, the phasing out of methyl bromide as a control measure for Verticillium wilt will have a great impact on the accumulation of microsclerotia in soil (46). The pathogen has a broad host range which includes many important crops, such as strawberry, potato, and oilseed rape. Efficacious control methods for Verticillium wilt are urgently needed for commercial crop production.
An environmentally friendly alternative to protect roots against fungal pathogens is rhizobacterium-mediated biological control (6, 47). Numerous studies have demonstrated the ability of several rhizobacteria to suppress diseases caused by fungal plant pathogens (12, 22, 48). One of the difficulties in developing rhizobacteria as a viable alternative is that many biological control agents are found to be too variable in their performance. According to Raaijmakers and Weller (35), variable expression of genes involved in disease suppression and poor root colonization are the major factors contributing to this inconsistency. Mechanisms of bacterial antagonism toward plant-pathogenic fungi include the competition for nutrients and space, the production of antibiotics, and the production of fungal cell wall-degrading enzymes (8, 13, 24). The production of antifungal metabolites is subject to complex regulation, allowing the bacteria to sense their own population density and to respond to different environmental factors (6, 9). Successful biological control requires not only a better understanding of the complex regulation of antifungal metabolite production by antagonists in response to environmental factors but also a better picture of what triggers root colonization and of the dynamics and composition of bacterial rhizosphere communities. Thus, little is known about plant specificity of antagonistic root-associated bacteria, which are an important functional group of beneficial bacteria in the rhizosphere (44). A few studies have indicated a plant-dependent composition of culturable bacteria (5, 15, 16, 21, 23, 28). Recently, denaturing gradient gel electrophoresis (DGGE) fingerprints of PCR-amplified 16S ribosomal DNA (rDNA) genes from community DNA were used to study dominant bacterial populations in the rhizosphere of the three V. dahliae Kleb. host plants—strawberry, potato, and oilseed rape—over two growing seasons (43). Using this cultivation-independent approach, a plant-dependent abundance of dominant bacterial populations could be shown for most of the sampling times. To examine the rhizosphere effect and the impact of the plant species on the abundance and diversity of Verticillium antagonists, in the same study fallow soil and rhizosphere samples of strawberry (Fragaria × ananassa [Duchense] Decaisne and Naudin [family, Rosaceae]), oilseed rape (Brassica napus L. [family, Brassicaceae]), and potato (Solanum tuberosum L. [family, Solanaceae]) were analyzed by a cultivation-dependent approach. Bacterial isolates obtained after plating on R2A and enrichment in microtiter plates containing high-molecular-weight substrates were screened by dual culture for antagonistic activity against V. dahliae. A comprehensive phenotypic and genotypic characterization of the antagonists provided new data on plant-dependent diversity of Verticillium antagonists.
This field study performed over two growth periods showed that different bacterial populations which are potentially antagonistic towards V. dahliae were enriched in the rhizosphere of different Verticillium host plants. The enormous phenotypic and genotypic diversity revealed at the subspecies level and an improved understanding of the plant-dependent bacterial diversity will contribute to the development of improved biological control strains.
MATERIALS AND METHODS
Experimental design.
Three different crop plants, potatoes (cultivar Element), oilseed rape (cultivar Express), and strawberries (cultivar Elsanta), were grown in a randomized block design with six replicates per crop plant and six unplanted plots. Field design and sampling were carried out according to the method of Smalla et al. (43).
Isolation of bacteria.
Roots with adhering soil from five plants from one plot were sampled into sterile petri dishes and then transported to the laboratory. Three grams of each sample was transferred into a sterile stomacher bag. To extract the rhizosphere microorganisms from the roots, 27 ml of demineralized water was added and samples were homogenized in a stomacher laboratory blender for 60 s (BagMixer; Interscience, St. Nom, France). This procedure was repeated three times for each sample. Samples were serially diluted with sterile 0.85% NaCl and plated onto R2A (Difco, Detroit, Mich.). Plates were incubated for 5 days at 20°C, and CFU were counted after 5 days to calculate the means of colonies (log10 CFU) based on fresh weight (FW). Per treatment (strawberry, potato, oilseed rape, and fallow) and sampling time, 48 colonies with different colony morphologies were picked from dilution plates with 20 to 100 colonies. To enrich bacteria with hydrolytic activities, microtiter plates with high-molecular-weight substrates were used (AZO-CM-cellulose and AZO-xylan [Megazym, Sydney, Australia], casein [Gibco, Paisley, United Kingdom], and chitin [Sigma, Deisenhofen, Germany]). The plates were filled with the following contents: 1.5 g of peptone of casein (Gibco); 0.5 g of peptone of soy (Gibco); 0.5 g of NaCl liter−1 (pH 7.3); and azurine-dyed, cross-linked (AZCL) substrates, chitin, and casein (0.5 g liter−1). The combined supernatants of six replicates per treatment were inoculated in serial dilution. The contents of wells of the last completely grown row were combined after 5 days' incubation, and 100 μl of this solution was plated after serial dilution on R2A. Between 15 and 20 colonies were isolated per high-molecular-weight substrate, purified, and stored at −70°C in broth containing 15% glycerol. Isolates obtained by plating were purified and stored at −70°C in broth containing 15% glycerol.
Screening of antagonistic bacteria.
Bacterial isolates were screened for their activity towards V. dahliae Kleb. by a dual-culture in vitro assay on Waksman agar containing 5 g of proteose peptone (Merck, Darmstadt, Germany), 10 g of glucose (Merck), 3 g of meat extract (Chemex, Munich, Germany), 5 g of NaCl (Merck), 20 g of agar (Difco), and distilled water (to 1 liter), pH 6.8. Zones of inhibition were measured after 5 days of incubation at 20°C according to the method of Berg (3). All strains were tested in three independent replicates with (i) V. dahliae Kleb. V16 (isolated from S. tuberosum L.), (ii) V. dahliae V25 (isolated from Brassica napus L.), (3), and V. dahliae V35 (isolated from Fragaria × ananassa [Duchense] Decaisne and Naudin). Only bacterial isolates which showed antagonistic activity towards V. dahliae were tested for their antagonism towards Rhizoctonia solani Kühn and Sclerotinia sclerotiorum Lib. (culture collection of the University of Rostock, Department of Microbiology) and Phytophthora cactorum (Lebert and Cohn) J. Schröt. PF8 (from the culture collection of the Federal Biological Research Center for Agriculture and Forestry, Darmstadt, Germany). These fungi were routinely grown on Sabouraud medium (Gibco) and stored at −70°C in broth containing 15% glycerol.
Identification of bacterial antagonists.
All antagonists were identified based on whole-cell cellular fatty acids; derivatized to methyl esters, i.e., fatty acid methyl esters (FAMEs); and analyzed by gas chromatography using the MIDI system (Microbial Identification System, Inc., Newark, N.J.). In addition, some strains were identified by 16S rDNA sequencing and aligned with the reference 16S rRNA gene sequence using the BLAST algorithm according to the method of Altschul et al. (1). Species richness, expressed as the number of species as a function (ratio) of the total number of individuals, was determined by the index proposed by Menhinick (27).
Screening for strains with endo-digesting hydrolytic activity.
Chitinase activity (β-1,4-glucosamine polymer degradation) was tested in chitin minimal medium, according to the method of Chernin et al. (7). Clearing zones were detected 5 days after incubation at 30°C. β-Glucanase activity was tested using chromogenic AZCL substrates (Megazym). Formation of blue haloes was recorded until 5 days after incubation. Protease activity (casein degradation) was determined from clearing zones in skim milk agar (50 ml of sterilized skim milk mixed at 55°C with 50 ml of one-fifth volume of tryptic soy agar and 4% agar) after 5 days of incubation at 30°C.
Production of secondary metabolites.
The ability of bacterial isolates to produce indole-3-acetic acid (IAA) was checked using the microplate method developed by Sawar and Kremer (41). The direct proof of cyanide production was made with an Aquaquant-14417-Testsystem (Merck) with culture broth (48 h) of the isolates. The ability of isolates to produce fluorescent siderophores was tested by plating bacteria on King's medium B (20) and incubating for 2 days at 25°C. Plates were inspected under 366-nm UV light.
BOX-PCR genomic fingerprints.
Bacterial DNA was prepared following the protocol of Andersen and McKay (2) modified for genomic DNA. BOX-PCR was done as described by Rademaker and De Bruijn (37) using the BOXA1R primer 5′-CTA CGG CAA GGC GAC GCT GAC G-3′. PCR amplification was performed with a Peltier thermal cycler (PTC-200; Biozym Diagnostic, Hessisch Oldendorf, Germany) using an initial denaturation step at 95°C for 6 min and subsequently 35 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min, and extension at 65°C for 8 min followed by final extension at 65°C for 16 min. A 10-μl aliquot of amplified PCR product was separated by gel electrophoresis on 1.5% agarose gels in 0.5× Tris-borate-EDTA buffer for 6 h, stained with ethidium bromide, and photographed under UV transillumination. The reproducibility of the results was verified in three independent experiments.
PCR detection of phlD and chiA genes.
Amplifications with gene-specific primer Phl2 (36) were performed in the following mix: 1 μl of target DNA, a 10 pM concentration of each primer, and 17 μl of PCR SuperMix High Fidelity (Gibco). PCR was performed under the following conditions: 3 min at 95°C followed by 29 cycles consisting of 1 min at 94°C, 45 s at 48°C, and 45 s at 72°C for 45 s. PCR was finished by a primer extension step at 72°C for 5 min. For chiA gene detection the following primers were aligned from Serratia marcescens (accession no. A25090) for amplification: ChiA1 (5′-ATG CGC AAA TTT AAT AACC-3′) and ChiA2 (3′-CCG ATT GAA CGCG-5′). PCR was performed in the same mix listed above using a 0.1 pM concentration of each primer. PCR was performed with 29 cycles consisting of 1 min at 95°C, 1 min at 55°C, and 55 s at 48°C subsequently followed by a 10-min final extension step at 72°C. PCR-amplified DNAs were detected by using a 1.5% agarose gel. The gels were stained with ethidium bromide for 30 min, and the PCR products were visualized with a UV transilluminator. The reproducibility of the results was verified in two independent experiments, and S. marcescens was used as a positive control.
Characterization of AHLs produced by rhizosphere isolates.
Production of N-acylhomoserine lactones (AHLs) by bacterial isolates was investigated with the aid of the bioluminescent sensor plasmid pSB403 (49). This sensor plasmid contains the Photobacterium fischeri luxR gene together with the luxI promoter region as a transcriptional fusion to the bioluminescence genes luxCDABE of Photorhabdus luminescens. The quorum-sensing system of P. fischeri relies on N-(3-oxo-hexanoyl)-homoserine lactone (3-oxo-C6-HSL), and the sensor plasmid consequently exhibits the highest sensitivity for this AHL molecule. However, several other AHL molecules are detected by the sensor, albeit with somewhat-reduced sensitivity (49). We also employed the green fluorescent protein-based biosensor Pseudomonas putida F117(pKR-C12) (45) for the detection of long-chain AHLs. Plasmid pKR-C12 (40) contains an AHL sensor cassette which is based on components of the las quorum-sensing system of Pseudomonas aeruginosa. Specifically, this cassette consists of a PlasB-gfp(ASV) translational fusion together with the lasR gene placed under control of Plac. Expectedly, as the cognate AHL of the las system is 3-oxo-C12-HSL (31), this system is most sensitive for 3-oxo-C12-HSL and other long-chain AHL molecules. In this case, production of AHLs was monitored by the expression of green fluorescence. This was accomplished by illuminating plates with blue light using an HQ 480/40 filter (F44-001; AHF-Analysentechnik, Tübingen, Germany) in combination with a halogen lamp (Intralux 5000-1; Volpi, Schlieren, Switzerland) as a light source. Illumination took place in a dark box equipped with the C2400-40 camera connected to a Pentax CCTV camera lens and an HQ 535/20 filter (F42-001; AHF-Analysentechnik).
Computer-assisted cluster analysis.
Computer-assisted evaluation of BOX-PCR-generated fingerprints was made using the GelCompare program (version 4.1; Applied Math, Kortrijk, Belgium). The cluster analysis was performed with Ward's algorithm and the unweighted pair-group method using arithmetic averages (UPGMA) algorithm. The physiological analysis data were converted to a binary code, and interisolate relationships were measured by the Euclidian metric algorithm and UPGMA in the program STATISTICA (StatSoft, Hamburg, Germany).
RESULTS
Isolation of bacteria from the rhizospheres and from soil.
At all sampling times the CFU counts were approximately 1 order of magnitude higher for rhizosphere samples than for soil from fallow plots. The CFU numbers determined for rhizosphere samples were rather similar for the different plants (strawberry and potato, log10 8.3 ± 2.5 (strawberry) or 3.1 (potato) CFU g−1 root FW; oilseed rape, log10 8.1 ± 1.7 CFU g−1 root FW) and showed no significant seasonal changes. A total of approximately 120 isolates per treatment (strawberry, potato, oilseed rape, and bulk soil) and per sampling time was used in the initial screening of antagonistic activity towards V. dahliae Kleb.
Screening for isolates antagonistic to V. dahliae.
A total of 5,854 bacterial isolates were screened for their ability to suppress V. dahliae in an in vitro dual-culture assay. Initially 334 isolates were found which were active against V. dahliae, of which 67 were strongly active, with inhibition zones larger than 10 mm. Although similar numbers of isolates from each of the treatments were tested, the proportion of isolates with antagonistic activity was different. The proportion of isolates with antifungal activity was highest for strawberry rhizosphere (9.5%), followed by oilseed rape rhizosphere (6.3%), potato rhizosphere (3.7%), and fallow soil (3.3%). The proportion of Verticillium antagonists isolated from R2A plates after previous enrichment in high-molecular-weight substrate plates (7 to 8%) was significantly higher than that for isolates obtained after direct plating on R2A, of which only 3% showed antagonistic activity.
Diversity of Verticillium antagonists.
The majority of the in vitro antagonists (n = 286) were identified by fatty acid analysis (Table 1). Based on their fatty acid profiles, 46 different bacterial species were identified. The richness of antagonistic species was plant species dependent. The highest number of different species with antagonistic activity was isolated from oilseed rape rhizosphere (n = 30), while only 18 or 12 different species were found in the rhizosphere of strawberry and potato, respectively. Interestingly, antagonists isolated after previous enrichment in high-molecular-weight substrates belonged mainly to the fast-growing γ-subdivision of the proteobacteria. The species composition did not depend on the kind of high-molecular-weight substrate. The diversity of antagonists obtained after direct plating onto R2A was higher, and gram-positive bacteria such as Bacillus spp. and Streptomyces spp. were only obtained by the direct plating approach. Gram-positive antagonistic isolates accounted only for a rather-small proportion of the Verticillium antagonists (11 of 286), and with one exception these isolates originated from bulk soil.
TABLE 1.
List of bacterial species with antagonistic properties isolated from the rhizosphere of strawberry, potato, and oilseed rape, and uncultivated soil in the vegetation period 1998 to 1999
| Bacterial speciesa | No. of isolatesb obtained from:
|
|||
|---|---|---|---|---|
| Rhizosphere
|
Soil | |||
| Strawberry | Potato | Oilseed rape | ||
| Acidovorax avenea | 1 | |||
| Acinetobacter baumannii | 1 | |||
| Acinetobacter calcoaceticus | 1 (1) | |||
| Agrobacterium rhizogenes | 1 | |||
| Agrobacterium tumefaciens | 1 | |||
| Bacillus circulans | 1 | |||
| Bacillus laterosporus | 1 | |||
| Bacillus megaterium | 1 (1) | |||
| Bacillus mycoides | 1 | |||
| Bacillus pumilus | 1 | |||
| Burkholderia cepacia | 1 | 1 | ||
| Brevibacterium acetylicum | 2 (2) | |||
| Chryseobacterium balustinum | 1 | |||
| Chryseobacterium indologenes | 1 | |||
| Comamonas acidovorans | 2 (2) | 1 (1) | ||
| Cytophaga johnsonae | 1 (1) | |||
| Enterobacter agglomerans | 1 | |||
| Enterobacter intermedius | 3 | |||
| Janthinobacterium lividum | 1 | |||
| Kluyvera cryorescens | 1 (1) | |||
| Kocuria kristinae | 1 | |||
| Pantoea agglomerans | 3 | 1 | ||
| Pasteurella anatipestifer | 1 (1) | |||
| Proteus vulgaris | 1 | 2 | ||
| Pseudomonas chlororaphis | 4 (1) | 8 | ||
| Pseudomonas corrugata | 3 | 3 | 1 | 1 |
| Pseudomonas fluorescens | 7 (3) | 2 | 9 (4) | 4 |
| Pseudomonas marginalis | 1 | 1 | 3 | 2 |
| Pseudomonas putida A | 6 | 6 | 13 | 10 |
| Pseudomonas putida B | 86 (2) | 24 (1) | 9 (1) | |
| Pseudomonas syringae | 7 | 3 (2) | 1 | |
| Pseudomonas tolaasii | 1 (1) | 3 (1) | ||
| Pseudomonas viridiflava | 1 | |||
| Salmonella enterica Typhimurium | 2 (2) | 1 (1) | ||
| Serratia fonticola | 1 | |||
| Serratia grimesii | 2 | 3 | 1 | |
| Serratia odorifera | 1 | |||
| Serratia plymuthica | 3 | |||
| Serratia proteamaculans | 2 | 1 | ||
| Stenotrophomonas maltophilia | 1 | 2 | 2 | |
| Streptomyces albidoflavus | 2 (1) | |||
| Streptomyces hygroscopicus | 1 (1) | |||
| Streptoverticillium reticulum | 2 | |||
| Weeksella zoohelcum | 2 (1) | |||
| Xenorhabdus nematophilia | 1 (1) | 1 (1) | ||
| Xenorhabdus luminescens | 1 (1) | |||
| Total no. of species | 18 | 12 | 30 | 15 |
| Total no. of isolates | 125 | 49 | 81 | 31 |
| Richness (d) | 1.61 | 1.71 | 3.33 | 2.69 |
Identification by FAME analysis of isolates identified with a similarity index of <0.5.
Numbers in parentheses indicate how many strains were isolated.
Only four species were obtained from the rhizospheres of all three plants and from soil—Pseudomonas corrugata, Pseudomonas fluorescens, Pseudomonas marginalis, and P. putida A—while P. putida B, Pseudomonas syringae, and Stenotrophomonas maltophilia were isolated from the rhizosphere of all three plants but not from soil. The highest number by far of antagonistic isolates from the rhizosphere and from soil belonged to the P. fluorescens intrageneric cluster (strawberry, 111 of 125 antagonists; potato, 43 of 49 antagonists; oilseed rape, 48 of 81 antagonists; soil, 17 of 31 antagonists), with a high proportion of isolates from the P. putida lineage. The proportion of antagonists belonging to this P. fluorescens cluster was particularly high for strawberry and potato plants (approximately 90%), while their proportion in the rhizosphere of oilseed rape and soil accounted only for 59 and 55%, respectively. The frequent isolation of P. putida B from the strawberry rhizosphere (86 of 125 antagonists) was striking, leading to a rather low evenness of antagonistic isolates from strawberry. The diversity indices calculated for species richness were 3.3 for oilseed rape, 2.7 for soil, 1.7 for potato, and 1.6 for strawberry. The highest number of antagonists belonging to different species was observed for antagonists from the rhizosphere of oilseed rape. Different species of enterobacterial genera (Serratia, Enterobacter, Pantoea, and Weeksella) with antagonistic activity were isolated only from the rhizosphere of oilseed rape. Thus, enterobacterial species seem to be in particular enriched in the rhizosphere of oilseed rape. However, a large proportion of species (21 of 66 antagonists) was isolated from one plant species, often being isolated only once.
Antifungal activity and production of antifungal metabolites of Verticillium antagonists.
All 286 Verticillium antagonists were tested in vitro for their activity against the plant pathogens R. solani (basidiomycete with a chitin-glucan-containing cell wall), S. sclerotiorum (ascomycete with a chitin-glucan-containing cell wall), and P. cactorum (oomycete with a cellulose-containing cell wall) and for the production of hydrolytic enzymes. Generally, the fungi grew as well as the bacterial isolates on Waksman agar. Inhibition was clearly discerned by limited growth or the complete absence of fungal mycelium in the inhibition zone surrounding a bacterial colony. Verticillium antagonists assigned to the same species often showed different patterns of antagonistic activity. While the majority of P. putida B isolates (mainly isolates from the strawberry rhizosphere) showed activity only against V. dahliae, a few P. putida B isolates also antagonized R. solani, S. sclerotiorum, and/or P. cactorum. Verticillium antagonists which showed a broad range activity and also suppressed R. solani, S. sclerotiorum, and P. cactorum originated from the rhizosphere of oilseed rape (Serratia spp. [n = 7]; P. fluorescens [n = 1]) and bulk soil (Bacillus circulans [n = 1]; P. marginalis [n = 2]). Altogether, more isolates with antagonistic activity against R. solani than against S. sclerotiorum and P. cactorum were found.
Strawberry.
A large proportion of the antagonists isolated from the rhizosphere of strawberry showed antifungal activity only against V. dahliae (81 of 125 antagonists; 65%), most of them identified as P. putida B. About 30% of isolates also suppressed R. solani (37 of 125 antagonists), while activity against S. sclerotiorum and P. cactorum was found only for 9 of 125 antagonists (7%) and 8 of 125 antagonists (6.4%), respectively. For most of the Verticillium antagonists (121 of 125) proteolytic activity was detected, while only three of the isolates had chitinolytic activity and none showed glucanolytic activity. Isolates with chitinolytic activity were identified by FAME analysis as Serratia (n = 2) and P. fluorescens (n = 1).
Potato.
Approximately 60% of the Verticillium antagonists isolated from the potato rhizosphere also antagonized R. solani, 20% had antagonistic activity towards S. sclerotiorum, and 18% had antagonistic activity towards P. cactorum. Fifteen isolates were suppressive towards three of the pathogens tested. Similarly to the strawberry isolates almost all isolates had proteolytic activity, while only two isolates (S. maltophilia and Serratia proteamaculans) showed chitinolytic activity.
Oilseed rape.
Antagonistic activity against R. solani was observed for 53% of the Verticillium antagonists, while approximately 47% antagonized S. sclerotiorum and 23% were active against P. cactorum. Isolates which showed activity against R. solani and S. sclerotiorum most often belonged to the Enterobacteriaceae. Seven Serratia isolates were active against all pathogens tested here. Almost all Verticillium antagonists from oilseed rape showed clearing zones on skim milk agar plates, suggesting proteolytic activity. The proportion of antagonists with chitinolytic activity (19 of 81 antagonists; 23%) was higher than that for the strains from the rhizosphere of strawberry and potato plants. All strains with chitinolytic activity belonged to the Enterobacteriaceae. β-1,3-Glucanolytic activity was observed for five isolates belonging to taxonomically different groups.
Fallow soil.
In contrast to the rhizosphere isolates, a higher proportion of Verticillium antagonists also suppressed R. solani (24 of 31; 77%) and S. sclerotiorum (19 of 31; 61%). Isolates displaying antagonistic activity towards these pathogens were most often identified as P. putida A. Two P. marginalis strains and one B. circulans strain were able to antagonize all pathogens tested here.
Characterization of Verticillium antagonists belonging to the Enterobacteriaceae.
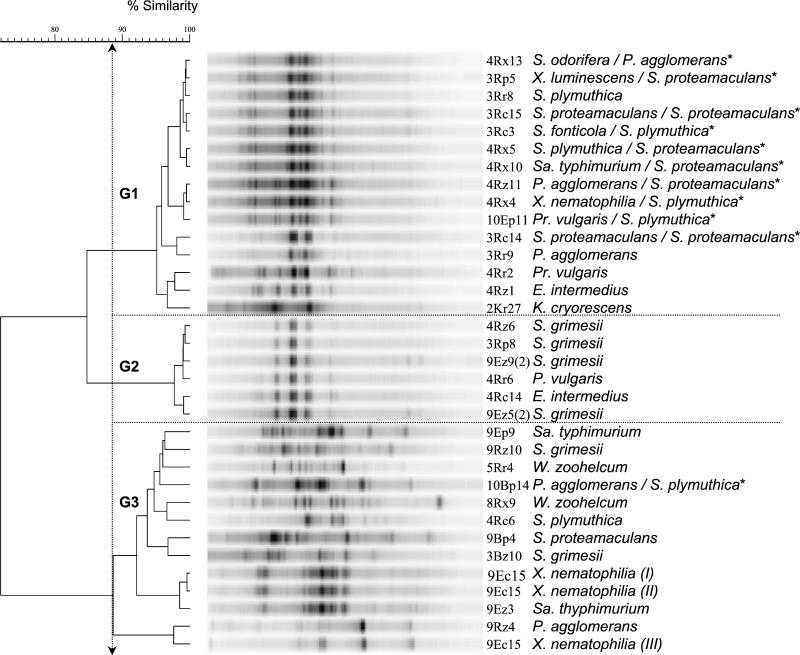
Verticillium antagonists assigned by FAME analysis to 13 different species belonging to the Enterobacteriaceae were mainly isolated from the rhizosphere of oilseed rape. BOX-PCR performed with genomic DNA yielded fingerprints with 12 to 35 amplification products, ranging from 100 to 3,000 bp. Isolates identified by FAME analysis as different species often displayed very similar BOX patterns (Fig. 1). This observation was confirmed when GelCompare was used for comparison of BOX patterns. Intraspecies diversity of BOX patterns analyzed in three independent replicates of isolate 9Ec15 (replicates I to III) was shown to be 89% similarity. Analysis of BOX patterns with more than 89% similarity resulted in three different cluster or genotype groups. Group 1 contained isolates identified with rather high similarity (of >0.8) as Enterobacter, Salmonella, Serratia, Proteus, and Xenorhabdus. The BOX patterns of the isolates which belong to this group are very similar or nearly identical (e.g., Serratia odorifera 4Rx13, Xenorhabdus luminescens 3Rp5, Serratia plymuthica 3Rr8). Cluster group 2 included isolates from Serratia and Proteus with highly homogeneous BOX patterns. Group 3 showed a more heterogeneous pattern than the other groups and contained 11 isolates belonging to four genera or seven species. To clarify these ambiguous results of the FAME analysis, partial 16S rDNA sequencing was done for 12 of the enteric isolates, each representing a BOX cluster. 16S rDNA sequencing revealed that 7 of the 12 isolates were most similar to S. proteamaculans, 4 were most similar to S. plymuthica, and 1 was most similar to Pantoea agglomerans (Table 2). Thus, the number of different species identified by FAME analysis, and consequently the diversity index calculated for richness of oilseed rape (Table 1), is an overestimate, and the corrected richness index would be 2.2. Although Serratia isolates which were active against all pathogens tested belonged to different BOX clusters, they all showed in vitro lytic activity. The majority of isolates were able to degrade chitin in plate assays. Additionally, when the molecular approach was used, the chiA gene was found in most of the chitinolytic strains. With the exception of four strains, enterics were proteolytic, while only three strains showed glucanolytic activity. Production of short-chain AHL signal molecules was detected for 15 out of 32 strains tested in cross-streaks against the sensor strain Escherichia coli MT102(pSB403). We also investigated the synthesis of long-chain AHLs with the aid of the biosensor P. putida F117(pKR-C12). However, only one strain (9Ep9) of all the Enterobacteriaceae tested gave a positive result, indicating that synthesis of long-chain AHL molecules is rare among members of this family. No obvious correlation between the production of AHL signal molecules and chitinolytic activity, proteolytic activity, or antagonism against fungi was observed.
FIG. 1.
Dendrogram showing the relationship of 32 Enterobacteriaceae isolates identified by FAME analysis from strawberry, potato, and oilseed rape rhizospheres and uncultivated soil based on BOX-PCR fingerprints using cluster analysis. An asterisk indicates a second identification by 16S rDNA sequencing. K., Kluyvera; P., Pantoea; S., Serratia; Pr., Proteus; Sa., Salmonella; W., Weeksella; X., Xenorhabdus.
TABLE 2.
Taxonomic classification and characterization of bacterial isolates with antagonistic properties belonging to Enterobacteriaceae
| Genotype groupa | Strain | FAME analysis
|
16S rDNA sequencing
|
Activity againstb:
|
Lytic enzyme productionc
|
AHL production
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identification | ID | Identification | % Similarity (strain) | V. dahliae | R. solani | S. sclerotiorum | P. cactorum | Glucanase | Chitinase | chiA product | Protease | MT 102d | FI17e | ||
| G1 | 4Rx13 | Serratia odorifera | 0.735 | Pantoea agglomerans | 99 (100-860) | +++ | ++ | +++ | − | − | − | − | + | + | − |
| Pantoea agglomerans | 0.701 | ||||||||||||||
| 3Rp5 | Xenorhabdus luminescens | 0.014 | S. proteamaculans | 99 (DSM4543) | +++ | + | ++++ | + | − | + | + | + | − | − | |
| 3Rr8 | Serratia plymuthica | 0.890 | ++ | +++ | + | + | − | + | + | + | − | − | |||
| 3Rc15 | Serratia proteamaculans | 0.839 | S. proteamaculans | 99 (DSM4543) | +++ | ++ | ++++ | − | − | + | + | + | − | − | |
| 3Rc3 | Serratia fonticola | 0.640 | S. plymuthica | 99 (DSM4540) | +++ | ++ | ++++ | + | − | + | + | + | − | − | |
| 4Rx5 | Serratia plymuthica | 0.715 | S. proteamaculans | 99 (DSM4543) | +++ | + | + | − | − | + | + | + | + | − | |
| 4Rx10 | Salmonella enterica serovar Typhimurium | 0.405 | S. proteamaculans | 99 (DSM4543) | ++ | +++ | + | − | − | + | + | + | − | ||
| 4Rz11 | Pantoea agglomerans | 0.723 | S. proteamaculans | 99 (DSM4543) | ++ | +++ | + | + | − | + | + | + | + | − | |
| 4Rx4 | Xenorhabdus nematophilia | 0.146 | S. plymuthica | 99 (DSM4540) | +++ | + | + | + | − | + | − | + | + | − | |
| 10Ep11 | Proteus vulgaris | 0.834 | S. plymuthica | 98 (124-822) | ++ | +++ | ++ | − | − | − | − | + | + | − | |
| 3Rc14 | Pantoea agglomerans | 0.871 | |||||||||||||
| Klebsiella pneumoniae | 0.829 | +++ | ++ | ++++ | − | + | + | + | − | − | − | ||||
| 3Rr9 | Serratia proteamaculans | 0.793 | S. proteamaculans | 99 (DSM4543) | ++ | ++ | ++++ | + | − | + | + | + | − | − | |
| 4Rr2 | Proteus vulgaris | 0.758 | + | +++ | ++++ | + | − | + | + | + | + | − | |||
| Serratia grimesii | 0.706 | ||||||||||||||
| 4Rz1 | Enterobacter intermedius | 0.591 | ++ | +++ | + | − | + | + | + | + | − | ||||
| 2Kr27 | Kluyvera cryorescens | 0.383 | S. proteamaculans | 99 (DSM4543) | + | − | − | − | − | + | + | + | − | − | |
| G2 | 4Rz6 | Serratia grimesii | 0.718 | ++ | +++ | + | − | − | + | + | + | + | − | ||
| Pantoea agglomerans | 0.714 | ||||||||||||||
| 3Rp8 | Serratia grimesii | 0.614 | +++ | +++ | ++++ | − | − | + | − | + | + | − | |||
| Pantoea agglomerans | 0.589 | ||||||||||||||
| 9Ez29 | Serratia grimesii | 0.768 | ++ | ++++ | + | − | − | + | + | + | + | − | |||
| Pantoea agglomerans | 0.765 | ||||||||||||||
| 4Rr6 | Proteus vulgaris | 0.721 | ++ | +++ | + | + | − | + | + | + | + | − | |||
| Pantoea agglomerans | 0.677 | ||||||||||||||
| 4Rc14 | Enterobacter intermedius | 0.736 | |||||||||||||
| 0.675 | + | +++ | + | − | − | + | + | + | + | − | |||||
| Serratia grimesii | |||||||||||||||
| 9Ez25 | Serratia grimesii | 0.777 | ++ | − | + | − | − | + | + | + | + | − | |||
| G3 | 9Ep9 | Salmonella enterica serovar Typhimurium | 0.496 | + | − | − | − | − | − | − | + | + | + | ||
| 9Rz10 | Serratia grimesii | 0.928 | ++++ | ++++ | + | − | − | − | − | − | − | − | |||
| 5Rr4 | Weeksella zoohelcum | 0.560 | ++ | +++ | +++ | − | + | − | − | − | − | ||||
| 10Bp14 | Pantoea agglomerans | 0.884 | S. plymuthica | 99 (DSM4540) | + | + | + | − | − | − | + | − | − | − | |
| 8Rx9 | Weeksella zoohelcum | 0.478 | +++ | − | − | − | + | − | − | + | − | − | |||
| 4Rc6 | Serratia plymuthica | 0.790 | ++ | ++ | ++++ | − | + | + | + | + | − | − | |||
| 9Bp4 | Serratia proteamaculans | 0.740 | + | − | − | − | − | − | + | + | − | − | |||
| 3Bz10 | Serratia grimesii | 0.928 | + | − | − | − | − | − | + | + | − | − | |||
| 9Ec15 | Xenorhabdus nematophilia | 0.096 | ++ | + | − | − | − | − | − | + | − | − | |||
| 9Ez3 | Salmonella enterica serovar Typhimurium | 0.398 | ++ | − | − | − | − | − | − | + | − | − | |||
| 9Rz4 | Pantoea agglomerans | 0.89 | ++++ | +++ | ++++ | − | − | − | − | + | − | − | |||
| Serratia grimesii | 0.872 | ++++ | +++ | ++++ | − | − | − | − | + | − | − | ||||
Grouping at 89% similarity.
Antagonism toward these four species was determined by dual-culture assay. Results indicate the width of the zone of inhibition as follows: +, 0 to 5 mm; ++, 5 to 10 mm; +++, 10 to 15 mm; ++++, >15 mm.
β-1,3-Glucanase, protease, and chitinase activities were demonstrated by plate assay (+, hydrolysis; −, no hydrolysis). The chiA gene was detected by a PCR approach.
E. coli MT 102(pSB403) for detection of short-chain AHLs.
P. putida F117(pKR-C12) for detection of long chain AHLs.
Characterization of P. putida A strains.
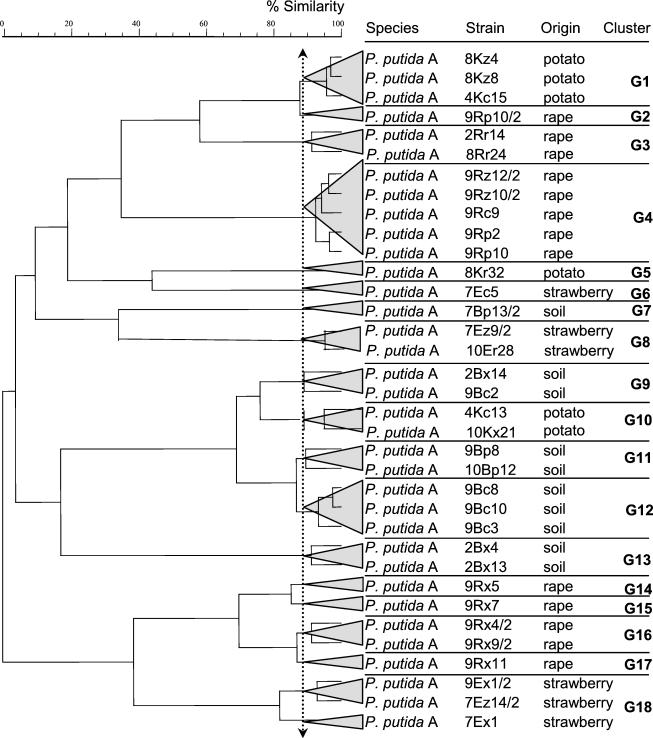
Approximately 12.2% of Verticillium antagonists were identified by FAME analysis as P. putida A. P. putida A isolates which antagonized V. dahliae originated from soil (n = 10), oilseed rape (n = 13), strawberry (n = 6), and potato (n = 6), indicating the absence of a plant-specific enrichment. P. putida A strains were mainly isolated in the second year (31 of 35 antagonists) and were obtained after enrichment or after direct plating onto R2A. Therefore, we analyzed for this subset of Verticillium antagonists a range of phenotypic and genotypic traits to find out whether a plant-specific enrichment of particular phenotypes and genotypes was detectable at a subspecies level (Table 3). The phenotypic characterization comprised the analysis of antifungal activity; the production of lytic enzymes (glucanases, chitinases, and proteases); fluorescence on King's B medium under UV light; and the production of cyanide, IAA, and AHL. BOX-PCR fingerprints were used to analyze the genomic relatedness between P. putida A isolates (Fig. 2). Furthermore, PCR was used to detect the presence of the phlD gene. While almost all P. putida A isolates showed in vitro proteolytic activity (33 of 35 antagonists; 94%), glucanolytic or chitinolytic activity was detected for none of them. Twenty P. putida A isolates were able to produce cyanide (HCN), which acts as an inducer of plant resistance. The production of the plant growth hormone IAA was detected for 71% of the P. putida A isolates. AHLs were detected for approximately 71% of the P. putida A isolates when the sensor strain E. coli MT102(pSB403) was used. With two exceptions a correlation of IAA and AHL production was detected. P. putida isolates which did not produce AHL and IAA fell into distinct genotype clusters (clusters G4, G5, G6, and G18). By employing the biosensor P. putida F117(pKR-C12), production of long-chain AHL molecules could be demonstrated for isolates of the clusters G14, G15, G16, and G17. Interestingly, all these strains were isolated from oilseed rape and grouped into one phenotype cluster (cluster 5). Only one soil isolate (7Bp13/2) was found to produce long-chain AHLs. This strain also belongs to phenotype cluster 5. The phlD gene was detected by PCR in isolates belonging to different genomic clusters originating from soil (4), oilseed rape (2), and strawberry (4).
TABLE 3.
Phenotypic characterization of P. putida A isolates belonging to the genotypic clusters defined in Fig. 2
| Genotype groupa | Strain | Origin | Activity againstb:
|
Lytic enzyme productionc
|
Secondary metabolite production
|
AHL production
|
Phenotype group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V. dahliae | R. solani | S. sclerotiorum | P. cactorum | Glucanase | Chitinase | Protease | King Bd | DAPGe | HCNf | IAAg | MT 102h | F117i | |||||
| G1 | 8Kz4 | Potato | Cellulose | ++ | ++ | ++ | − | − | − | + | − | − | + | + | + | − | 2 |
| 8Kz8 | Potato | Cellulose | ++ | ++ | + | − | − | − | + | − | − | + | + | + | − | 2 | |
| 4Kc15 | Potato | Chitin | + | +++ | + | − | − | − | + | − | − | + | + | + | − | 1 | |
| G2 | 9Rp10/2 | Rape | Casein | + | +++ | + | − | − | − | + | − | − | − | + | + | − | 5 |
| G3 | 2Rr14 | Rape | R2A | + | +++ | + | − | − | − | + | − | − | − | + | + | − | 5 |
| 8Rr24 | Rape | R2A | +++ | ++ | + | − | − | − | − | − | − | + | + | + | − | 3 | |
| G4 | 9Rz12/2 | Rape | Cellulose | + | − | − | − | − | − | + | + | − | − | − | − | − | 5 |
| 9Rz10/2 | Rape | Cellulose | + | − | − | − | − | − | + | + | − | − | − | − | − | 5 | |
| 9Rc9 | Rape | Chitin | + | +++ | +++ | − | − | − | + | + | + | − | − | − | − | 7 | |
| 9Rp2 | Rape | Casein | + | − | − | + | − | − | + | + | − | − | − | − | − | 7 | |
| 9Rp10 | Rape | Casein | + | − | − | + | − | − | + | + | − | − | − | − | − | 5 | |
| G5 | 8Kr32 | Potato | R2A | + | ++ | − | − | − | − | + | + | − | − | − | − | ND | 7 |
| G6 | 7Ec5 | Strawberry | Chitin | ++ | + | − | − | − | − | + | − | + | + | − | − | ND | 6 |
| G7 | 7Bp13/2 | Soil | Casein | ++ | + | − | + | − | − | + | + | + | − | − | + | + | 5 |
| G8 | 7Ez9/2 | Strawberry | Cellulose | ++ | − | − | − | − | − | + | − | − | − | ||||
| 10Er28 | Strawberry | R2A | ++ | ++ | + | − | − | − | + | − | − | + | + | + | − | 1 | |
| G9 | 2Bx14 | Soil | Xylan | + | +++ | + | − | − | − | + | − | − | + | + | + | − | 1 |
| 9Bc2 | Soil | Chitin | ++ | +++ | ++ | − | − | − | − | − | − | + | + | + | − | 3 | |
| G10 | 4Kc13 | Potato | Chitin | + | +++ | ++ | − | − | − | + | − | − | + | + | + | − | 1 |
| 10Kx10/2 | Potato | Xylan | + | +++ | + | − | − | − | + | − | − | + | + | + | − | 1 | |
| G11 | 9Bp8 | Soil | Casein | + | +++ | + | − | − | − | + | − | − | + | + | + | − | 2 |
| 10Bp12 | Soil | Casein | ++ | +++ | + | − | − | − | + | − | − | + | + | + | − | 1 | |
| G12 | 9Bc8 | Soil | Chitin | ++ | +++ | ++ | − | − | − | + | − | + | + | + | + | − | 1 |
| 9Bc10 | Soil | Chitin | + | ++ | + | − | − | − | + | − | + | + | + | + | + | 1 | |
| 9Bc3 | Soil | Chitin | ++ | +++ | + | − | − | − | + | − | − | + | + | + | − | 1 | |
| G13 | 2Bx4 | Soil | Xylan | + | +++ | + | − | − | − | + | − | + | + | + | + | − | 1 |
| 2 Bx13 | Soil | Xylan | + | +++ | + | − | − | − | + | − | − | + | + | + | − | 1 | |
| G14 | 9Rx5 | Rape | Xylan | + | − | − | + | − | − | + | + | + | − | + | + | + | 5 |
| G15 | 9Rx7 | Rape | Xylan | + | − | − | − | − | − | + | + | + | − | + | + | + | 5 |
| G16 | 9Rx24 | Rape | Xylan | + | − | − | + | − | − | + | + | − | − | + | + | + | 5 |
| 9Rx29 | Rape | Xylan | ++ | − | +++ | + | − | − | + | + | − | − | + | + | + | 5 | |
| G17 | 9Rx11 | Rape | Xylan | ++ | − | − | − | − | − | + | + | − | − | + | + | + | 5 |
| G18 | 9Ex1/2 | Strawberry | Xylan | + | − | − | − | − | − | + | + | + | + | + | − | ND | 4 |
| 7Ez14/2 | Strawberry | Cellulose | +++ | − | − | − | − | − | + | − | + | + | − | − | − | 6 | |
| 7Ex1 | Strawberry | Xylan | +++ | − | − | − | − | − | + | − | + | + | − | − | ND | 6 | |
Grouping at 89% similarity
Antagonism toward these four species was determined by dual-culture assay. Results indicate the width of the zone of inhibition as follows: +, 0 to 5 mm; ++, 5 to 10 mm; +++, 10 to 15 mm; ++++, >15 mm.
β-1,3-Glucanase, protease, and chitinase activities were determined by plate assay (+, hydrolysis; −, no hydrolysis).
Fluorescence on King's B agar.
DAPG, 2,4-diacetylphloroglucinol; PCR approach.
Production of cyanide as shown by the Merck Schnelltest: (+, > 0.001 mg liter−1).
Microplate method of Sawar and Kremer (41) (+, > 0.1 μg ml−1).
E. coli MT 102(pSB403) for detection of short-chain AHLs.
P. putida F117(pKR-C12) for detection of long-chain AHLs.
Grouping at 75% similarity.
FIG. 2.
Dendrogram showing the relationship of 35 P. putida A isolates from strawberry, potato, and oilseed rape rhizospheres and uncultivated soil based on BOX-PCR fingerprints and using cluster analysis.
Characterization of Verticillium antagonists identified as P. putida B.
More than one-third of all Verticillium antagonists isolated were identified by FAME analysis as P. putida B. P. putida B strains antagonistic towards V. dahliae were isolated from the rhizosphere of strawberries at all sampling times. Thus, P. putida B strains clearly resembled the most-abundant group of Verticillium antagonists which were isolated from all plant rhizospheres. To explore the diversity of P. putida B isolates, a total of 98 isolates from the rhizospheres of strawberry, oilseed rape, and potato was characterized by BOX-PCR fingerprints. BOX-PCR fingerprints revealed an enormous diversity of P. putida B at the subspecies level (data not shown). Analysis of the BOX fingerprints by UPGMA using GelCompare resulted in a cutoff level of 85% in 38 groups, of which 19 groups contained only one isolate. While 14 of the 19 groups with more than one isolate consisted only of isolates from one plant species, five groups had isolates from potato and strawberry rhizosphere (n = 3) or strawberry and oilseed rape rhizosphere (n = 2). Eight groups contained isolates which were isolated from only one substrate. Six of the 19 groups with more than one isolate contained strains isolated at different sampling points in both years, six groups consisted of isolates from different sampling times but all isolated in the same year, and seven groups contained only isolates from the same sampling time. In a subset of P. putida B (44) isolates analyzed for AHL production, 11 isolates were found to be positive. Interestingly, only 3 of 27 tested strains from strawberry rhizosphere produced AHLs. These three strains activated both AHL sensor strains used in this study. In contrast, three out of seven oilseed rape rhizosphere strains and 5 out of 10 potato rhizosphere strains tested were AHL positive in cross-streaks against E. coli MT102(pSB403), and most of these strains (two of the three oilseed rape rhizosphere strains and four of the five potato rhizosphere strains) also activated the long-chain AHL sensor P. putida F117(pKR-C12). The phlD gene was detected by PCR in a considerable proportion of the isolates from the strawberry (64 of 86 antagonists) and the potato (15 of 24 antagonists) rhizospheres, while no PCR product was obtained from the nine P. putida B isolates from the oilseed rape rhizosphere.
DISCUSSION
Root exudates such as amino acids, sugars, and organic acids are an important nutritional source for bacteria colonizing the roots. The composition of root exudates was shown to vary depending on the plant species and the stage of plant development (17). Thus, the plant is supposed to profoundly influence the relative abundance of indigenous rhizobacteria as well as the population dynamics of introduced biological control strains. Recently Rainey (39) showed that P. fluorescens genes involved in nutrient acquisition, stress response, or secretion had elevated levels of expression during rhizosphere colonization. To come to an improved understanding of factors affecting the ability of bacteria to colonize the rhizosphere, the plant should be taken into account. To explore the rhizosphere effect of different plant species on abundance and diversity of antagonistic bacteria, isolates originating from the rhizosphere of field-grown host plants of V. dahliae—strawberry, potato, and oilseed rape—and from fallow soil were analyzed for their antagonistic properties. Verticillium antagonists selected by dual-culture tests were identified by FAME analysis and characterized for their phenotypic and genotypic properties. To enrich bacteria with hydrolytic enzyme activities, the bacterial cells recovered from rhizosphere or bulk soil were incubated in microtiter plates with high-molecular-weight substrates. The previous incubation in high-molecular-weight-substrate microtiter plates resulted in an enrichment of fast-growing γ-proteobacteria. Thus, gram-positive bacteria were only isolated after direct plating onto R2A. DGGE analysis of DNA extracted from the cells recovered from the highest dilution row, which completely scored positive for growth, showed similar profiles for all substrates (data not shown). Obviously, a specific substrate-dependent enrichment of bacteria did not occur due to the presence of more easily degradable substrate (1/10 volume of tryptic soy agar). A reduction of the number of dominant DGGE bands was observed during incubation. Similar observations were made when rhizosphere communities were incubated in BIOLOG plates (42). Since Verticillium antagonists were isolated from the highest dilutions, they represent a considerable proportion of the culturable bacterial fraction. While no differences in the bacterial plate counts (CFU on R2A) were found between the different rhizospheres, the abundance, taxonomic composition, and diversity of Verticillium antagonists differed for the different treatments. Although we isolated the highest number of Verticillium antagonists from the rhizosphere of strawberry, their diversity in terms of richness was surprisingly low. A lower number of dominant bands was recently also found for the DGGE patterns of eubacterial populations from the strawberry rhizosphere compared to the more complex DGGE patterns of oilseed rape and potato rhizospheres (43). However, while the cultivation-independent approach indicated that the patterns were more similar between potato and oilseed rape compared to strawberry, this finding could not be confirmed for isolates with antagonistic activity towards V. dahliae. The proportion and taxonomic composition of the isolates were found to be specific for each of the plant species and soil. The most-remarkable findings were the high proportion of P. putida B isolates from the rhizosphere of strawberry and a high number of Verticillium antagonists belonging to the Enterobacteriaceae from the rhizosphere of oilseed rape. The widespread occurrence of Serratia species with in vitro antagonistic activity towards V. dahliae in the rhizosphere of oilseed rape was already reported by Kalbe et al. (18). The majority of Verticillium antagonists from all treatments belonged to the P. fluorescens intrageneric cluster (29), with a substantially lower number of Pseudomonas isolates from soil. Strains belonging to the genus Pseudomonas are the biological control agents which are best characterized at the molecular level (6, 30). Although P. putida A strains were isolated from all rhizospheres and soil, a clustering dependent on the origin of the isolates was observed when BOX-PCR profiles were compared. P. putida B isolates, which represented 42% of the collection of Verticillium antagonists, were exclusively isolated from the rhizosphere and not from bulk soil. Obviously, P. putida B isolates are enriched from soil by root exudates, in particular those from strawberry plants. Again BOX-PCR fingerprints revealed a great diversity, and several of the genomic clusters contained only isolates isolated from one plant species. Several recently published studies used repetitive extragenic palindromic (REP)-PCR fingerprints (38), such as those obtained by BOX-, REP-, or enterobacterial repetitive intergenic consensus (ERIC)-PCR, to explore the diversity of pseudomonads originating from rhizospheres and soils. Based on REP-PCR fingerprints all studies found an enormous genomic diversity of Pseudomonas spp. at the subspecies level (14, 26). Fromin et al. (14) reported that the genotypic structure of Pseudomonas brassicacearum populations analyzed by REP-PCR fingerprints are significantly influenced by the Arabidopsis thaliana genotype.
The proportion of Verticillium antagonists which were also suppressive to other pathogens tested here to some extent reflected the species composition of the collections obtained from each treatment. The proportion of Verticillium antagonists which were active also against R. solani, S. sclerotiorum, and P. cactorum was particularly high for isolates from oilseed rape rhizosphere and soil. Although significant differences were found in the production of hydrolytic enzymes, which is known to be an important mode of action in antagonism (8), no correlation was observed between the production of lytic enzymes and the range of fungal pathogens antagonized in vitro. Since antibiotics such as 2,4-diacetyl-phloroglucinol (encoded by phl) are major determinants of biological control of fungal pathogens and the phlD gene was shown to be conserved among Phl producers of worldwide origin (36), we have used a PCR screening approach to analyze the presence of the phlD gene in the P. putida A and B isolates. The phlD gene was detected in a surprisingly high number of P. putida B isolates from the rhizospheres of strawberry (74.4%) and potato (62.5%) but not in isolates from oilseed rape rhizosphere. The proportion of potential Phl producers among our P. putida B collection is considerably higher than previously reported frequencies of fluorescent Pseudomonas isolates from rhizospheres grown in disease-suppressive soils (19). In P. putida A the phlD gene was found in isolates from soil and strawberry, oilseed rape, and potato rhizospheres but was much less frequently detected. For many antagonists it was shown that the expression of genes involved in disease suppression (antifungal metabolites such as antibiotics or extracellular enzymes) is regulated in response to their own population densities, a phenomenon termed quorum sensing (10, 34). One prominent example is Pseudomonas aureofaciens, which is capable of protecting wheat from take-all disease, caused by the ascomycete fungus Gaeumannomyces graminis var. tritici. Disease suppression is due to the production of phenazine antibiotics, the synthesis of which is regulated by a quorum-sensing circuit (33). Evidence that accumulated over the past few years showed that AHL-mediated cell-cell communication is a widespread phenomenon among plant-associated bacteria (11, 34, 50). In this study all P. putida A strains, a subset of the P. putida B strains, and a subset of the Serratia and Pantoea strains were analyzed for the production of AHL. In contrast to the findings of Elasri et al. (11), who suggested that AHL production is more common among plant-associated bacteria than among pseudomonads originating from soil, we observed AHL production for P. putida A strains isolated from the rhizosphere and from soil. Recent work has shown that AHL signal molecules serve not only as population density sensors but also for communication between cells of different species colonizing the plant rhizosphere (32, 45). It has been speculated that AHL molecules may be important for coordinating the various functions of the different populations within the rhizosphere. Although AHLs were detected in a considerable proportion of the P. putida A and P. putida B strains tested, the functions regulated by AHL remain to be elucidated. About 50% of the Serratia and Pantoea strains were shown to produce AHLs, but neither exoenzyme production nor antifungal activity seemed to be associated with the production of signal molecules.
Sixty randomly selected isolates from this study were further characterized with regard to their plant growth-promoting activity in a strawberry seedling assay, and three selected isolates from each plant were characterized in greenhouse experiments (4). In this in vitro study, isolates from all plants were able to enhance plant growth in strawberries. However, the success of biological approaches to control plant diseases and enhance growth must be judged by their performance under field conditions. Raaijmakers and Weller (35) suggested that by matching rhizobacterium genotypes with crops for which they have colonization preference, root colonization could be increased. This study supports the notion that the rhizosphere of different plants might provide conditions (e.g., nutritional sources) differently supportive for biological control strains. The phenotypic and genotypic diversity found in natural populations and which was observed in the collection of Verticillium antagonists isolated in this study offers a tremendous resource for the improvement of biological control strains.
Acknowledgments
We thank Hella Goschke (Rostock, Germany) for valuable technical assistance and Jessica Parzy (Braunschweig, Germany) for performing FAME analysis. Jens Frankowski (Rostock, Germany) was helpful in chiA gene detection.
This study was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, G. 1996. Rhizobacteria of oil seed rape antagonistic to Verticillium dahliae. J. Plant Dis. Protect. 103:20-30. [Google Scholar]
- 4.Berg, G., A. Fritze, N. Roskot, and K. Smalla. 2001. Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. J. Appl. Microbiol. 156:75-82. [DOI] [PubMed] [Google Scholar]
- 5.Berg, G., A. Buchner, E. H. M. Wellington, and K. Smalla. 2000. Successful strategy for the selection of new strawberry-associated rhizobacteria antagonistic to Verticillium wilt. Can. J. Microbiol. 46:1128-1137. [DOI] [PubMed] [Google Scholar]
- 6.Bloemberg, G. V., and B. J. J. Lugtenberg. 2001. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 7.Chernin, L., Z. Ismailov, S. Haran, and I. Chet. 1995. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 61:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chet, I., A. Ordentlich, R. Shapira, and A. Oppenheim. 1990. Mechanisms of biocontrol of soil-borne plant pathogens by rhizobacteria. Plant Soil 129:85-92. [Google Scholar]
- 9.Duffy, B. K., and G. Défago. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberl, L. 1999. N-acyl homoserinelactone-mediated gene regulation in gram-negative bacteria. Syst. Appl. Microbiol. 22:493-506. [DOI] [PubMed] [Google Scholar]
- 11.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emmert, E. A. B., and J. Handelsman. 1999. Biocontrol of plant disease: a (Gram+) positive perspective. FEMS Microbiol. Lett. 171:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Fravel, D. R. 1988. Role of antibiosis in the biocontrol of plant diseases. Annu. Rev. Phytopathol. 26:75-91. [Google Scholar]
- 14.Fromin, N., W. Achouak, J. M. Thiéry, and T. Heulin. 2001. The genotypic diversity of Pseudomonas brassicacearum populations isolated from roots of Arabidopsis thaliana: influence of plant genotype. FEMS Microbiol. Ecol. 37:21-29. [Google Scholar]
- 15.Germida, J. J., S. D. Siciliano, J. Renato de Freitas, and A. M. Seib. 1998. Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol. Ecol. 26:43-50. [Google Scholar]
- 16.Grayston, S. J., S. Wang, C. D. Campbell, and A. C. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 17.Jaeger, C. H., III, S. E. Lindow, W. Miller, E. Clark, and M. K. Firestone. 1999. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalbe, C., P. Marten, and G. Berg. 1996. Members of the genus Serratia as beneficial rhizobacteria of oilseed rape. Microbiol. Res. 151:4433-4440. [DOI] [PubMed] [Google Scholar]
- 19.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 21.Kremer, R. J., M. F. T. Begonia, L. Stanlay, and E. T. Lanham. 1990. Characterization of rhizobacteria associated with weed seedlings. Appl. Environ. Microbiol. 56:1649-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurze, S., R. Dahl, H. Bahl, and G. Berg. 2000. Biological control of soil-borne pathogens in strawberry by Serratia plymuthica HRO-C48. Plant Dis. 85:529-534. [DOI] [PubMed] [Google Scholar]
- 23.Lemanceau, P., T. Corberand, L. Gardan, X. Latour, G. Laguerre, J. M. Boeufgras, and C. Alabouvette. 1995. Effect of two plant species, flax (Linum usitatissinum [sic] L.) and tomato (Lycopersicum esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl. Environ. Microbiol. 61:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 25.Maas, J. L. 1998. Compendium of strawberry diseases. APS Press, St. Paul, Minn.
- 26.McSpadden Gardener, B. B., K. L. Schroeder, S. E. Kalloger, J. M. Raajmakers, L. S. Thomashow, and D. M. Weller. 2000. Genotypic and phenotypic diversity of phlD-containing Pseudomonas strains isolated from the rhizosphere of wheat. Appl. Environ. Microbiol. 66:1939-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menhinick, E. F. 1964. A comparison of some species-individuals diversity indices applied to samples of field insects. Ecology 45:859-861. [Google Scholar]
- 28.Miller, H. J., G. Henken, and J. A. van Veen. 1989. Variation and composition of bacterial populations in the rhizospheres of maize, wheat, and grass cultivars. Can. J. Microbiol. 35:656-660. [Google Scholar]
- 29.Moore, E. R. B., M. Mau, A. Arnscheidt, E. C. Böttger, R. A. Hutson, M. D. Collins, Y. van der Peer, R. de Wachter, and K. N. Timmis. 1996. The determination and comparison of 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of natural intrageneric relationships. Syst. Appl. Microbiol. 19:478-492. [Google Scholar]
- 30.O'Sullivan, D. J., and F. O'Gara. 1992. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol. Rev. 56:662-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierson, E. A., D. W. Wood, J. A. Cannon, F. M. Blachere, and L. S. Pierson III. 1998. Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant-Microbe Interact. 11:1078-1084. [Google Scholar]
- 33.Pierson, L. S., III, V. D. Keppenne, and D. W. Wood. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierson, L. S., III, D. W. Wood, and E. A. Pierson. 1998. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 36:207-225. [DOI] [PubMed] [Google Scholar]
- 35.Raaijmakers, J. M., and D. M. Weller. 2001. Exploiting genotypic diversity of 2,4-diacetylphloroglucinol-producing Pseudomonas spp.: characterization of superior root-colonizing P. fluorescens strain Q8r1-96. Appl. Environ. Microbiol. 67:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raaijmakers, J. M., D. M. Weller, and L. S. Thomashow. 1997. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 63:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rademaker, J. L. W., and F. J. De Bruijn. 1997. Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer-assisted pattern analysis, p. 151-171. In G. Caetano-Anollés and P. M. Gresshoff (ed.), DNA markers: protocols, applications and overviews. J. Wiley & Sons, Inc., New York, N.Y.
- 38.Rademaker, J. L. W., F. J. Louws, U. Rossbach, and F. J. de Bruijn. 1999. Computer assisted pattern analysis of molecular fingerprints and data base construction, p. 1-33. In A. D. L. Akkermans, J. D. van Elsas, and F. J. DeBruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers. Dordrecht, The Netherlands.
- 39.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 40.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Hoiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 41.Sawar, M., and R. J. Kremer. 1995. Determination of bacterially derived auxins using a microplate method. Lett. Appl. Microbiol. 20:282-285. [Google Scholar]
- 42.Smalla, K., U. Wachtendorf, H. Heuer, W.-T. Liu, and L. Forney. 1998. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl. Environ. Microbiol. 64:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, N. Roskot, H. Heuer, and G. Berg. 2001. Bacterial bulk and rhizosphere communities studied by denaturing gradient gel electrophoresis of PCR-amplified fragments of 16S rRNA genes: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sørensen, J. 1997. The rhizosphere as a habitat for soil microorganisms, p. 21-45. In J. D. Van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 45.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tjamos, E. C., R. C. Rowe, J. B. Heale, and D. R. Fravel. 2000. Advances in Verticillium research and disease management. APS Press, St. Paul, Minn.
- 47.Weller, D. M. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379-407. [Google Scholar]
- 48.Whipps, J. M. 1997. Ecological considerations involved in commercial development of biological control agents for soil-borne diseases, p. 525-545. In J. D. Van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 49.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jørgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl-homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Z., and L. S. Pierson III. 2001. A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 67:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]