Abstract
The community structure of bacterioplankton was studied at different depths (0 to 25 m) of a temperate eutrophic lake (Lake Plußsee in northern Germany) by using comparative 5S rRNA analysis. The relative amounts of taxonomic groups were estimated from 5S rRNA bands separated by high-resolution electrophoresis. Comparison of partial 5S rRNA sequences enabled detection of changes in single taxa over space and during seasons. Overall, the bacterioplankton community was dominated by 3 to 14 abundant (>4% of the total 5S rRNA) taxa. In general, the number of 5S rRNA bands (i.e., the number of bacterial taxa) decreased with depth. In the fall, when thermal stratification and chemical stratification were much more pronounced than they were in the spring, the correlation between the depth layers and the community structure was more pronounced. Therefore, in the fall each layer had its own community structure; i.e., there were different community structures in the epilimnion, the metalimnion, and the hypolimnion. Only three 5S rRNA bands were detected in the hypolimnion during the fall, and one band accounted for about 70% of the total 5S rRNA. The sequences of individual 5S rRNA bands from the spring and fall were different for all size classes analyzed except two bands, one of which was identified as Comamonas acidivorans. In the overall analysis of the depth profiles, the diversity in the epilimnion contrasted with the reduced diversity of the bacterioplankton communities in the hypolimnion, and large differences occurred in the composition of the epilimnion at different seasons except for generalists like C. acidivorans.
A fundamental concept in limnology is that the water column of a lake is divided into three horizontal layers, the epilimnion, the metalimnion, and the hypolimnion (22). These strata are shaped by solar radiation and wind. In temperate regions, they are characterized by gradients of temperature and oxygen, and they have different functions for the lake as an ecosystem. The epilimnion is well mixed and acts as a bioreactor for primary production, and the hypolimnion is a sink where biomass accumulates and is remineralized (43). These two parts are connected by the metalimnion or thermocline, often characterized by a steep decrease in temperature. This basic view of the structure of a lake illustrates that depth is a key factor for all limnic life and that depth profiles are key features for understanding aquatic ecosystems.
The depth dependence of the structure of phyto- and zooplankton communities in lakes and oceans has been studied since the beginning of aquatic ecology and has contributed enormously to the understanding of the functioning of freshwater and marine ecosystems. Despite the recognition of the important role of bacteria in the microbial food web and the contribution of bacteria to the functioning of aquatic ecosystems, knowledge about the depth distribution of these organisms is still scarce (1, 5). Pioneering work has been done on the depth-dependent community structure of bacterioplankton in the oceans by DeLong (6), Giovannoni et al. (15), and Fuhrman et al. (13). These studies were based on analysis of 16S ribosomal DNA (rDNA) clone libraries from deep ocean samples (500 to 2,000 m) and provided insight into the depth distribution of single taxa. Field et al. (12) obtained higher resolution of the variation in bacterial communities with depth by using group-specific 16S rRNA-targeted oligonucleotide probes and environmental bulk DNA-RNA for assessment of the abundance of specific taxa belonging to the α subgroup of the Proteobacteria in the Sargasso Sea. However, community structure can be assessed most rapidly by community fingerprint methods that are based on electrophoretic separation of 16S rDNA amplicons or 5S rRNA obtained from natural microbial communities (27, 30, 34, 35). The first community fingerprinting analysis of bacterioplankton with depth was done by using amplicons of the V3 region of 16S rRNA and archaeal and bacterial primers (29). Øvreås et al. (29) found that the diversity of the bacterial community decreased from the surface to the bottom in an estuary with a strong vertical gradient of salinity and oxygen. However, a major problem of PCR-based methods is that they can be biased for some phylogenetic groups, and quantitative assessment of community diversity is difficult (39). Therefore, we have used 5S rRNA fingerprinting to obtain a direct molecular fingerprint from natural communities and have calculated, based on the relative densities of individual 5S rRNA bands, the diversity of the bacterioplankton during the year (20).
The goals of the present study were to compare changes in physical, chemical, and biological parameters with depth at a high spatial resolution (i.e., every meter) with changes in the structure of the bacterioplankton community and to extend the previous seasonal study in terms of depth (20). The ecosystem chosen was Lake Plußsee, a naturally eutrophic lake in northern Germany which has been examined extensively (28). We analyzed the 5S rRNA fraction of environmental rRNA obtained directly (i.e., without PCR) from the bacterioplankton. The structure of the bacterioplankton community as a function of depth was analyzed based on high-resolution 5S rRNA community fingerprints and partial or full sequences of individual 5S rRNA.
MATERIALS AND METHODS
Sampling.
Water samples were collected from the central part of Lake Plußsee (Schleswig-Holstein, Germany; 54°10.0′N, 10°0.23′E) on 25 April and 19 September 1994 by using a 2.5-liter Ruttner sampler at the following depths: 0, 1, 2, 3, 4, 5, 6, 7, 8, 10, 15, 20, and 25 m. The bacterioplankton fraction was filtered onto a sandwich consisting of a glass fiber filter (type GF/F; diameter, 90 mm; Whatman Corp.) on top of a polycarbonate filter (pore size, 0.2 μm; Nuclepore Corp.) and was stored frozen (−70°C) for later RNA analysis. The samples had been prefiltered through a polycarbonate filter (Nuclepore Corp.) with a pore size of 3 μm to eliminate larger organisms.
Biological background parameters, such as total bacterial counts, total heterotrophic nanoflagellate (HNF) counts, and total particulate chlorophyll a concentrations, were measured by standard techniques; details are given elsewhere (4).
5S rRNA fingerprints of bacterioplankton.
5S rRNA analysis of bacterioplankton started with extraction of the total RNA directly from the frozen filter sandwiches. This extraction was based on mechanical extraction with glass beads in a bead beater in combination with chemical extraction with phenol and sodium dodecyl sulfate (SDS) (8, 16). In short, total environmental RNA was extracted from the sandwiches by using 4 g of 2- and 3-mm-diameter glass beads, a high-speed cell disrupter (Microdismembrator; Braun-Diessel Corp.) with a Teflon extraction cell, and a mixture of 5 ml of RNA extraction buffer and 5 ml of buffer-saturated phenol (17). Extraction buffer consisted of 50 mM sodium acetate, 10 mM EDTA, and 1% SDS (pH 4.2). Phenol was saturated with extraction buffer lacking 1% SDS until a final pH of 4.2 to 4.4 was reached. A thin slurry (total volume, about 12 ml) of the two filters and the extraction mixture was generated by vibration for 2 min with the cell disrupter. This slurry was then centrifuged for 10 min at 8,500 × g. After repeated phenol (pH 4.2) extraction of the pellet, the supernatants were pooled and treated twice with 5 ml of chloroform. Total RNA was precipitated from the pooled supernatant by addition of ethanol (2.5 times the volume of the supernatant) and storage at −20°C overnight.
The total RNA obtained by the filter extraction procedure was 3′ end labeled with cytidine 3′,5′-[5′-32P]bisphosphate (specific activity, 3,000 Ci/mmol) by using RNA ligase with an efficiency of about 62% (7, 10). The radioactively labeled RNA was subjected to denaturing high-resolution electrophoresis on a 10% polyacrylamide gel (Sequicel; 0.4 mm by 38 cm by 80 cm; Bio-Rad) by using running conditions consisting of a stepwise increase over a 5-h period from 100 to 300 W. After electrophoresis, the gel was exposed to X-ray film or a storage phosphor screen (Molecular Probes Corp.) for 30 min to several hours. More details concerning RNA extraction and electrophoretic analysis are given elsewhere (8, 16, 17, 19, 40).
Comparative analysis of 5S rRNA fingerprints.
The 5S rRNA fraction was evaluated quantitatively by scanning the autoradiograms with an optical gel scanner (Hirschmann Corp.) and by using a phosphorimager (Molecular Probes Corp.). In this way the abundance of single 5S rRNA bands was quantified in terms of baseline corrected peak areas and in terms of number of pixels per band, respectively. These arbitrary units were used to calculate the relative abundance of every individual 5S rRNA band and its contribution to the total 5S rRNA.
The diversity of the community was determined by determining the number of 5S rRNA bands per sample and by calculating the Shannon diversity index (H) with the following equation:
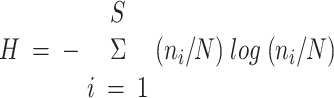 |
, where ni is the relative abundance of a single taxon (single 5S RNA band), N is the total abundance of all taxa, and S is the total number of abundant species. Only bands accounting for more than 4% of the total 5S rRNA were included in the calculation of the diversity index and the total number of bands. This 4% threshold is not based on the detection limit of the 5S rRNA analysis, which is less than 0.1%. Rather, it is an arbitrary threshold set to make our data analysis comparable with analyses in earlier studies (18).
Sequence analysis of single environmental 5S rRNA.
After high-resolution electrophoresis, 32P-labeled 5S rRNA bands were excised from the gel, eluted with 150 μl of elution buffer (0.5 M sodium acetate, 0.1% SDS, 1 mM magnesium chloride, 100 μM EDTA; pH 7.8), and ethanol precipitated at −20°C overnight. Before application to the sequencing gel, 5S rRNA was enzymatically digested with base-specific RNases (9, 16). The cleaved 5S rRNA of a single excised band was run on a high-resolution sequencing gel (80 by 40 cm; thickness, 0.4 mm;10% acrylamide LongRanger [FMC Corp.] prepared according to the protocol of the manufacturer) to determine the positions of the bases in comparison to the bases of totally hydrolyzed 5S rRNA of Escherichia coli, which was used as the molecular size marker. This sequencing gel provided information about the positions of almost all bases of the 5S rRNA from the 5′ end. Only around 2% of the molecule (i.e., two or three nucleotides at the 3′ end) could not be read. These sequences could lead to classification or identification of the taxon represented by a single band provided that the reference sequence of a known bacterial species is available in the 5S rRNA sequence database (33). Also, this sequence information was used to determine the homogeneity of a band (i.e., if one or more taxa formed the band). Furthermore, it allowed comparison of 5S rRNA bands of the same length for samples from different depths and times of the year. Such comparisons were also possible with partial sequences (e.g., by using only RNase T1 to examine the positions of all guanine residues within a 5S rRNA band of a specific size).
RESULTS
Environmental conditions in Lake Plußsee.
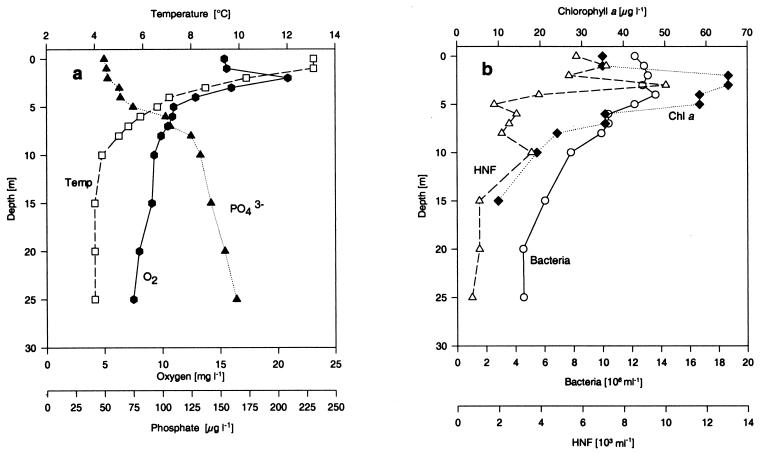
Physical, chemical, and biological background data for spring 1994 for Lake Plußsee are given in Fig. 1. The temperature steadily decreased with depth from 1 to 10 m, and thus there was not an extended epilimnion. The oxygen content peaked at 2 m, corresponding to the top of the chlorophyll a maximum (Fig. 1b), indicating that the maximum primary production occurred at this depth. The phosphate concentrations decreased to about 50 μg/liter in the upper 5 m, indicating that the phytoplankton was most active in this layer. Below 5 m, the phosphate and inorganic nitrogen contents (7; data not shown) increased rapidly to the high hypolimnic concentrations. The chlorophyll a concentrations reached 65 μg/liter, indicating that there was a well-developed spring phytoplankton bloom and that the lake was eutrophic. The phytoplankton community during this spring bloom was composed of a variety of diatoms and green algae (28). The total bacterial numbers (Fig. 1b) in the upper 5 m were rather constant (more than 1.2 × 107 cells/ml) and decreased steadily with depth. The concentration of HNF peaked at 3 m at about 1 × 105 cells/ml; the concentration then dropped rapidly to less than 4 × 103 cells/ml. These background data indicate that there was a fairly normal spring situation for a temperate eutrophic lake with a small, unstable epilimnion (0 to 1 m), an extended metalimnion (1 to 8 m) with maximum primary production close to the surface (2 m), and a hypolimnion below 8 m which was still fully oxic.
FIG. 1.
Physical, chemical, and biological background data for depth profiles of Lake Plußsee during the spring (25 April 1994). (a) Temperature (□), oxygen content ( ), and phosphorus content (▴); (b) chlorophyll a content (♦), total bacterial number (○), and number of HNF (▵).
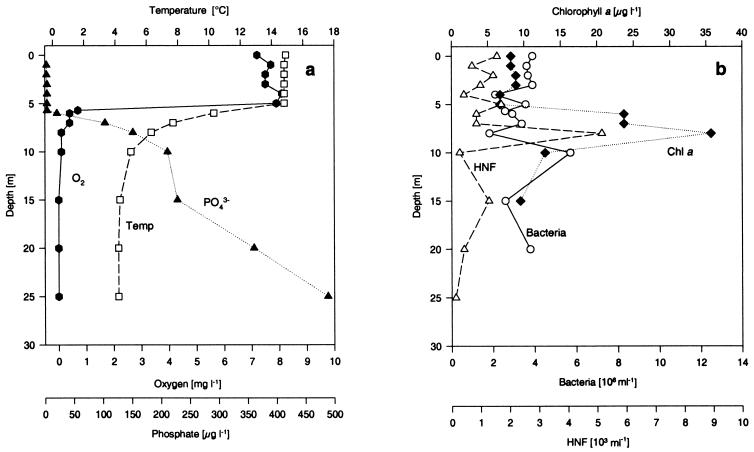
The fall data (Fig. 2) revealed a homogenous epilimnion (0 to 5 m), a metalimnion with very steep decreases in oxygen concentration and temperature (5 to 8 m), and a fully anoxic hypolimnion at depths below 8 m. The phosphate concentrations were below the detection limit in the epilimnion and increased to rather high levels in the metalimnion. The chlorophyll a concentration was low in the epilimnion and peaked at 8 m (i.e., in the anoxic hypolimnion). The bacterial concentration did not vary significantly with depth, but the concentration of HNF peaked together with the chlorophyll a concentration, as it did in the spring. As in the spring, in the fall of 1994 Lake Plußsee had the very stable and well-structured depth profile of a naturally eutrophic lake during autumn (22).
FIG. 2.
Physical, chemical, and biological background data for depth profiles of Lake Plußsee during the fall (19 September 1994). (a) Temperature (□), oxygen content ( ), and phosphorus content (▴); (b) chlorophyll a content (♦), total bacterial number (○), and number of HNF (▵).
Changes in the overall structure of the bacterioplankton community with depth.
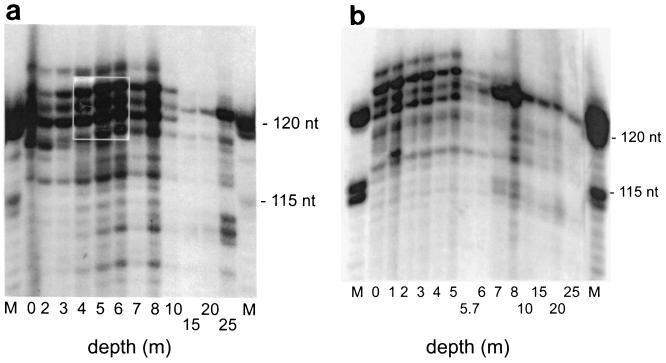
In the spring, the community structure for the depth profile showed three different banding patterns of the 5S rRNA that corresponded with the three layers of the lake (Fig. 3a). The epilimnion (0 to 3 m) had six or seven abundant bands ranging in size from 116 to 123 nucleotides (nt), the metalimnion (4 to 8 m) had the highest number of bands with additional bands in the size range from 109 to 115 nt, and the hypolimnion (10 to 25 m) had the lowest number of bands, with one band comprising more than 50% of the total 5S rRNA. At greater depths the community structure changed the most. For example, at 15 to 20 m the two most abundant bands comprised more than 70% of the total 5S rRNA and the 5S rRNA bands at 122 and 123 nt had disappeared. The amount of small 5S rRNA (111 to 113 nt), which comprised about 20% of the total 5S rRNA, increased in the deepest water (25 m).
FIG. 3.
Autoradiograms of 5S rRNA from bacterioplankton obtained at different depths. Samples of the water column were taken at different depths. For detection the 5S rRNA was 3′ end labeled with 32P. (a) Depth profile for the spring (25 April 1994). Lanes M contained a molecular weight standard consisting of hydrolyzed 5S rRNA from E. coli. The intensity of the bands in the boxed area was electronically reduced for better visibility of the single bands. Each lane is labeled with the depth (in meters) at which the sample examined was obtained. (b) Depth profile for the autumn (19 September 1994): Lanes M contained a molecular weight standard consisting of hydrolyzed 5S rRNA from E. coli. Each lane is labeled with the depth (in meters) at which the sample examined was obtained.
During thermal stratification in the autumn the number of 5S rRNA bands was high in the epilimnion and was greatly reduced below the thermocline (Fig. 3b). Also, in the upper 5 m the number of bands, their relative positions, and their relative amounts were very stable. Only at 1 m was an additional band at 117 nt observed. The bacterioplankton community of the anoxic hypolimnion (7 to 25 m) was dominated by a single 5S rRNA band at 121 nt (on average, this band accounted for 63% of the total 5S rRNA from 10 m and below).
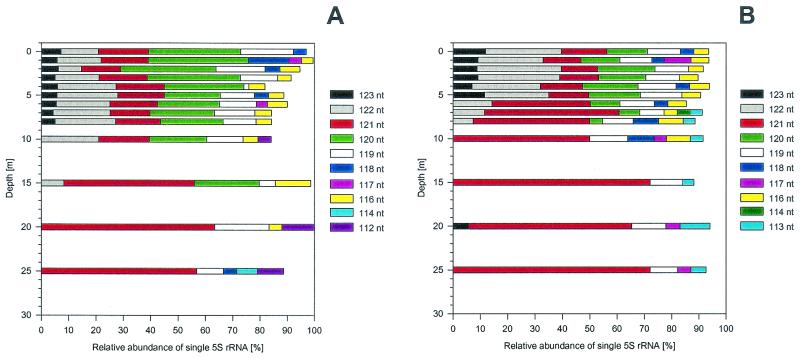
The results of a detailed quantitative analysis of the amount of 5S rRNA per abundant band in the autoradiograms, obtained by using a phosphorimager for quantification, are shown in Fig. 4. In both depth profiles the number of abundant bands was rather limited, ranging from three to seven. Also, these abundant bands (i.e., bands that accounted for more than 4% of the total 5S rRNA) comprised more than 90% of the total 5S rRNA in most samples (Fig. 4B). The spring profile was slightly more variable in this respect than the fall profile due to the larger number of less abundant bands occurring in the metalimnion. Another general trend for both depth profiles was that in the epilimnion no single band exceeded a relative abundance of 34% and a small number of bands (three to five bands), each comprising 10 to 30% of the total, represented the majority of the bacterioplankton community. With increasing depth one band became dominant in both depth profiles; i.e., it had a relative abundance of more than 50%. Furthermore, in both profiles for depths below 15 m the dominant band was the same size (121 nt) despite the anoxic conditions in the fall. The two depth profiles did differ, however; when anoxic conditions were reached at depths below 6 m, the 123-nt band disappeared and a 113-nt band appeared. In general, changes in the community composition were more gradual in the spring profile than in the fall profile.
FIG. 4.
(A) Relative amounts of the single abundant 5S rRNA bands for the spring shown in Fig. 3a. Different colors indicate 5S rRNA bands of different sizes that account for more than 4% of the total 5S rRNA calculated as detailed in Materials and Methods. (B) Relative amounts of single abundant 5S rRNA bands for the fall shown in Fig. 3b.
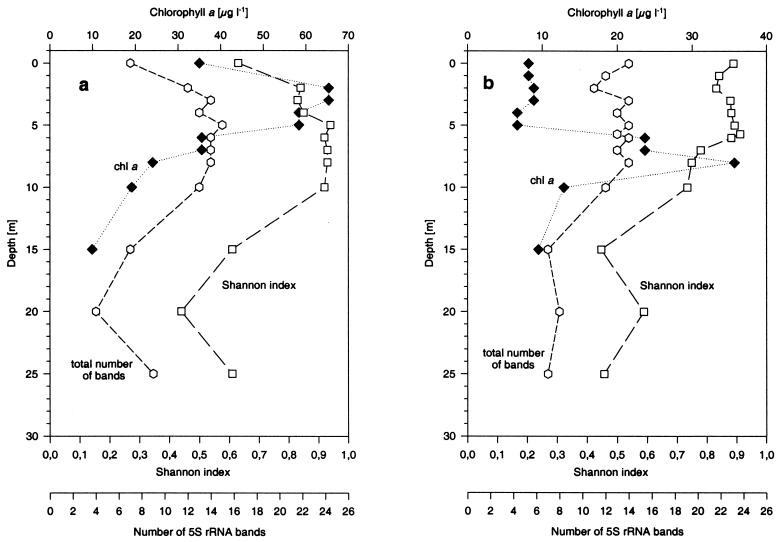
To examine overall diversity, the Shannon indices were calculated for all samples based on the total number of 5S rRNA bands, not just the number of abundant 5S rRNA bands, and their relative amounts (Fig. 5). The total number of bands was determined by counting every band that had a relative abundance of more than 0.1% of the total 5S rRNA. This threshold reflects the level of resolution of the 5S rRNA fingerprinting technique when a phosphorimager is used. In contrast, the 4% threshold for the abundant 5S rRNA is an arbitrary threshold that was introduced earlier to make community fingerprints more comprehensible (18). Additionally, the following assumptions for calculation of the Shannon diversity index based on the relative amounts of individual 5S rRNA bands were made: (i) RNA was extracted with comparable efficiencies from all taxa, (ii) 5S rRNA was 32P labeled with comparable efficiencies, and (iii) sequence analysis of individual 5S rRNA shows homogeneity. The validity of these assumptions was demonstrated in calibration experiments with reference cultures (7, 8, 40) or direct sequencing (see below). During the spring, the overall diversity increased until 5 m and then stayed maximal until 10 m. Below 10 m, the diversity decreased by a factor of two until 25 m (Fig. 5a). During the fall, the diversity was highest in the epilimnion and then declined uniformly in the anoxic part of the water column (Fig. 5b). In both profiles the Shannon index generally followed the total number of 5S rRNA bands. The most striking difference was that the maximum diversity occurred in the spring below the maximum chlorophyll a concentration, whereas in the fall the maximum diversity was found above the maximum chlorophyll a concentration.
FIG. 5.
(a) Total number of bands ( ) (bands containing more than 0.1% of the total 5S rRNA) and Shannon diversity index (□) calculated from all 5S rRNA bands at each depth during the spring. The chlorophyll a content (♦) is shown for comparison. (b) Total number of bands ( ) and Shannon diversity index (□) calculated from all 5S rRNA bands at each depth during the fall. The chlorophyll a content (♦) is shown for comparison.
Detailed community structure as revealed by 5S rRNA sequence analysis.
A sequence analysis of single 5S rRNA bands was performed by using base-specific RNases in order to compare 5S rRNA bands from different depths that were the same size and to identify the closest taxonomic relatives by comparison with the international sequence database (33). To examine 5S rRNA bands with depth, only partial sequences were determined by using RNase T1 and sometimes RNase U2 digests to obtain a G and/or A ladder of the 5S rRNA band for about 80% of the molecule, as described previously (17, 20). In the spring, 5S rRNA bands at 116, 118, 120, 121, and 123 nt were compared. All of the bands analyzed had the same partial sequences from 0 to 8 m. Below 8 m, only a few bands could be sequenced due to the much lower amount of radioactivity per band (Fig. 3a). The partial sequence of the 121-nt band from 25 m, which dominated the community at this depth, was different from the sequence of the 121-nt band obtained from the upper 8 m.
In the fall, 5S rRNA bands at 116, 120, 121, 122, and 123 nt were compared for the upper 5 m. In addition, the 121-nt band was sequenced from all samples of the depth profile. There were no sequence differences detected in any of the fall 5S rRNA bands with depth; i.e., all bands sequenced in the upper 5 m and the 121-nt band found throughout the water column were the same. A comparison of the sequences from the spring with the sequences from the fall showed significant differences for all of the single size classes analyzed except the 116- and 122-nt bands. These bands had identical partial sequences; i.e., all As and Gs were at the same position in the 5S rRNA for both seasons. The 121-nt band, which was the most abundant 5S rRNA in the hypolimnion in both depth profiles, had different sequences in the two seasons, despite the identical sizes. These sequence data indicate that different taxa dominated the hypolimnion in the spring and in the fall.
The only 5S rRNA for which a definite phylogenetic relative could be found was the 116-nt band, which had a sequence identical to a Comamonas acidovorans sequence (accession number AJ131594). The presence of C. acidovorans has been documented in samples from Lake Plußsee before (11, 20). For all other bands except the 123-nt band either the sequence information obtained was not complete enough for alignment with sequences from the international database or no close relative could be found; the closest phylogenetic neighbor of the 123-nt band was in the genera Thiovolum and Campylobacter of the ε subgroup of the Proteobacteria.
DISCUSSION
Overall community structure and activity of bacterioplankton at different depths and in different seasons.
In contrast to PCR-driven community fingerprints, 5S rRNA fingerprints of bacterioplankton reflect directly the amounts of different 5S rRNA from different members of the bacterial community. Therefore, we have defined a single 5S rRNA band with a homogenous sequence as an operational taxonomic unit (OTU) (20). This OTU definition enables us to calculate the Shannon diversity index for each community based on the relative abundance of each 5S rRNA in comparison to the total amount of 5S rRNA for each depth. Two technical prerequisites for assessment of the relative amounts of 5S rRNA have to be fulfilled; the efficiencies of extracting RNA from natural bacterioplankton should be similar for all taxa, and the efficiencies of 32P labeling the 3′ ends of the 5S rRNA should be similar. It has been demonstrated that both prerequisites are fulfilled with the methodology used here (7, 8, 20, 40). Another theoretical and technical consideration for calculation of the Shannon index is to set a lower limit for the relative abundance of a single OTU. Such a lower limit has been set at a relative abundance of 4% for clarity (Fig. 4) and comparability with past studies (18, 20). On the other hand, the detection limit for single 32P-labeled 5S rRNA bands is about 0.1% if adjacent bands are not so large that they completely cover the weak bands next to them in the autoradiogram. This analytical problem can be overcome by using a phosphorimager that enables detection of bands next to each other whose band intensities are more than 4 orders of magnitude different. A 0.1% detection limit means that we were able to detect taxa whose concentrations were as low as 0.5 × 106 to 1 × 106 cells per liter in the 5S rRNA fingerprints of bacterioplankton from Lake Plußsee with the described methodology if an average cell density of 5 × 109 to 10 × 109 cells per liter was assumed (Fig. 1b and 2b). This detection limit is about 1 order of magnitude lower than the 1% limit commonly reported for PCR-based fingerprint methods, such as denaturing gradient gel electrophoresis or single-strand conformation polymorphism analysis (27, 31). In addition, not only does the presence of a 5S rRNA band reflect the abundance of an OTU, but the band also represents an active member of the bacterioplankton community since the amount of rRNA is correlated with the growth of bacteria (3).
Vertical gradients of physical and chemical parameters strongly affect the biology and ecology of all pelagic ecosystems (22). This universal finding is also reflected in the structure of the bacterioplankton community. The most striking feature of both depth profiles was the much lower diversity in the hypolimnion than in the upper layers. The decrease can be seen in the reduction of the Shannon index by about one-half, as well as in the number of bands, which decreased from about 14 to 7 in both depth profiles (Fig. 5). One exception to this overall trend was the 25-m sample collected in the spring, for which the number of bands increased. Conceivably, the diatoms which dominate the phytoplankton of Lake Plußsee in early spring were still being degraded in the deep hypolimnion after they sank in late March (28, 32).
When the diversity data for the depth profiles were compared with the data from the seasonal study of bacterioplankton in Lake Plußsee, it was obvious that the low diversity, as indicated by a Shannon index of around 0.4, in the hypolimnion was encountered only during the three minima occurring in the epilimnion at special times of the year (20). During most of the year the Shannon index remained in the range found for the epilimnion for both depth profiles. The first minimum for the diversity index in the seasonal study could be explained by strong grazing by cladocerans, whereas grazing by HNF did not have a significant effect on diversity. Grazing by HNF did not directly affect the high diversity in the upper layers of depth profiles, where HNF grazing is the major mortality factor in the epilimnion (41). On the other hand, viral lysis was demonstrated to be the key mortality factor in the anoxic hypolimnion of Lake Plußsee, where all grazing was strongly reduced (41). This result corroborates the finding that only very few members of the bacterioplankton community were active in the hypolimnion because low-diversity bacterial communities can be controlled more easily by viral lysis than high-diversity communities (37). The major reason for the low diversity in the hypolimnion for both depth profiles might be the decrease in the dissolved organic carbon content, which not only decreased by about 50% with depth but also accounted for a far higher proportion of refractory organic compounds in the hypolimnion than in the epilimnion of Lake Plußsee (26).
Depth-dependent and seasonal changes in detailed structure of the bacterioplankton community.
The presence and depth distribution of C. acidovorans in Lake Plußsee have been demonstrated previously by immunofluorescence microscopy (11, 42). Additionally, a seasonal study in which 5S rRNA profiling was used showed that C. acidovorans is abundant at a depth of 1 m during all seasons (20). Both depth profiles obtained by using 5S rRNA analysis confirmed the presence of C. acidovorans in the whole profile, with an average abundance of about 6.0%. (The levels in deeper samples were around 3% and therefore are not shown in Fig. 4). This finding is in contrast to the results obtained by immunofluorescence microscopy that indicated that C. acidovorans cells disappeared at depths below 10 m in the spring of 1993 and that there was a significant lack of C. acidovorans cells in the metalimnion during the fall of 1996 (11, 42). The most likely explanation for this finding is that the monoclonal antibodies used for detection of C. acidovorans cells are specific for a certain subspecies type, whereas the 5S rRNA bands represent the whole taxon. Therefore, C. acidovorans was present in abundance in Lake Plußsee not only during all seasons but also at all depths, including the anoxic hypolimnion. Furthermore, the presence of significant amounts of rRNA indicated that this group of bacteria was metabolically active at all depths year round. This supports the hypothesis, put forward earlier (42), that C. acidovorans is a K-strategist well adapted to the pelagic environment of a eutrophic lake because of its relative resistance to protozoan grazing and its ability to utilize refractory dissolved organic matter.
Partial sequencing revealed differences in all bands except the 116- and 122-nt bands in the different seasons. The strong seasonal differences make sense considering the fact that the phytoplankton communities during the two seasons were completely different and that in the fall the hypolimnion was anoxic. Detection of members of the ε subgroup of the Proteobacteria in the anaerobic water column is unusual but has been observed with clone libraries from coastal marine water samples obtained during an algal bloom and from the chemocline of a meromictic lake in the Swiss Alps (2, 21).
Comparison of bacterioplankton community structure with other depth distributions.
A variety of studies have been performed with different molecular methods to understand the depth distribution of the bacterioplankton community (14, 25). Most of these studies were based on clone libraries of 16S rDNA genes (see reference 14 for a review), followed by hybridization studies and community fingerprinting. While clone library studies provide insight into community composition and diversity, fingerprinting studies reveal the overall structure of the whole community and enable direct comparisons of a variety of samples. Fingerprinting studies of the bacterioplankton of a brackish meromictic lake and the central Baltic Sea indicated that there was higher diversity in the upper layer than in the anoxic bottom layer, as found for Lake Plußsee (18, 29). Recent studies of bacterioplankton in a deep ultraoligotrophic lake and the eastern Mediterranean Sea showed that there were distinct changes with depth but did not reveal substantial decreases in diversity (24, 38). Several studies performed with group-specific rRNA-targeted probes revealed the depth dependence of these groups in the water column of the oceans (12, 23). The amounts of some of these groups correlated well with microbial processes, such as the intensity of primary production (36). In general, the following picture appears to emerge for the structure of the bacterioplankton community with depth. Pelagic environments have specific bacterioplankton community structures at specific depths, and the overall diversity and composition might vary from layer to layer depending on the trophic status and the availability of electron acceptors.
Conclusions.
We demonstrated that there were changes in bacterioplankton community structure with depth in a eutrophic lake. These changes corresponded well with the three layers, epilimnion, metalimnion, and hypolimnion, of the lake. The more the lake was stratified, the better the three different communities were separated. Whereas the community composition generally varied with the season, some members of the community, such as C. acidovorans, were abundant at all depths during both seasons. Another common feature was the reduction in the overall diversity of the bacterioplankton with depth. This decrease in diversity means that the diversity of the bacterioplankton community in the hypolimnion was quite low compared to the diversity of the community in the epilimnion, where the diversity is low only two or three times during the year (20).
Acknowledgments
We thank D. Albrecht from the MPI of Limnology in Plön for providing background data. The valuable comments of Ingrid Brettar and Markus G. Weinbauer on an earlier version of the manuscript are greatly acknowledged. Improvement of the English by David Kirchman is greatly appreciated.
This project was supported by funds from Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie grant BEO-0319433B and Deutsche Forschungsgemeinschaft grant Ho 930/2-1.
REFERENCES
- 1.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer-Reil, and F. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 2.Bossard, P. P., Y. Santini, D. Grüter, R. Stettler, and R. Bachofen. 2000. Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbiol. Ecol. 31:173-182. [DOI] [PubMed] [Google Scholar]
- 3.Bremer, H., and H. H. Dennis. 1996. Modulation in chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 4.Brettar, I., and M. G. Höfle. 1992. Influence of ecosystematic factors on survival of Escherichia coli after large-scale release into lake water mesocosms. Appl. Environ. Microbiol. 58:2201-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, B. C., and F. Azam. 1990. Biogeochemical significance of bacterial biomass in the ocean's euphotic zone. Mar. Ecol. Prog. Ser. 63:253-259. [Google Scholar]
- 6.DeLong, E. F. 1992. Archaea in coastal marine waters. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominik, K. 1998. Vergleichende 5S rRNA-Analyse der zeitlichen und räumlichen Dynamik von Bakterioplankton aus dem Plußsee und anderen ostholsteinschen Seen. Ph.D. thesis. Technical University, Braunschweig, Germany.
- 8.Dominik, K., and M. G. Höfle. 1999. Extraction of total RNA from bacterioplankton, p. 1.2.2.1-1.2.2.9. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 9.Donis-Keller, H. 1979. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 7:179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.England, T. E., A. G. Bruce, and O. C. Uhlenbeck. 1980. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 65:65-85. [DOI] [PubMed] [Google Scholar]
- 11.Faude, U., and M. G. Höfle. 1997. Development and application of monoclonal antibodies for in situ detection of indigenous bacterial strains in aquatic ecosystems. Appl. Environ. Microbiol. 63:4534-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field, K. G., D. Gordon, T. Wright, M. Rappé, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archaebacterial group from marine plankton. Nature (London) 356:148-149. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 15.Giovannoni, S. J., T. B. Brigtschgi, C. L. Moyer, and G. K. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature (London) 345:60-63. [DOI] [PubMed] [Google Scholar]
- 16.Höfle, M. G. 1992. Aquatic microbial community structure and dynamics during large-scale release of bacteria as revealed by low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 58:3387-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höfle, M. G. 1998. Genotyping of bacterial isolates from the environment using low-molecular-weight RNA fingerprints, p. 3.3.7.1-3.3.7.23. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Höfle, M. G., and I. Brettar. 1995. Taxonomic diversity and metabolic activity of microbial communities in the water column of the central Baltic Sea. Limnol. Oceanogr. 40:868-874. [Google Scholar]
- 19.Höfle, M. G., and I. Brettar. 1996. Genotyping of heterotrophic bacteria from the central Baltic Sea by use of low-molecular-weight RNA profiles. Appl. Environ. Microbiol. 62:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höfle, M. G., K. Dominik, and H. Haas. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as detected by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerkhof, L. J., M. A. Voytek, R. M. Sherrell, D. Millie, and O. Schofield. 1999. Variability in bacterial community structure during upwelling in the coastal ocean. Hydrobiologia 401:139-148. [Google Scholar]
- 22.Lampert, W., and U. Sommer. 1999. Limnoökologie, 2nd ed. Georg Thieme Verlag, Stuttgart, Germany.
- 23.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moeseneder, M. M., C. Winter, and G. J. Herndl. 2001. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol. Oceanogr. 46:95-107. [Google Scholar]
- 25.Mullins, T. D., T. B. Britschgi, R. L. Kerst, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:145-158. [Google Scholar]
- 26.Münster, U., and D. Albrecht. 1994. Dissolved organic matter: analysis of composition and function by a molecular-biochemical approach, p. 24-62. In J. Overbeck and R. J. Chróst (ed.), Microbial ecology of Lake Plußsee. Springer, New York, N.Y.
- 27.Muyzer, G., E. C. De Waal, and A. G. Unterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overbeck, J., and R. J. Chróst (ed.). 1994. Microbial ecology of Lake Plußsee. Springer-Verlag, New York, N.Y.
- 29.Øvreås, L., L. Forney, L. F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 8:1-55. [Google Scholar]
- 31.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sommer, U. 1993. Phytoplankton competition in Plußsee: a field test of the resource-ratio hypothesis. Limnol. Oceanogr. 38:838-845. [Google Scholar]
- 33.Specht, T., M. Szymansji, M. Z. Barciszewska, J. Barciszewski, and V. A. Erdmann. 1997. Compilation of 5S rRNA and 5S rRNA gene sequences. Nucleic Acids Res. 25:96-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl, D. A. 1997. Molecular approaches for the measurement of density, diversity, and phylogeny, p. 102-114. In C. J. Hurst (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 35.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1985. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl. Environ. Microbiol. 49:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, M. T., C. M. Preston, F. P. Chavez, and E. D. DeLong. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat. Microbiol. Ecol. 24:117-127. [Google Scholar]
- 37.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 38.Urbach, E., K. L. Versin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 39.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 40.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Höfle. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinbauer, M. G., and M. G. Höfle. 1998. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl. Environ. Microbiol. 64:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinbauer, M. G., and M. G. Höfle. 1998. Distribution and life strategies of two bacterial populations in a eutrophic lake. Appl. Environ. Microbiol. 64:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wetzel, R. G. 2001. Limnology, lake and river ecosystems, 3rd ed. Academic Press, San Diego, Calif.