Abstract
We isolated a bacterial strain, Agrobacterium radiobacter P230, which can hydrolyze a wide range of organophosphate (OP) insecticides. A gene encoding a protein involved in OP hydrolysis was cloned from A. radiobacter P230 and sequenced. This gene (called opdA) had sequence similarity to opd, a gene previously shown to encode an OP-hydrolyzing enzyme in Flavobacterium sp. strain ATCC 27551 and Brevundimonas diminuta MG. Insertional mutation of the opdA gene produced a strain lacking the ability to hydrolyze OPs, suggesting that this is the only gene encoding an OP-hydrolyzing enzyme in A. radiobacter P230. The OPH and OpdA proteins, encoded by opd and opdA, respectively, were overexpressed and purified as maltose-binding proteins, and the maltose-binding protein moiety was cleaved and removed. Neither protein was able to hydrolyze the aliphatic OP malathion. The kinetics of the two proteins for diethyl OPs were comparable. For dimethyl OPs, OpdA had a higher kcat than OPH. It was also capable of hydrolyzing the dimethyl OPs phosmet and fenthion, which were not hydrolyzed at detectable levels by OPH.
Synthetic organophosphates (OPs) are used widely as insecticides in agriculture. OPs contain three phosphoester linkages and are hence often termed phosphotriesters. The phosphorus is also linked by a double bond to either an oxygen (P=O) in oxon OPs or a sulfur (P=S) in thion OPs. These insecticides are potent acetylcholinesterase (AchE) inhibitors, and various clinical effects can occur due to OP poisoning in humans. In general, hydrolysis of one of the phosphoester bonds reduces the toxicity of an OP, and in the case of parathion (O,O-diethyl p-nitrophenyl phosphorothioate), a 100-fold reduction in toxicity occurs (37, 38).
Enzymatic detoxification of OPs has become the focus of many studies because other means of removing OP residues are impractical or costly or are themselves environmentally hazardous. Enzymes from insect species resistant to OPs have been identified and considered for use in bioremediation (27). However, these enzymes are capable of hydrolyzing only oxon OPs and function at rates several orders of magnitude below the diffusion-limited maximum rate (28). Bacterial enzymes have also received considerable attention and may have advantages in terms of broader substrate specificities (both oxon and thion OPs) and superior kinetics (10, 13). The most widely studied bacterial enzyme is the OPH (organophosphorus-hydrolyzing) protein.
OPH is a zinc-containing homodimeric protein found in the membrane of Flavobacterium sp. strain ATCC 27551 and Brevundimonas diminuta MG (5, 13). OPH is capable of hydrolyzing a wide range of oxon and thion OPs (13) and hydrolyzes paraoxon at a rate approaching the diffusion limits (6, 35). The OPH enzyme is encoded by the opd gene. Other opd-containing organisms (for example, a Pseudomonas strain) have been identified by using this gene in Southern hybridization analysis (8), while other OP-hydrolyzing organisms clearly do not contain the opd gene (10, 11). Flavobacterium sp. strain ATCC 27551 and B. diminuta MG contain identical opd genes, but it is not clear how this has occurred as the genes are on very different plasmids (17, 25). Another class of OPs are the prolidase-type enzymes identified in Alteromonas spp. These enzymes are better at detoxifying the nerve gas reagents sarin and soman than the insecticidal OPs (10), and therefore OPH and homologues of this enzyme are better suited to bioremediation of insecticidal OPs.
We previously isolated a bacterium capable of hydrolyzing coumaphos (3-chloro-7-diethoxy phosphino thioloxy-4-methyl coumarin) from an enrichment culture containing OPs as the sole phosphorus source (16). In this paper we describe identification of this organism and characterization of the gene-enzyme system responsible for its OP-hydrolyzing activity. We also demonstrate that the protein produced by this strain is well suited for bioremediation of dimethyl OPs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown on a modified Luria-Bertani medium (16). Escherichia coli was grown at 37°C, and Agrobacterium species were grown at 28°C. When used in the medium for E. coli, ampicillin, kanamycin, and tetracycline were used at concentrations of 100, 25, and 10 μg/ml, respectively. When included in the medium for growth of Agrobacterium, rifampin, kanamycin, and tetracycline were included at concentrations of 100, 25, and 1 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH10β | Host for α-complementation cloning vectors | Gibco BRL |
| S17-1 | Mobilizing strain; carries chromosomally integrated derivative of RP4 | 40 |
| JM109 λpir | Host for α-complementation cloning vectors; lysogenized with phage lambda carrying the pir gene; allows replication of R6K-based suicide vectors | 29 |
| S17-1 λpir | S17-1 lysogenized with λpir | 30 |
| Agrobacterium strains | ||
| A. tumefaciens C58 | Wild type, OP− Rifr | 44 |
| A. radiobacter P230 | Wild type, OP+ Rifr | 16 |
| A. radiobacter Par− | OpdA− | This study |
| Plasmids | ||
| pBluescript KS+ (pBS) | Cloning vector, Apr | Stratagene |
| pJP5603 | R6K-based suicide vector | 29 |
| p65 | 12-kb Sau3AI partial fragment of A. radiobacter P230 genome in pBS | This study |
| pH2 | 4-kb HindIII fragment of p65 in pBS | This study |
| pB1 | 8-kb BamHI fragment of p65 in pBS | This study |
| pP1 | 6-kb PstI fragment of pB1 in pBS | This study |
| pJPH2 | 4-kb HindIII fragment from pH2 in pJP5603 | This study |
| pJPH2Sp | pJPH2 with NotI Ω Spr from pUI1188Sp in NotI site in opdA | This study |
| pUI1188Sp | ΩSpr in pBS | A. Suwanto |
| pMAL-c | Overexpression vector for MBP fusion proteins | New England Biolabs |
| pMAL-c2X | As for pMAL-c, with different cloning sites | New England Biolabs |
| pmal-opdA | malE::opdA translational fusion in pMAL-c | This study |
| pFmal | malE::opd translational fusion in pMAL-c2X | This study |
| pR751::Tn813 | Conjugative, cointegrative plasmid | 3 |
Ap, ampicillin; Rif, rifampin; ΩSpr, spectinomycin-resistant cassette bordered by transcription terminators.
Bacterial identification.
Isolate P230 is a gram-negative, catalase-positive, oxidase-positive, rod-shaped bacterium (tests were performed by using standard methods [19, 20]). Its 16S rRNA gene was PCR amplified from chromosomal DNA (extracted by the method of Rainey et al. [32]) by using the 27f and 1492r universal primers (23). Sequencing of PCR fragments was performed by using the 27f, 530f, 1100r, and 1492r universal primers (23) after purification of the PCR products with a QIAquick PCR purification kit (Qiagen).
DNA manipulation.
Routine DNA manipulations were carried out as described by Sambrook et al. (34). Isolation of chromosomal DNA from Agrobacterium radiobacter P230 was performed as described by Gardiner et al. (14). A size-fractionated genomic library was constructed from 10- to 15-kb DNA fragments (obtained from a partial Sau3AI digest) cloned into the BamHI site of plasmid pBluescript KS+ (pBS) and transformed into E. coli DH10β. Triparental matings of the library clones were performed by using the method of Bonnett et al. (2) with the conjugative cointegrative plasmid pR751::Tn813. Insertional mutants were detected by PCR by using opdA-specific primers listed below with extracted chromosomal DNA (32). The GenBank accession number for the opdA gene is AY043245.
Assays and biochemical techniques.
Cell extracts were prepared as previously described (16). Protein concentrations were determined by the method of Bradford (4) by using bovine serum albumin as a standard. For more accurate determinations of purified protein concentrations, the method of Gill and von Hippel (15) was employed. Hydrolysis of coumaphos, hydrolysis of coroxon, and hydrolysis of O,O-dimethyl 4-methyl umbelliferyl phosphate (dMUP) were measured by monitoring the formation of fluorescent products (16, 33; A. L. Devonshire, R. Heidari, K. L. Bell, P. M. Campbell, B. E. Campbell, W. A. Odgers, J. G. Oakeshott, and R. J. Russell, submitted for publication). Hydrolysis of parathion, hydrolysis of methyl parathion, and hydrolysis of paraoxon were measured spectrophotometrically by monitoring the production of p-nitrophenol at 405 nm (13). Hydrolysis of fenthion was measured spectrophotometrically by determining the loss of fenthion (at 252 nm) (18), while hydrolysis of phosmet and hydrolysis of malathion was measured by quantifying the formation of thiol groups produced during hydrolysis by using Ellman's reagent (22). Hydrolysis of diazinon and hydrolysis of chlorfenvinphos (CVP) were measured by using the radiometric partition assay of Campbell et al. (7). All assays were performed in 50 mM Tris-HCl (pH 8.0) at 25°C. Sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (ratio of acrylamide to bisacrylamide, 30:1) was performed by the method of Laemmli (21).
Construction of plasmids for OPH and OpdA overexpression and purification.
OPH and OpdA were overexpressed in E. coli DH10β by using the pMAL protein fusion and purification system, which results in expression of a maltose-binding protein (MBP) fusion protein (New England Biolabs). To construct an MBP-OPH overexpression plasmid, the opd gene (26) without the signal peptide domain was amplified by a PCR. The upstream and downstream oligonucleotide primers, 5′GATCGTGGATCCTCGATCGGCACAGGCGATCGG and 5′GATCGTAAGCTTTCATGACGCCCGCAAGGTCGG, respectively, were designed to contain a BamHI restriction site at the opd start codon and a HindIII restriction site at the stop codon (underlined bases). The PCR fragment was subsequently cloned into the BamHI-HindIII restriction sites of pMAL-c2X (New England Biolabs) to generate the recombinant plasmid pFmal. An MBP-OpdA overexpression plasmid was constructed in a similar way. The opdA gene was amplified by PCR by using upstream and downstream primers, 5′GATCGTCTGCAGCCAATCGGTACAGGCGATCTG and 5′GATCGTAAGCTTTCATCGTTCGGTATCTTGACGGGGAAT, respectively, with a PstI site at the start codon and a HindIII site at the stop codon (underlined bases). The PCR fragment was subsequently cloned into the PstI-HindIII sites of pMAL-c (New England Biolabs) to generate the recombinant plasmid pmal-opdA.
Optimal production of MBP fusion proteins was obtained when mid-log-phase cells (optical density at 600 nm, 0.6) were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 5 h at 37°C. Harvested cells were disrupted by sonication, and the soluble fraction was loaded onto an amylose resin (New England Biolabs) equilibrated with 50 mM Tris-HCl (pH 7.5). MBP fusion proteins were eluted with 10 mM maltose in 50 mM Tris-HCl (pH 7.5). Fractions containing coumaphos-hydrolyzing activity were pooled and cleaved with Xa protease (10 μg/ml; New England Biolabs) for 5 h. The cleaved fractions were then placed on a DEAE-Sepharose ion-exchange column. Cleaved OPH and OpdA proteins were not bound to the resin and eluted with the void volume. This purification process removed the Xa protease, as well as the MBP moiety. Fractions from the collected sample appeared to be homogeneous, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (data not shown). Approximately 35 μg of purified protein was obtained from 500 ml of culture.
Chemicals.
Radiolabeled (specific activity, 306.5 MBq/mmol) CVP {2-chloro-1-(2,4-dichlorophenyl)vinyl di-[14C]ethyl phosphate} was obtained from Internationale Isotope München. Fenthion {O,O-dimethyl O-[3-methyl 4-(methylthio)phenyl] phosphorothioate} was a gift from G. W. Levot, Department of Agriculture, New South Wales, Australia. Malathion [O,O-dimethyl S-(1,2-dicarbethoxyethyl) phosphorodithioate], methyl parathion (O,O-dimethyl O-p-nitrophenyl phosphorothioate), and parathion (O,O-diethyl p-nitrophenyl phosphorothioate) were obtained from Riedel-de Haan AG, Seelze, Germany. Coumaphos (3-chloro-4-methyl-7-coumarinyl diethyl phosphorothioate) was a gift from Bayer, Bayerwerk, Germany. Coroxon (3-chloro-4-methyl-7-coumarinyl diethyl phosphate), radiolabeled (14.8 MBq/mmol) diazinon {O,O-di-[14C]ethyl-O-(2-isopropyl-4-methyl-6-pyrimidinyl)-phosphorothioate}, and phosmet {S-[(1,3-dihydro-1,3-dioxo-2H-isoindol-2-yl)methyl] O,O-dimethyl phosphorodithioate} were purchased from Alltech. dMUP was synthesized by Alan Devonshire (unpublished data).
RESULTS AND DISCUSSION
Identification of isolate P230.
The sequence of approximately 1,320 bp of the 16S rRNA gene of P230 was 100% identical to that of the 16S rRNA gene of A. radiobacter LMG383 (GenBank accession no. ara130719), 99.7% similar to that of the 16S rRNA gene of Agrobacterium sp. strain LMG11936 (GenBank accession no. asp130721), 99.5% similar to that of the 16S rRNA gene of Agrobacterium sp. strain MSMC211 (GenBank accession no. asaj4859), and 99.3% similar to that of the 16S rRNA gene of Agrobacterium sp. strain LMG11915 (GenBank accession no. asp130720) (calculated by using the FASTA algorithm [29]). Use of the Biolog system (Oxoid) showed that isolate P230 was capable of using glucose, sucrose, and ornithine as carbon sources, suggesting that it was most similar to either Agrobacterium tumefaciens biovar 1 or A. radiobacter biovar 1, depending on the presence of a tumor-inducing plasmid. Attempts to isolate a plasmid(s) from isolate P230 were not successful. Tumor-inducing ability was also tested on tomato seedlings by using the method of Lippincott and Heberlein (24), and there was no evidence of tumors after a period of 4 weeks. Isolate P230 was therefore designated a strain of A. radiobacter biovar 1.
Functional cloning of the gene encoding an OP-hydrolyzing enzyme.
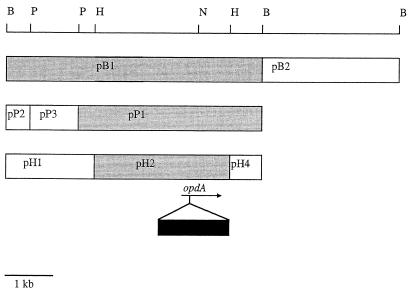
As it was not known whether the gene of interest would be expressed in E. coli, a library of A. radiobacter P230 genomic DNA fragments was constructed in pBS in E. coli and transferred via the conjugative cointegrative plasmid pR751::Tn813 (contained in E. coli JM109 λpir) into A. tumefaciens strain C58, which possessed negligible parathion- and coumaphos-hydrolyzing activity. Approximately 350 transformants from the genomic library of A. radiobacter P230 were transferred by using this triparental mating procedure. The mating mixtures were then assayed for coumaphos-hydrolyzing activity. One transformant (p65) exhibited coumaphos-hydrolyzing activity in the mating mixture. This clone also conferred coumaphos-hydrolyzing activity on E. coli DH10β; the activity was 3.30 ± 0.07 nmol of chlorferon produced/min/mg of protein (mean ± standard error), compared with 0.78 ± 0.04 nmol of chlorferon produced/min/mg of protein in control extracts of E. coli DH10β(pBS). While clone p65 contained approximately 12 kb of P230 chromosomal DNA, coumaphos-hydrolyzing activity was confined to a 4-kb HindIII fragment (Fig. 1). The activity associated with the 4-kb HindIII fragment in E. coli depended on the orientation of the fragment in pBS (Fig. 1), suggesting that it was dependent on the lacZ promoter in pBS for expression. This fragment was sequenced.
FIG. 1.
Subcloning of the opdA gene and insertional inactivation. The positions of restriction enzyme sites used for subcloning are shown at the top, and the boxes represent fragments generated by the restriction enzymes. The shaded boxes represent fragments that conferred OP-hydrolyzing activity on E. coli when they were cloned into pBluescript; the left side is closest to the T7 sequencing primer in pBluescript. The arrow represents the opdA gene and indicates its direction of transcription. The solid box represents the spectinomycin resistance cassette placed into the NotI site in opdA in pJPH2Sp. B, BamHI; P, PstI; N, NotI; H, HindIII.
Sequence of the gene encoding the OP-hydrolyzing enzyme.
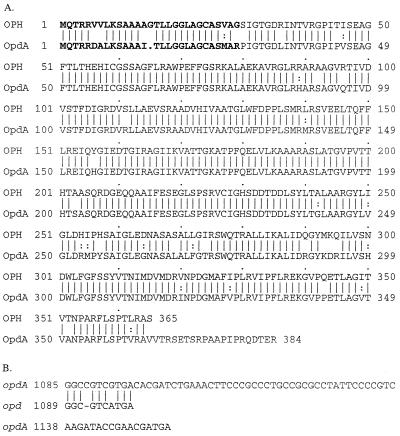
One open reading frame in the 4-kb HindIII fragment was identified that was 88.4% identical at the nucleotide level to the opd phosphotriesterase gene previously identified in Flavobacterium sp. strain ATCC 27551 and B. diminuta MG (26). The amino acid sequence of its putative translation product was 94.2% similar and 90.0% identical to the OPH sequence (26). This gene was termed opdA (opd from Agrobacterium), and an alignment of its inferred amino acid sequence with that of the Flavobacterium-B. diminuta OPH is shown in Fig. 2A. Some notable differences were observed between the Flavobacterium-B. diminuta OPH sequence and the sequence of OpdA from A. radiobacter P230. The first difference is that there is one less amino acid in the putative signal sequence of OpdA. The second is that there is a frameshift near the 3′ end of the opdA gene (Fig. 2B), resulting in an additional 16 amino acids in OpdA. This region has been sequenced five times in both directions with multiple sequencing primers to ensure that the extra base in opdA is not a sequencing error. Our data thus suggest that OpdA is a 384-amino-acid protein with a molecular mass of approximately 35 kDa when it is cleaved from its signal peptide.
FIG. 2.
(A) Alignment of the amino acid sequences of OPH and OpdA. The secretion signals are indicated by boldface type. The vertical lines indicate identical residues, while the colons indicate similar residues. (B) Alignment of DNA sequences at the C termini of the two genes.
One obvious sequence difference between OPH and OpdA is an extended C terminus. The C terminus of OPH was previously shown not to interact with the substrate analogue 4-methylbenzylphosphonate in a crystallized structure of OPH, but rather to lie on the outside of the protein structure (43). It therefore seems unlikely that the extended C terminus in OpdA contributes directly to differences in substrate selectivity. It may, however; confer different physical properties on OpdA.
Plasmid pJPH2 (4-kb HindIII fragment in pJP5603) conferred coumaphos-hydrolyzing activity of 3.7 ± 0.4 nmol/min/mg on E. coli JM109 λpir. Insertion of the spectinomycin-resistant cassette into the NotI site of opdA (Fig. 1) reduced the OP-hydrolyzing activity to that seen for the vector-only control (0.031 ± 0.003 nmol/min/mg). Note that this activity is lower than the background phosphotriesterase activity for E. coli strain DH10β described above; we and others have observed variable activities in different E. coli strains (37), but the values are well within the range of variation for background phosphotriesterase activity. The opdA gene is at one end of the 4-kb HindIII fragment, and no open reading frames have been identified downstream of opdA. We therefore interpret the loss of activity as the result of an insertion in opdA and not as the result of a loss of expression of any other gene. The construct was then used to inactivate opdA in A. radiobacter P230, to create A. radiobacter Par−. This mutant also exhibited reduced parathion-, methyl parathion-, coumaphos-, coroxon-, and paraoxon-hydrolyzing activities (approximately 0.42% ± 0.07% of the wild-type activities). This demonstrates that the opdA gene is the only gene in A. radiobacter P230 encoding an enzyme capable of hydrolyzing OPs.
The opd and opdA genes have been identified in organisms isolated from different geographic locations, from the Philippines (39) to the United States (8, 36) and Australia (16). Also, they have been found in four taxonomically distinct organisms. Presumably, these genes were acquired from the same ancestral organism. Since distantly related opd-like genes have been identified in organisms in genome-sequencing projects (1, 31), the native host of the opd and opdA genes may well be a ubiquitous soil organism. Although no plasmids could be identified in A. radiobacter P230 and the opdA gene appears to be chromosomally located, the inability of A. tumefaciens C58 to hydrolyze OPs and the lack of hybridization of an opd probe to A. tumefaciens C58 bulk DNA suggest that A. radiobacter P230 acquired opdA by a lateral gene transfer mechanism. This is not uncommon, as other plasmid-borne genes (for example, the TOL genes involved in toluene catabolism [41]) have also been found chromosomally. Movement of the TOL genes through soil populations has been suggested to occur via transposition (42). This may be the means by which the opd genes having the same DNA sequence and encoding the same protein sequence were acquired by B. diminuta MG and Flavobacterium sp. strain ATCC 27551 (17), as well as the means by which a related gene was acquired by A. radiobacter P230.
Analysis of the substrate specificity of OpdA and comparison with OPH.
To assess the substrate range of OpdA, the enzyme was purified and analyzed in parallel with its counterpart from Flavobacterium sp. strain ATCC 27551, OPH. The substrates tested were paraoxon, parathion, methyl parathion, malathion, CVP, phosmet, diazinon, fenthion, coumaphos, coroxon, and dMUP (Fig. 3). Note that the kcat values for OPH reported here are much lower than those reported previously (12, 13), probably because we did not add metals to the assay mixtures or during growth, both of which can increase the specific activity (12). No activity against malathion or CVP could be detected for either OPH or OpdA. In general, OpdA appeared to have a slightly higher Km for the diethyl substrates, but the kcat values of the two enzymes against the diethyl substrates were comparable. All four dimethyl substrates tested were hydrolyzed by OpdA, while OPH hydrolyzed methyl parathion and dMUP but apparently did not hydrolyze fenthion or phosmet. The inability of OPH to hydrolyze fenthion has been reported previously (5). Furthermore, the kcat values for OpdA against methyl parathion and dMUP were significantly higher than those for OPH; in the case of dMUP the values were approximately four times higher, and in the case of methyl parathion the values were almost 20 times higher. If the ratio for phosmet was similar to the latter value, only 7.2 mol of phosmet per mol of OPH would be hydrolyzed in a 24-h period. Given the amounts of enzyme used in assays in this study (in the nanomolar to micromolar range), this amount of hydrolysis would not be observed by our methods.
FIG. 3.
Kinetic constants for hydrolysis of OPs by OPH and OpdA. nd, not detected.
Chen-Goodspeed et al. (9) identified three essential substrate binding sites in OPH. These were termed the small and large subsites, which bound the diethyl or dimethyl moieties of the substrate, and the leaving group subsite, which bound the aromatic leaving group. OpdA retains all the amino acid residues important in substrate binding in OPH in the small subsite and the leaving group subsite. However, OpdA differs in three of the four residues in the large subsite. These residues are H254R, H254Y, and L271F. It is conceivable that the change of histidine to the much larger residue tyrosine and the change of leucine to the larger residue phenylalanine might close in the binding site, enhancing the activity of OpdA with dimethyl substrates compared to the activity of OPH. The kinetics of an H254R mutation of OPH made by diSioudi and coworkers are consistent with this possibility (12). The OPH mutant had an almost 20-fold decrease in the ability to hydrolyze diisopropyl fluorophosphate (DFP) (12). The leaving group of DFP contains two isopropyl groups and is considerably larger than the dimethyl leaving group of methyl parathion. It could be anticipated that OpdA also has a reduced capacity to hydrolyze DFP as a result of a smaller large subsite. By inference, OpdA may not be as effective in degrading nerve agents as OPH since these agents contain bulkier alkyl groups (e.g., isopropyl chains in the case of soman) than the insecticidal OPs suitable for detoxification by OpdA.
Constriction of the large subsite in OpdA compared to the OPH large subsite might also affect the stereospecificity of the enzyme. While most insecticidal OPs have either dimethyl or diethyl substituents, a few have a mixture of methyl and ethyl or, in exceptional cases, propyl groups. The stereospecificity of OPH for these asymmetric OPs has been attributed to the size difference between the small and large subsites (9). This specificity might be reduced in OpdA because of the proposed constriction in its large subsite.
Conclusions.
OPH is believed to be an ideal enzyme for bioremediation of insecticidal OPs because of its ability to hydrolyze the compounds at a rate approaching the diffusion limits (35), and it is by far the best enzyme of the phosphotriesterases identified (10, 24). In this study we isolated a naturally occurring variant of OPH that has a broader substrate range for insecticidal OPs and kinetics superior to those of OPH for some substrates. OpdA is capable of hydrolyzing dimethyl substrates at a higher rate than OPH, and the rate of hydrolysis of diethyl OPs is not affected; therefore, OpdA is a better enzyme for practical bioremediation of insecticidal OPs.
Acknowledgments
We thank Susan Dorrian and Michelle Williams for their technical assistance.
This research was supported by Orica Australia Ltd. and the Australian Horticulture Research and Development Corporation.
REFERENCES
- 1.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collador-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 2.Bonnett, T. C., P. Cobine, R. E. Sockett, and A. G. McEwan. 1995. Phenotypic characterization and genetic complementation of dimethylsulfoxide respiratory mutants of Rhodobacter sphaeroides and Rhodobacter capsulatus. FEMS Microbiol. Lett. 133:163-168. [DOI] [PubMed] [Google Scholar]
- 3.Bowen, A. R. S., and J. M. Pemberton. 1985. Mercury resistance transposon Tn813 mediates chromosome transfer in Rhodopseudomonas sphaeroides and intergeneric transfer in pBR322, p. 105-115. In D. R. Helsinki, S. N. Cohen, D. B. Clewell, D. A. Jackson, and A. Hollaender (ed.), Plasmids in bacteria. Plenum Press, New York, N.Y.
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brown, K. A. 1980. Phosphotriesterases of Flavobacterium sp. Soil Biol. Biochem. 12:105-112. [Google Scholar]
- 6.Caldwell, S. R., J. R. Newcomb, K. A. Schlecht, and F. M. Raushel. 1991. Limits of diffusion in the hydrolysis of substrates by the phosphotriesterase from Pseudomonas diminuta. Biochemistry 30:7438-7444. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, P. M., R. D. Newcomb, R. J. Russell, and J. G. Oakeshott. 1998. Two different amino acid substitutions in the ali-esterase, E3, confer alternative types of organophosphorus insecticide resistance in the sheep blowfly, Lucilia cuprina. Insect Biochem. Mol. Biol. 28:139-150. [Google Scholar]
- 8.Chaudhry, G. R., A. N. Ali, and W. B. Wheeler. 1988. Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the opd gene from a Flavobacterium sp. Appl. Environ. Microbiol. 54:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen-Goodspeed, M., M. A. Sogorb, F. Wu, S.-B. Hong, and F. M. Raushel. 2001. Structural determinants of the substrate and stereochemical specificity of phosphotriesterase. Biochemistry 40:1325-1331. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, T. C., S. P. Harvey, and A. N. Stroup. 1993. Purification and properties of a highly active organophosphorus acid anhydrolase from Alteromonas undina. Appl. Environ. Microbiol. 59:3138-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dave, K. I., C. E. Miller, and J. R. Wild. 1993. Characterization of organophosphorus hydrolases and the genetic manipulation of the phosphotriesterase from Pseudomonas diminuta. Chem.-Biol. Interact. 87:55-68. [DOI] [PubMed] [Google Scholar]
- 12.diSioudi, B., J. K. Grimsley, K. Lai, and J. R. Wild. 1999. Modification of near active site residues in organophosphorus hydrolase reduces metal stoichiometry and alters substrate specificity. Biochemistry 38:2866-2872. [DOI] [PubMed] [Google Scholar]
- 13.Dumas, D. P., S. R. Caldwell, J. R. Wild, and F. M. Raushel. 1989. Purification and properties of the phosphotriesterase from Pseudomonas diminuta. J. Biol. Chem. 264:19659-19665. [PubMed] [Google Scholar]
- 14.Gardiner, A. T., R. C. MacKenzie, S. J. Barrett, K. Kaiser, and R. G. Cogdell. 1996. The purple photosynthetic bacterium Rhodopseudomonas acidophila contains multiple puc peripheral antenna complex (LH2) genes: cloning and initial characterization of four α/β pairs. Photosynth. Res. 49:223-235. [DOI] [PubMed] [Google Scholar]
- 15.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 16.Harcourt, R. L., I. Horne, T. D. Sutherland, B. D. Hammock, R. J. Russell, and J. G. Oakeshott. 2002. Development of a simple and sensitive fluorimetric method for isolation of coumaphos-hydrolysing bacteria. Lett. Appl. Microbiol. 34:263-268. [DOI] [PubMed] [Google Scholar]
- 17.Harper, L. L., S. McDaniel, C. E. Miller, and J. R. Wild. 1988. Dissimilar plasmids isolated from Pseudomonas diminuta MG and Flavobacterium sp. (ATCC 27551) contain identical opd genes. Appl. Environ. Microbiol. 54:2586-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim, F. B., and J. C. Cavagnol. 1966. Ultraviolet spectrophotometric method for fenthion. J. Agric. Food Chem. 14:369-371. [Google Scholar]
- 19.Kovac, N. 1956. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703.. [DOI] [PubMed] [Google Scholar]
- 20.Krieg, N. R., and J. G. Holt (ed.). 1984. Bergey's manual of systematic bacteriology, vol. 1. The Williams and Wilkins Co., Baltimore, Md.
- 21.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lai, K., N. J. Stolowich, and J. R. Wild. 1995. Characterization of P-S bond hydrolysis in organophosphorothioate pesticides by organophosphorus hydrolase. Arch. Biochem. Biophys. 318:59-64. [DOI] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 24.Lippincott, J. A., and G. T. Heberlein. 1965. The quantitative determination of the infectivity of Agrobacterium tumefaciens. Am. J. Bot. 52:856-863. [PubMed] [Google Scholar]
- 24a.Mulbry, W. W. 1992. The aryldialkylphosphatase-encoding gene adpB from Nocardia sp. strain B-1: cloning, sequencing and expression in Escherichia coli. Gene 121:149-153. [DOI] [PubMed] [Google Scholar]
- 25.Mulbry, W. W., P. C. Kearney, J. O. Nelson, and J. S. Karns. 1987. Physical comparison of parathion hydrolase plasmids from Pseudomonas diminuta and Flavobacterium sp. Plasmid 18:173-177. [DOI] [PubMed] [Google Scholar]
- 26.Mulbry, W. W., and J. S. Karns. 1989. Parathion hydrolase specified by the Flavobacterium opd gene: relationship between the gene and the protein. J. Bacteriol. 171:6740-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newcomb, R. D., P. M. Campbell, D. L. Ollis, E. Cheah, R. J. Russell, and J. G. Oakeshott. 1997. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. USA 94:7464-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oakeshott, J. G., C. Claudianos, R. J. Russell, and G. C. Robin. 1999. Carboxyl/cholinesterases: a case study of the evolution of a successful multigene family. Bioessays 21:1031-1042. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 31.Philipp, W. J., S. Poulet, K. Eiglmeier, L. Pascopella, V. Balasubramanian, B. Heym, S. Bergh, B. R. Bloom, W. R. Jacobs, Jr., and S. T. Cole. 1996. An integrated map of the genome of the tubercule bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc. Natl. Acad. Sci. USA 93:3132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rainey, F. A., M. Dorsch, H. W. Morgan, and E. Stackebrandt. 1992. 16S rDNA analysis of Spirochaeta thermophila: phylogenetic position and implications for the systematics of the order Spirochaetales. Syst. Appl. Microbiol. 15:197-202. [Google Scholar]
- 33.Roth, M. 1969. Fluorimetric assay of enzymes. Methods Biochem. Anal. 17:189-285. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Scanlan, C. S., and R. C. Reid. 1995. Evolution in action. Chem. Biol. 2:71-75. [DOI] [PubMed] [Google Scholar]
- 36.Serdar, C. M., D. T. Gibson, D. M. Munnecke, and J. H. Lancaster. 1982. Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Appl. Environ. Microbiol. 44:246-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serdar, C. M., and D. T. Gibson. 1985. Enzymatic hydrolysis of organophosphates: cloning and expression of a parathion hydrolase gene from Pseudomonas diminuta. Bio/Technology 3:567-571. [Google Scholar]
- 38.Serdar, C. M. December 1996. Hydrolysis of cholinesterase inhibitors using parathion hydrolase. U.S. patent 5,589,386.
- 39.Sethunathan, N., and T. Yoshida. 1973. A Flavobacterium that degrades diazinon and parathion. Can. J. Microbiol. 19:873-875. [DOI] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 41.Sinclair, M. I., P. C. Maxwell, B. R. Lyon, and B. W. Holloway. 1986. Chromosomal location of TOL plasmid DNA in Pseudomonas putida. J. Bacteriol. 168:1302-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuda, M., and T. Iino. 1987. Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid, pWW0. Mol. Gen. Genet. 210:270-276. [DOI] [PubMed] [Google Scholar]
- 43.Vanhooke, J. L., M. M. Benning, F. M. Raushel, and H. M. Holden. 1996. Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methyl benzyl phosphonate. Biochemistry 35:6020-6025. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerer, R. P., R. H. Hamilton, and C. Pootjes. 1966. Isolation and morphology of temperate Agrobacterium tumefaciens bacteriophage. J. Bacteriol. 92:746-750. [DOI] [PMC free article] [PubMed] [Google Scholar]