Abstract
The nucleotide sequence encoding the crystalline bacterial cell surface (S-layer) protein SbpA of Bacillus sphaericus CCM 2177 was determined by a PCR-based technique using four overlapping fragments. The entire sbpA sequence indicated one open reading frame of 3,804 bp encoding a protein of 1,268 amino acids with a theoretical molecular mass of 132,062 Da and a calculated isoelectric point of 4.69. The N-terminal part of SbpA, which is involved in anchoring the S-layer subunits via a distinct type of secondary cell wall polymer to the rigid cell wall layer, comprises three S-layer-homologous motifs. For screening of amino acid positions located on the outer surface of the square S-layer lattice, the sequence encoding Strep-tag I, showing affinity to streptavidin, was linked to the 5′ end of the sequence encoding the recombinant S-layer protein (rSbpA) or a C-terminally truncated form (rSbpA31-1068). The deletion of 200 C-terminal amino acids did not interfere with the self-assembly properties of the S-layer protein but significantly increased the accessibility of Strep-tag I. Thus, the sequence encoding the major birch pollen allergen (Bet v1) was fused via a short linker to the sequence encoding the C-terminally truncated form rSpbA31-1068. Labeling of the square S-layer lattice formed by recrystallization of rSbpA31-1068/Bet v1 on peptidoglycan-containing sacculi with a Bet v1-specific monoclonal mouse antibody demonstrated the functionality of the fused protein sequence and its location on the outer surface of the S-layer lattice. The specific interactions between the N-terminal part of SbpA and the secondary cell wall polymer will be exploited for an oriented binding of the S-layer fusion protein on solid supports to generate regularly structured functional protein lattices.
Crystalline bacterial cell surface layers (S-layers) represent the outermost cell envelope component of many bacteria and archaea (35, 37, 38). S-layers are composed of identical protein or glycoprotein subunits, and they completely cover the cell surface during all stages of bacterial growth and division. The S-layer subunits assemble into either oblique, square, or hexagonal lattices. In the case of Bacillaceae, the N-terminal part is involved in anchoring the S-layer subunits via a distinct type of secondary cell wall polymer (SCWP) to the rigid cell wall layer (2, 5, 21, 25, 34, 35).
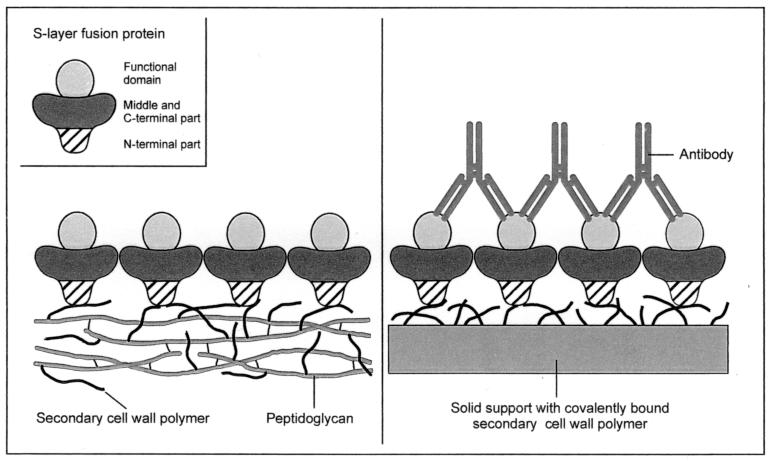
S-layers are unique biomaterials with properties most relevant for applications in molecular nanotechnology, nanobiotechnology, and biomimetics (38). Many S-layer proteins recrystallize into regularly structured monolayers on solid supports, such as silicon wafers, gold chips, and silanized glass or plastic materials, as well as on Langmuir lipid films, on liposomes (20, 23), and at the air-water interface. Pores passing through S-layer lattices are of identical size and morphology, and functional groups show a regular distribution and high density. For production of S-layer-based biosensors (38), affinity microparticles (45), and solid-phase immunoassays (3, 4, 39), functional groups in the S-layer lattice were exploited as covalent binding sites for biologically active macromolecules, such as enzymes, antibodies, or ligands. As alternatives to the existing technology, namely, immobilization by chemical methods, genetic approaches are particularly attractive for incorporation of functional peptide sequences into S-layer proteins, which must be done at positions that do not interfere with their self-assembly properties and their interaction with SCWP. To guarantee that the integrated or fused functional sequence remains located on the outer surface of the S-layer lattice and available for further binding reactions, the specific interactions with the SCWP shall be exploited in future developments to achieve an oriented binding of S-layer fusion proteins on artificial supports. The concept of generating oriented functional protein lattices implies that SCWPs are attached to solid supports or that appropriate glycolipids comprising the smallest functional units of SCWPs are synthesized and incorporated into liposomes or Langmuir lipid films (Fig. 1).
FIG. 1.
Schematic drawing showing the formation of oriented functional S-layer lattices on peptidoglycan-containing sacculi and artificial supports coated with secondary cell wall polymer. In the case of peptidoglycan-containing sacculi, only the outer S-layer is shown.
Bacillus sphaericus represents a strictly aerobic group of mesophilic endospore-forming bacteria. The S-layer lattice of B. sphaericus CCM 2177 shows square symmetry with a center-to-center spacing of the morphological units of 13.1 nm. It is composed of identical subunits with an estimated relative molecular weight of 127,000. In a previous study, it was demonstrated that this S-layer protein recognizes the SCWP withthe structure →3)-[4,6-O-(1-carboxyethylidene)]∼0.5-β-d-ManpNAc-(1→4)-β-d-GlcpNAc-(1→ as a binding site in the rigid cell wall layer. The polymer chains consist of eight or nine disaccharide repeating units, and they are covalently linked to the peptidoglycan backbone (13).
So far, the genes encoding the S-layer proteins of B. sphaericus P-1 (6) and B. sphaericus 2362 (1) have been cloned and sequenced. In the present study, the sbpA gene, encoding the S-layer protein of B. sphaericus CCM 2177, was sequenced, cloned, and expressed in Escherichia coli. Furthermore, the accessibility of the C-terminal amino acids of the recombinant S-layer protein (rSbpA) and a C-terminally truncated form (rSbpA31-1068) was investigated by fusion of the sequence encoding the short affinity peptide Strep-tag I (AWRHPQFGG) (36) to the sequences of the appropriate S-layer gene forms. Strep-tag I confers binding activity towards streptavidin and Strep-Tactin. The latter is a genetically modified streptavidin with increased affinity to Strep-tag I and Strep-tag II (44). Exploiting the improved accessibility of the C-terminal amino acids of the C-terminally truncated form, the chimeric gene encoding an S-layer fusion protein comprising the sequence of Bet v1, the major birch pollen allergen, was cloned and expressed in E. coli. The obtained fusion protein, rSbpA31-1068/Bet v1, will be used for preparing Bet v1-specific immunoglobulin E (IgE)-binding monolayers, as required for biochip development.
MATERIALS AND METHODS
Growth of B. sphaericus CCM 2177, preparation of cell wall fragments, and isolation of the S-layer protein and peptidoglycan-containing sacculi.
B. sphaericus CCM 2177 was grown in continuous culture as described in a previous study (13). Preparation of cell wall fragments, isolation of the S-layer protein, purification of peptidoglycan-containing sacculi, and preparation of self-assembly products were done as described previously (7). Peptide mapping and N-terminal sequencing of the whole S-layer protein as well as N-terminal sequencing of selected proteolytic cleavage fragments were performed as described previously (8).
Other strains, plasmids, culture conditions, and DNA manipulations.
For cloning, E. coli TG1 was transformed with the plasmid pBluescript II KS(+) or pET28a. For expression, E. coli HMS174(DE3) was chosen as a host strain for derivatives of pET28a (Table 1) as described previously (41). E. coli was grown on Luria-Bertani medium (Gibco BRL Life Technologies) or on modified M9ZB medium (41) at 37°C. For selection of transformants harboring KS(+) or pET28a, kanamycin was added to the medium to a final concentration of 30 μg/ml. Chromosomal DNA of B. sphaericus CCM 2177 was prepared by using Genomic Tips 100 (Qiagen) according to the manufacturer's instructions. Digestion of DNA with restriction endonucleases, separation of DNA fragments by agarose gel electrophoresis, ligation of DNA fragments, and transformation procedures were performed as described previously (32). DNA fragments were recovered from agarose gels by using the Qiaex II Gel Extraction Kit (Qiagen).
TABLE 1.
Plasmid pET28a with various derivatives of the sbpA gene inserted
| pE28a derivative | Sequence encoded by insert | Cloning site |
|---|---|---|
| pSbpA1 | rSbpA | NdeI/BamHI |
| pSbpA2 | rSbpA/Strep-tag I | NcoI/XhoI |
| pSbpA3 | rSbpA31-1068/Strep-tag I | NcoI/XhoI |
| pSbpA4 | rSbpA31-1068/Bet v1 | NcoI/XhoI |
Isolation of the sbpA gene.
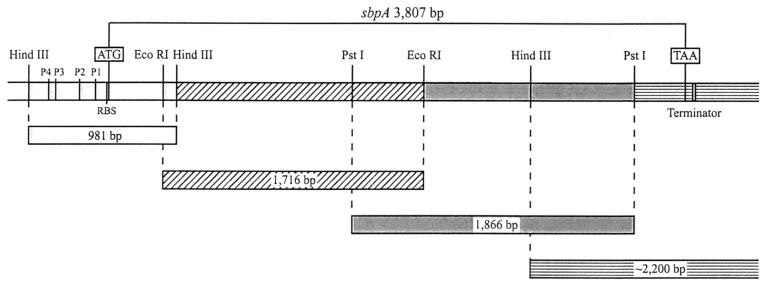
Twenty-four bases of the conserved DNA sequence of the secretion signals of the S-layer proteins of B. sphaericus P-1 (6) and B. sphaericus 2362 (1) were chosen for construction of the oligonucleotide primer sbpA1 (5′-ATC AGC AAC AGC TGC ATT AGT TGC-3′), which was used as a probe in hybridization studies to detect the sbpA gene. Isolation of the sbpA gene was performed by a PCR-based technique according to procedures described previously (17) (Fig. 2). PCR product sequencing of the four overlapping sbpA gene fragments was performed twice, once from each strand, by applying the dideoxy chain termination method described previously (33) with a Perkin-Elmer Applied Biosystems 377 DNA sequencer and the BigDye Terminator Cycle Sequencing Kit (Perkin-Elmer).
FIG. 2.
Schematic drawing showing the four chromosomal DNA fragments from B. sphaericus CCM 2177 used for isolation of the sbpA gene by a PCR-based technique. RBS, ribosome binding site.
Isolation of RNA and DNA and RNA hybridization.
Total RNA was isolated from exponentially growing cells of B. sphaericus CCM 2177 harvested from shaking flask cultures at an optical density of 0.78 under conditions described previously (17). Northern and Southern blotting were performed as described previously (32). For hybridization of Southern blots, oligonucleotides were 3′ tailed with digoxigenin-11-dUTP (DIG-dUTP) (Roche Molecular Biochemicals) and DNA probes were labeled with DIG-dUTP by incorporation during PCR. Hybridization was carried out according to the manufacturer's recommendations. Hybrids were detected using the DIG Luminescent Detection Kit (Roche Molecular Biochemicals). Northern blotting of total RNA from B. sphaericus CCM 2177 was performed as described previously (17). Hybridization of Northern blots was carried out with a 4,045-bp DNA fragment comprising the coding region of the sbpA gene randomly labeled with DIG-dUTP. The size of the sbpA mRNA was estimated from its mobility relative to those of the RNAs in the DIG-dUTP-labeled RNA Molecular Weight Marker II (size range, 1.5 to 6.9 kb; Roche Molecular Biochemicals).
Cloning of the genes encoding rSbpA, rSbpA/Strep-tag I, rSbpA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1.
For PCR amplification of the sbpA gene with chromosomal DNA of B. sphaericus CCM 2177 as the template, the oligonucleotide primers sbpA28 (5′-CGC GGA TCC CAT ATG GCG CAA GTA AAC GAC TAT AAC AAA ATC-3′) and sbpA29 (5′-CGC GGA TCC TTA TTT TGT AAT TGT TAC TGT TAA TTC AGC-3′), which introduced restriction sites (in boldface) for NdeI (at the 5′ end of the coding sequence) and BamHI (at the 3′ end), respectively, were constructed. For cloning, the gel-purified PCR fragment was inserted into the corresponding restrictions sites of plasmid pET28a, and the derivative pSbpA1 (Table 1) was established in E. coli TG1.
The genes encoding the SbpA derivatives rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I were amplified by PCR from chromosomal DNA of B. sphaericus CCM 2177 by using the oligonucleotide primer sbpA37 (forward primer for both genes) (5′-CGG AAT TCC ATG GCG CAA GTA AAC GAC TAT AAC AAA ATC-3′) and sbpA39 (reverse primer for the gene encoding rSbpA/Strep-tag I) (5′-GA CCG CTC GAG TTA ACC ACC GAA CTG CGG GTG ACG CCA CGC ACC ACC TTT TGT AAT TGT TAC TGT TAA TTC AGC-3′) or sbpA2a (reverse primer for the gene encoding rSbpA31-1068/Strep-tag I) (5′-GA CCG CTC GAG TTA ACC ACC GAA CTG CGG GTG ACG CCA CGC ACC ACC TTC TGA ATA TGC AGT AGT TGC TGC-3′). The primers sbpA39 and sbpA2a introduced the sequence of Strep-tag I (underlined) at the 3′ end of the coding sequence of sbpA. For cloning, NcoI and XhoI restriction sites (boldface) were introduced during PCR at the 5′ and 3′ ends, respectively. The resulting PCR fragments were cloned into pET28a to generate the new plasmids pSbpA2 and pSbpA3 (Table 1).
To obtain the desired chimeric PCR product encoding the S-layer fusion protein rSbpA31-1068/Bet v1, overlap extension PCR (11) was applied. For amplification of the sbpA derivative encoding rSbpA31-1068, DNA isolated from whole cells of B. sphaericus CCM 2177 was used as a template and the oligonucleotide primers sbpA37 (5′-CGG AAT TCC ATG GCG CAA GTA AAC GAC TAT AAC AAA ATC-3′) (primer 1) and sbpA6a (5′-GTA ATT GAA AAC ACC CAT ACC ACC TTC TGA ATA TGC AGT AGT TGC TGC C-3′) (primer 2) were chosen. Primer 1 introduced a NcoI restriction site (boldface) at the 5′ end of the coding sequence, and primer 2 contained an overlapping complementary Bet v1 gene sequence (underlined) at the 3′ end. For PCR amplification of the Bet v1 gene from plasmid pMW175 (12), the nucleotide primers sbpA7a (5′-ACT ACT GCA TAT TCA GAA GGT GGT ATG GGT GTT TTC AAT TAC GAA ACT GAG-3′) (primer 3) and spbA5a (5′-GAC CGC TCG AGT TAG TTG TAG GCA TCG GAG TGT G-3′) (primer 4), which introduced a complementary sbpA overlap (underlined) at the 5′ end and the restriction site XhoI (boldface) at the 3′ end of the coding sequence, were used. In primers containing the overlapping sequences, the sequence encoding two glycine residues (GGT GGT) was introduced as a spacer between rSbpA31-1068 and Bet v1. For amplification of the chimeric gene encoding the fusion protein rSbpA31-1068/Bet v1, the gel-purified PCR products from the PCRs described above were used as templates together with primer 1 and primer 4. For cloning in pET28a, the obtained PCR product was used to generate the new plasmid pSbpA4 (Table 1).
Expression of rSbpA, rSbpA/Strep-tag I, rSpbA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1.
The plasmid stability test and heterologous expression in E. coli HMS174(DE3) were performed as described previously (17). Samples (1.5 ml) of the E. coli HMS174(DE3) cultures were taken 1 to 5 h after induction of expression of the sbpA gene or its derivatives. Preparation of biomass samples and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were carried out as described previously (17). For electron microscopic investigation, whole cells of E. coli HMS174(DE3) were fixed and embedded in Spurr resin and subjected to ultrathin sectioning according to procedures described previously (28). Immunoblotting with a polyclonal rabbit antiserum raised against the S-layer protein of B. sphaericus CCM 2177 was performed as described previously (8). The presence of Bet v1 epitopes was checked by immunoreactivity with BIP1, a monoclonal mouse anti-Bet v1 antibody (15). The presence of Strep-tag I was checked by dot blot assays. For this purpose, 5 μl of solutions containing 600 μg of either rSbpA/Strep-tag I or rSbpA31-1068/Strep-tag I per ml in 0.1 M Tris-buffered saline (TBS) were dried on a nitrocellulose membrane. After blocking with 3% bovine serum albumin (BSA) in 0.1 M TBS for 1 h at 20°C, the membrane was incubated with a polyclonal rabbit antiserum raised against the Strep-tag (IBA-Strep-tag Detection System) (diluted 1:2,000 in wash buffer [0.1% Tween 20 in 0.1 M TBS]) for 1 h at 20°C and washed three times. Incubation with anti-rabbit IgG-alkaline phosphatase (ALP) conjugate (Sigma) (diluted 1:4,000 in wash buffer) was done for 1 h at 20°C. After three further wash steps, detection was accomplished by treatment with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium chloride (Roche). In addition, a Strep-Tactin-ALP conjugate (IBA-Strep-tag Detection System) was used to detect the presence of Strep-tag I.
Isolation and purification of rSbpA, rSbpA/Strep-tag I, rSbpA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1.
Isolation of all rSbpA forms was performed as described previously (16). For purification, aliquots (120 mg) of the lyophilized recombinant S-layer protein forms were suspended in 5 ml of 2 M guanidine hydrochloride (GHCl) in 50 mM Tris-HCl buffer (pH 7.2). After centrifugation at 16,000 × g for 5 min at 4°C, the supernatants were filtered through a 0.45-μm-pore-size RC membrane (Minisart RC 25), and the clear solutions were subjected to gel permeation chromatography (GPC) using a Superdex 200 column (Pharmacia) equilibrated in 2 M GHCl in 50 mM Tris-HCl buffer (pH 7.2) for separation. Fractions containing S-layer protein were pooled, dialyzed against distilled water for 18 h at 4°C, lyophilized, and stored at −20°C. In addition, rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I were purified with a Ready-to-Use Strep-Tactin Sepharose column (IBA-Strep-tag Purification System). The purities of rSbpA, rSbpA/Strep-tag I, rSbpA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1 were finally checked by SDS-PAGE.
Self-assembly properties of rSbpA, rSbpA/Strep-tag I, rSbpA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1 and recrystallization of soluble rSbpA forms on poly-l-lysine-coated electron microscopy (EM) grids
To investigate the ability of the various rSbpA forms to self-assemble, 3 mg of the GPC-purified rSbpA forms was dissolved in 1 ml of 5 M GHCl in 50 mM Tris-HCl buffer (pH 7.2) and the solutions were dialyzed against 10 mM CaCl2 in distilled water for 18 h at 4°C. Negative staining of the suspensions and freeze-drying were performed as described previously (29). To investigate the ability of soluble (monomeric and/or oligomeric) rSbpA to recrystallize on poly-l-lysine-coated copper grids, the GHCl extracts were dialyzed against distilled water for 18 h at 4°C. After centrifugation for 5 min at 16,000 × g, the clear supernatants containing nonassembled S-layer protein were incubated with poly-l-lysine (Sigma P2636)-coated copper grids by procedures described previously (29).
Recrystallization of rSbpA, rSbpA/Strep-tag I, rSbpA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1 on peptidoglycan-containing sacculi and studies on accessibility of the fused Strep-tag I or Bet v1 sequence in the square S-layer lattice
In all experiments rSbpA, rSbpA/Strep-tag I, rSbpA31-1068/Strep-tag I, and rSbpA31-1068/Bet v1 were recrystallized on native peptidoglycan-containing sacculi of B. sphaericus CCM 2177 according to the protocol described previously (7), except that samples were dialyzed against 10 mM CaCl2. Peptidoglycan-containing sacculi completely covered with outer and inner S-layer lattices were termed recrystallization products. For investigating the accessibility of Strep-tag I on the outer surface of the S-layer lattice, recrystallization products prepared with rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I (each corresponding to 1 mg of S-layer protein) were incubated with recombinant streptavidin (Sigma S0677) (500 μg/ml in 50 mM Tris-HCl buffer, pH 8.0) for 30 min at 20°C and washed twice with buffer. Samples were then prepared for SDS-PAGE, and bound streptavidin was visualized on immunoblots after incubation with a polyclonal antiserum raised against streptavidin (Sigma) (1:5,000 in 3% BSA in 0.15 M TBS buffer) and anti-rabbit IgG-ALP conjugate (Sigma) (diluted 1:4,000 in 3% BSA in 0.15 M TBS). The accessibility of Strep-tag I was derived from the binding capacity of recrystallization products for streptavidin.
For investigation of the functionality and accessibility of the Bet v1 portion at the C terminus of rSbpA31-1068/Bet v1, recrystallization products were incubated with the monoclonal mouse antibody BIP1. After centrifugation of the suspension at 16,000 × g at 4°C for 20 min and five washing steps with 0.15 M TBS, the pellet was suspended in 40 μl of anti-mouse IgG-colloidal gold conjugate (Amersham) and incubated for 60 min at 20°C. Unbound gold-labeled antibody was removed by centrifugation under the conditions described above and three washing steps with 0.15 M TBS. The pellet was resuspended in 40 μl of distilled water and subjected to negative staining. As a control, the same procedure was carried out with recrystallization products prepared with rSbpA recrystallized on peptidoglycan-containing sacculi.
RESULTS
Molecular characterization of the sbpA gene.
The entire sbpA sequence (GenBank accession no. AF211170) indicated one open reading frame of 3,804 bp encoding a protein of 1,268 amino acids with a theoretical molecular mass of 132,062 Da and a calculated isoelectric point of 4.69. The open reading frame started with ATG, preceded by a typical ribosome binding site (GGAGG) with an appropriate distance of 12 bp between the middle A of this sequence and the start codon. The upstream region (480 bp in front of the start codon) showed four putative promoter sequences characteristic of the promoter regions of prokaryotes. By applying the mfold program (24), a palindromic sequence forming a deduced stem-loop of 38 nucleotides 52 nucleotides downstream of the stop codon TAA was identified as a putative rho-independent transcriptional terminator.
The size of the mRNA transcribed from the sbpA gene was determined by Northern blotting using a DNA probe comprising the coding region of the sbpA gene. The probe detected a transcript of approximately 4.45 kb (Fig. 3), which was in good agreement with the size of the sbpA gene.
FIG. 3.
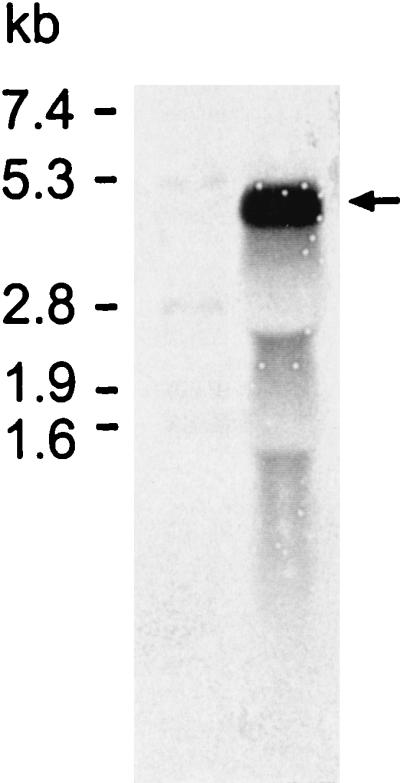
Northern blot of total RNA isolated from B. sphaericus CCM 2177 cells harvested in the exponential growth phase. The DNA probe comprising the coding region of the sbpA gene detected a transcript of approximately 4.45 kb (arrow). The size of the sbpA mRNA was estimated by using the DIG-dUTP-labeled RNA Molecular Weight Marker II (Boehringer Mannheim).
Cloning and expression of the sbpA gene in E. coli HMS174(DE3).
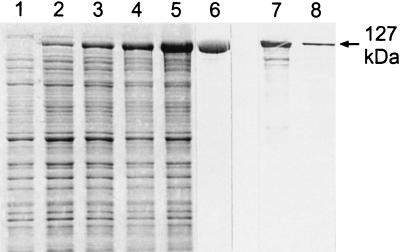
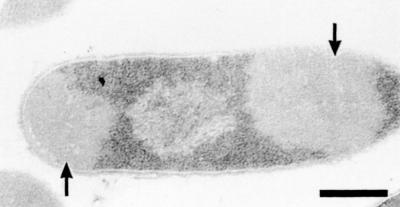
A PCR product comprising the DNA sequence which encoded the mature SbpA and was derived from PCR amplification using the primers sbpA28 and sbpA29 was ligated into the pET28a vector. The plasmid was cloned first in E. coli TG1 and then in E. coli HMS174(DE3) for expression. After induction of sbpA expression by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside), samples of cultures from E. coli HMS174(DE3) were taken at various time points and analyzed by SDS-PAGE. In SDS extracts from whole cells harvested 1 h after induction, an additional major protein band with an apparent molecular mass of 127,000 Da (Fig. 4, lane 2) was visible, and the intensity of this protein band increased with the time of induction (Fig. 4, lanes 3, 4, and 5). Noninduced E. coli HMS174(DE3) cells, which carried the pET28a vector including the sbpA gene, showed no protein band with the same molecular mass on SDS gels (Fig. 4, lane 1). Immunoblotting using the polyclonal rabbit antiserum raised against the S-layer protein of B. sphaericus CCM 2177 revealed a strong cross-reaction with the additional high-molecular-mass protein band (not shown). E. coli HMS174(DE3) cells containing the plain pET28a plasmid did not cross-react with the polyclonal rabbit antiserum (not shown). Ultrathin sections of E. coli HMS174(DE3) cells induced to express the sbpA gene revealed the presence of inclusion body-like structures with low electron density which occupied large areas of the cytoplasm of the host cells (Fig. 5). These findings are in contrast to data obtained with other recombinant S-layer proteins (16, 18, 19), which formed self-assembly products in the cytoplasm of the host cells.
FIG. 4.
Lanes 1 to 5, SDS-PAGE pattern of SDS-extracted whole cells of E. coli HMS174(DE3) harboring the pET28a/sbpA gene construct before (lane 1) and 1, 2, 3, and 5 h after (lanes 2 to 5, respectively) induction of sbpA expression. Lane 6, rSbpA purified by GPC. Lanes 7 and 8, immunoblot analysis using a polyclonal rabbit antiserum raised against the whole S-layer protein of B. sphaericus CCM 2177. In lanes 7 and 8, SDS extracts of the S-layer protein from B. sphaericus CCM 2177 or purified rSbpA were applied.
FIG. 5.
Electron micrograph of an ultrathin-section preparation of E. coli HMS174(DE3) induced to express the sbpA gene. Inclusion body-like structures are indicated by arrows. Bar, 400 nm.
Comparison of the sbpA gene with the S-layer genes of B. sphaericus strains P-1 and 2362 and comparison of the respective S-layer proteins.
For sequence comparison the CLUSTAL W multiple alignment program (42) was used. The sbpA gene and the S-layer genes of B. sphaericus strains P-1 and 2362 clearly had similarities in size and in G+C content, and they showed significant identities in their 5′ upstream regions. The sequence GGAGG, characteristic of a ribosome binding site, and the DNA sequences of the putative promoter regions comprising a −10 sequence (TATAAT) located 88 bp upstream of the translation start and a putative −35 sequence (AGTTTTG) starting 111 nucleotides in front of the start codon could be detected in the upstream regions of sbpA and the other S-layer genes. The two putative transcription start points were separated by an AT-rich region of 16 nucleotides. Furthermore, within the upstream regions (480 bp in front of the start codon) of sbpA and the S-layer gene of B. sphaericus P-1, three further putative promoter sequences with identical compositions and locations were detected (data for this part of the upstream region of B. sphaericus 2362 are not known). By contrast, the putative termination signals and the 3′ downstream regions of the three S-layer genes indicated no similarities. The first 90 bp encoding the signal peptide showed identities of 97%. However, the difference of 3 nucleotides did not affect the resulting protein sequences, which were 100% identical. The N-terminal region (amino acids 31 to 202) of SbpA showed high identity to the corresponding regions of the S-layer proteins of B. sphaericus P-1 (82.9% identity) and B. sphaericus 2362 (78.5% identity). Interestingly, beyond amino acid 202, the identities between SbpA and the S-layer proteins from B. sphaericus P-1 and B. sphaericus 2362 were only 35.4 and 15.4%, respectively. The identities between the entire SbpA sequence and those of the S-layer proteins from B. sphaericus P-1 and B. sphaericus 2362 were 44.8 and 25.4%, respectively.
Isolation and purification of rSbpA from host cells and characterization of its properties.
As derived from SDS-PAGE, rSbpA had accumulated in the insoluble fraction of the lysed E. coli HMS174(DE3) cells (data not shown). After purification of rSbpA by GPC, a single protein band with an apparent molecular mass of 127,000 Da was detected on SDS gels (Fig. 4, lane 6). By applying the isolation and purification procedure described above, about 120 mg of GPC-purified rSbpA was obtained from 5 g (wet pellet) of biomass from E. coli HMS174(DE3) harvested 5 h after induction of sbpA expression. Immunoblotting using the polyclonal rabbit antiserum raised against the S-layer protein of B. sphaericus CCM 2177 revealed a strong cross-reaction with the additional high-molecular-mass protein band representing purified rSbpA (Fig. 4, lane 8). The S-layer protein isolated from B. sphaericus CCM 2177 taken as a control showed a comparably strong cross-reaction with the polyclonal rabbit antiserum (Fig. 4, lane 7). When purified rSbpA was subjected to N-terminal sequencing, the first five amino acids (AQVND) were identical to those of the mature S-layer protein of B. sphaericus CCM 2177. Together with the data from SDS-PAGE and immunoblotting, these findings indicated that the sbpA gene encodes the S-layer protein SbpA of B. sphaericus CCM 2177.
Cloning and expression of the genes encoding rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I and characterization of their properties.
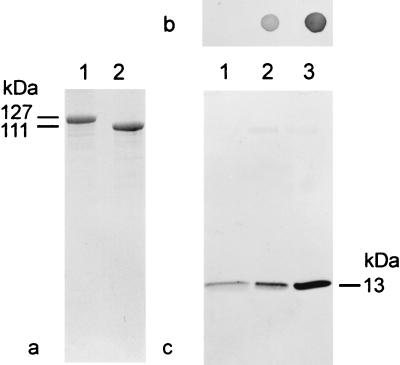
The PCR products encoding the SbpA derivatives rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I, ligated into the pET28a vector, were cloned in E. coli TG1 and expressed in E. coli HMS174(DE3). Whole cells of E. coli HMS174(DE3) induced to express these genes showed additional protein bands with apparent molecular masses of 127,000 or 111,000 Da on SDS gels (Fig. 6a). N-terminal sequencing of isolated and purified rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I confirmed that the first five amino acids of both SbpA derivatives were identical to those of the S-layer protein of B. sphaericus CCM 2177. The presence of the SbpA-specific part in both rSbpA forms was confirmed by immunoblotting using the polyclonal rabbit antiserum raised against the S-layer protein of B. sphaericus CCM 2177 (data not shown). For investigation of the presence of Strep-tag I, dot blot assays with rSbpA/Strep-tag I and rSbpA31-1068/Strep-tag I, purified via a Ready-to-Use Strep-Tactin Sepharose column, were performed. Independent of the use of a polyclonal rabbit antiserum raised against the Strep-tag (not shown) or the use of Strep-Tactin-ALP (Fig. 6b), rSbpA31-1068/Strep-tag I (Fig. 6b, spot 3) gave a significantly stronger reaction than rSbpA/Strep-tag I (Fig. 6b, spot 2), indicating a clearly improved accessibility of Strep-tag I in the C-terminally truncated form. The formation of self-assembly products and the ability to recrystallize on native peptidoglycan-containing sacculi demonstrated that neither the deletion of 200 C-terminal amino acids in rSbpA31-1068/Strep-tag I nor the presence of Strep-tag I in both rSbpA forms interfered with the self-assembly and cell wall binding properties.
FIG. 6.
(a) SDS-PAGE of GPC-purified SbpA derivatives. Lane 1, rSbpA/Strep-tag I; lane 2, rSbpA31-1068/Strep-tag I. (b) Dot blot assays indicating the accessibility of Strep-tag I to Strep-Tactin-ALP in the soluble S-layer protein fractions. Spot 1, rSbpA; spot 2, rSbpA/Strep-tag I; spot 3, rSbpA31-1068/Strep-tag I. (c) Immunoblot analysis for detection of recombinant streptavidin bound by recrystallization products prepared with rSbpA (lane 1), rSbpA/Strep-tag I (lane 2), and rSbpA31-1068/Strep-tag I (lane 3). Monomers of core streptavidin show a molecular mass of 13,000 Da on SDS gels. In the case of streptavidin, low nonspecific adsorption was observed for the square S-layer lattice formed by rSbpA.
Investigation of the accessibility of the C-terminally fused Strep-tag I on the outer surface of the S-layer lattice on recrystallization products prepared with rSbpA31-1068/Strep-tag I.
To determine whether the C-terminally fused Strep-tag I of rSbpA31-1068/Strep-tag I was accessible on the outer surface of the square S-layer lattice, recrystallization products were incubated with recombinant streptavidin (Sigma S0677). For comparison, recrystallization products prepared with rSbpA (Fig. 6c, lane 1) or rSbpA/Strep-tag I (Fig. 6c, lane 2) were used. Recrystallization products prepared with the C-terminally truncated form showed a considerably higher binding capacity for streptavidin (Fig. 6c, lane 3) than those prepared with rSbpA/Strep-tag I. Thus, rSbpA31-1068 was chosen as the base form to construct the S-layer fusion protein incorporating the Bet v1 sequence at the C-terminal end.
Cloning and expression of rSbpA31-1068/Bet v1.
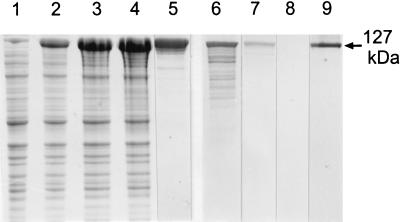
A PCR product comprising the chimeric gene encoding the S-layer fusion protein rSbpA31-1068/Bet v1 obtained by PCR using primers 1 and 4 was ligated into the pET28a vector. The plasmid was first cloned in E. coli TG1 and then established in E. coli HMS174(DE3) for expression. As shown by SDS-PAGE (Fig. 7), in comparison to uninduced E. coli HMS174(DE3) cells (Fig. 7, lane 1), an additional high-molecular-mass protein band was observed in whole cells from E. coli HMS174(DE3) cells induced to express the gene encoding rSbpA31-1068/Bet v1 (Fig. 7, lanes 2, 3, and 4). This additional protein band had an apparent molecular mass of 127,000 Da. On immunoblots, a cross-reaction was observed between the high-molecular-mass protein band and the polyclonal rabbit antiserum raised against the S-layer protein of B. sphaericus CCM 2177 (Fig. 7, lane 7), as well as with the monoclonal mouse antibody BIP1 recognizing epitopes of Bet v1 (Fig. 7, lane 9). The S-layer protein of B. sphaericus CCM 2177, which was used as a control, showed a strong cross-reaction with the polyclonal rabbit antiserum (Fig. 7, lane 6) and no reaction with BIP1 (Fig. 7, lane 8). E. coli HMS174(DE3) cells containing the plain pET28a plasmid showed a cross-reaction neither with the polyclonal rabbit antiserum raised against the S-layer protein SbpA nor with the monoclonal mouse antibody BIP1 recognizing Bet v1 (data not shown). Ultrathin sections of whole cells from E. coli HMS174(DE3) induced to express the gene encoding rSbpA31-1068/Bet v1 revealed the presence of inclusion bodies in the cytoplasm of the host cells (data not shown). N-terminal sequencing of the purified rSbpA31-1068/Bet v1 confirmed that the first five amino acids (AQVND) were identical to those of the S-layer protein of B. sphaericus CCM 2177. Isolation and purification of the S-layer fusion protein from the host cells were performed as described for rSbpA, leading to a single major protein band with a molecular mass of 127,000 Da on SDS gels (Fig. 7, lane 5).
FIG. 7.
Lanes 1 to 4, SDS-PAGE pattern of SDS-extracted whole cells of E. coli HMS174(DE3) containing pET28a with the sbpA derivative encoding rSbpA31-1068/Bet v1 before (lane 1) and 1, 3, and 5 h after (lanes 2 to 4, respectively) induction of S-layer fusion protein expression. Lane 5, rSbpA31-1068/Bet v1 purified by GPC. Lanes 6 and 7, immunoblot analysis using a polyclonal rabbit antiserum raised against SbpA; SDS extracts of the S-layer protein from B. sphaericus CCM 2177 or rSbpA31-1068/Bet v1 were applied. Lanes 8 and 9, immunoblot analysis using the monoclonal antibody BIP1 raised against Bet v1; the samples corresponding to lanes 6 and 7 were applied.
Self-assembly properties of rSbpA and rSbpA31-1068/Bet v1 and recrystallization on poly-l-lysine-coated copper grids.
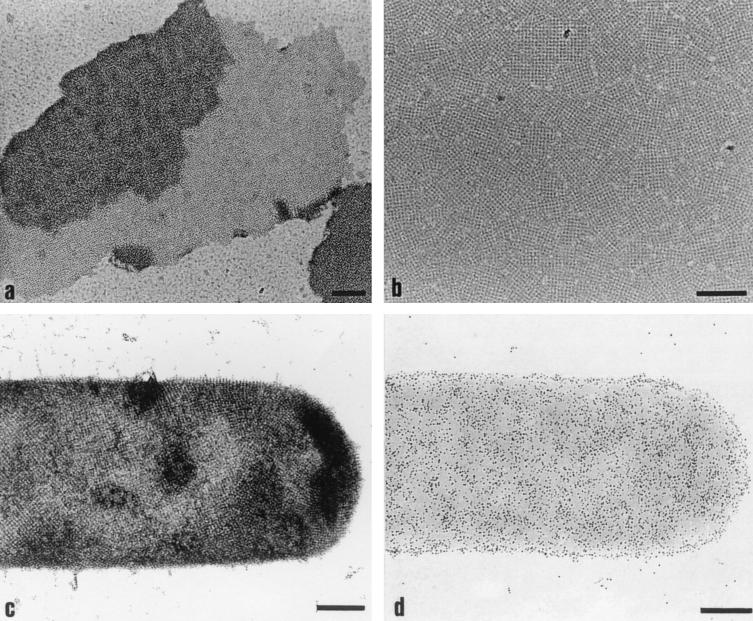
For investigating the self-assembly properties, purified rSbpA was disintegrated in 5 M GHCl, and the solution was dialyzed against 10 mM CaCl2 for 18 h at 4°C. As shown by negative staining, rSbpA reassembled into flat double-layer sheets with a maximum size of 2 μm, clearly exhibiting the square lattice structure (Fig. 8a). Freeze-drying and high-resolution shadowing revealed that the individual monolayers faced each other with the more corrugated inner surface (not shown), which had also been described for the wild-type S-layer protein (13). The lattice constants for self-assembly products formed by rSbpA were 13.1 nm and therefore identical to those determined for self-assembly products formed by the wild-type S-layer protein (17; A. Neubauer, personal communication). If GHCl-extracted rSbpA was dialyzed against distilled water, about 75% of the total amount of the S-layer protein remained in the soluble fraction, consisting of monomeric or oligomeric precursors (14). Soluble rSbpA was capable of recrystallizing into a closed monolayer on poly-l-lysine-coated EM grids. As shown in Fig. 8b, the monolayer consisted of numerous randomly orientated patches, clearly exhibiting the square lattice structure.
FIG. 8.
Electron micrographs of negatively stained preparations. (a) Self-assembly properties of rSbpA. (b) Ability of rSbpA to recrystallize on poly-l-lysine-coated EM grids. (c) Formation of the square S-layer lattice by rSbpA31-1068/Bet v1 on peptidoglycan-containing sacculi of B. sphaericus CCM 2177. (d) Immunogold labeling of recrystallization products obtained with rSbpA31-1068/Bet v1 using the Bet v1-specific monoclonal antibody BIP1. Bars, 200 nm.
For evaluating the self-assembly properties of rSbpA31-1068/Bet v1, GHCl extracts were dialyzed against 10 mM CaCl2 for 18 h at 4°C. As shown by negative staining, rSbpA31-1068/Bet v1 reassembled into flat sheets, which clearly exhibited the square lattice structure (data not shown). As described for rSbpA, the S-layer fusion protein was capable of recrystallizing into a closed monolayer consisting of numerous randomly oriented patches on poly-l-lysine-coated EM grids.
Immunogold labeling of recrystallization products obtained with rSbpA31-1068/Bet v1.
To investigate the location and functionality of the fused Bet v1 sequence, rSbpA31-1068/Bet v1 was recrystallized on peptidoglycan-containing sacculi of B. sphaericus CCM 2177 (Fig. 8c) and subsequently labeled with BIP1. The bound antibody was visualized with an anti-mouse IgG-colloidal gold conjugate. The dense labeling of recrystallization products provided evidence for the functionality of the fused Bet v1 portion and its location on the outer surface of the S-layer lattice (Fig. 8d). Recrystallization products prepared with rSbpA remained unlabeled (data not shown).
DISCUSSION
Previously, the nucleotide sequences of the genes encoding the S-layer proteins of B. sphaericus P-1 (6) and B. sphaericus 2362 (1) have been established. In the present study, the sbpA gene, encoding the S-layer protein SbpA of B. sphaericus CCM 2177, has been sequenced and cloned and the 5′ upstream region has been characterized. The existence of a typical rho factor-independent transcriptional terminator at the downstream noncoding sequence of the sbpA gene demonstrated the monocistronic nature of the S-layer gene, which could be confirmed by Northern blotting.
The amino acid composition of SbpA indicated a high content of acidic and hydrophobic amino acids; a low content of histidine, arginine, and methionine; and no cysteine. This composition is characteristic of S-layer proteins from Bacillaceae (35). Prediction of the secondary structure from sequence data as described previously (31) indicated the concentration of α-helices in the N-terminal part, whereas the middle and C-terminal parts are dominated by loops and short β-strands.
The N-terminal part of SbpA, with a calculated pI of 4.49, comprises three typical SLH motifs, each consisting of 50 amino acids. In a previous study, the N-terminal part of SbpA was found to be involved in anchoring the S-layer subunits via a pyruvylated SCWP to the rigid cell wall layer (13). Mesnage et al. (25) demonstrated that the mechanism of anchoring between SLH motifs and pyruvylated peptidoglycan-associated wall polymers is widespread among prokaryotes and has been conserved during evolution. In general, SLH motifs occur in a single copy or in two or three copies either at the N-terminal part of many S-layer proteins or at the C-terminal ends of cell-associated exoenzymes and other exoproteins of gram-positive bacteria (Table 2) (9, 22, 23). Comparison of the pIs of the SLH domains generally revealed an acidic nature, with pIs comparable to those of the remaining parts of the sequences. However, a few exceptions with calculated pIs of >7 were found, which are the SLH domains of the S-layer protein SbsB of Bacillus stearothermophilus PV72/p2 (18), the hexagonal wall protein HWP from Bacillus brevis HPD31 (43), and the cellulosome-anchoring protein and cell surface glycoprotein 2 of Clostridium thermocellum NCIMB 10682 (10) (Table 2). In these cell-associated exoproteins, the basic pIs of the SLH domains are in contrast to the acidic pIs of the remaining part of the proteins. In the case of SbpA, the acidic nature of the SLH domain may explain why bivalent cations are required for binding of the S-layer subunits to the rigid cell wall layer (13). On the other hand, neither monovalent nor bivalent cations are necessary for binding of the S-layer proteins with basic N-terminal parts, such as SbsB (pI 8.67), to the rigid cell wall layer (30).
TABLE 2.
Comparison of SLH domain-containing cell-associated exoproteins from gram-positive bacteria
| Species | Strain | Protein | GenBank accession no. | Number, localization, and order of SLH motifs | pI
|
||
|---|---|---|---|---|---|---|---|
| Leader peptide | SLH domaina | Remaining protein sequence | |||||
| Bacillus sphaericus | CCM 2177 | S-layer protein SbpA | AF211170 | 1, 2, 3 (N terminal) | 11.31 | 4.49 | 4.63 |
| P1 | S-layer protein | A45814 | 1, 2, 3 (N terminal) | 11.31 | 4.43 | 4.71 | |
| 2362 | S-layer protein | M28361 | 1, 2, 3 (N terminal) | 11.31 | 4.22 | 5.07 | |
| Bacillus stearothermophilus | PV72/p2 | S-layer protein SbsB | X98095 | 1, 2, 3 (N terminal) | 10.05 | 8.67 | 4.75 |
| Bacillus anthracis | Sterne derivative, substrain 9131 | S-layer protein EA1 | X99724 | 1, 2, 3 (N terminal) | 10.03 | 5.85 | 5.36 |
| S-layer protein SAP | Z36946 | 1, 2, 3 (N terminal) | 10.03 | 5.52 | 6.06 | ||
| Thermoanerobacterium kivui | DSM 2030 | S-layer protein Slp | M31069 | 1, 2, 3 (N terminal) | 10.31 | 6.37 | 4.77 |
| Bacillus brevis | HPD31 | S-layer protein (hexagonal wall protein HWP) | D90050 | 1, 2, 3 (N terminal) | 9.89 | 8.32 | 4.53 |
| 47 | S-layer protein (middle wall protein MWP) | M14238 | 1, 2, 3 (N terminal) | 10.01 | 5.21 | 4.38 | |
| Thermotoga maritima | Outer membran protein Omp-α | X68276 | 1 (N terminal) | 11.05 | 5.75 | 4.56 | |
| Thermus thermophilus | HB8 | S-layer protein SlpA | X57333 | 1 (N terminal) | 11.21 | 5.12 | 4.54 |
| Clostridium thermocellum | NCIMB 10682 | S-layer protein SlpA | U79117 | 1, 2, 3 (N terminal) | 10.31 | 4.21 | 4.36 |
| Cellulosome-anchoring protein | X67506 | 1/2 1, 2, 3, 1/2 1 (C terminal)b | 12.21 | 7.28 | 4.36 | ||
| S-layer protein 1 (cell surface glycoprotein 1) | X67506 | 1/2 1, 2, 3, 1/2 1 (C terminal) | 11.31 | 5.38 | 3.54 | ||
| S-layer protein 2 (cell surface glycoprotein 2) | X67506 | 1/2 1, 2, 3, 1/2 1 (C terminal) | 10.01 | 8.97 | 4.84 | ||
| Exoglucanase XYNX | M67438 | 1, 2, 3 (C terminal) | 10.01 | 4.69 | 4.34 | ||
| Thermoanaerobacterium thermosulfurigenes | EM1 | Endoxylanase | U50952 | 1, 2, 3 (C terminal) | 12.11 | 4.48 | 4.47 |
| Amylopullulanase | M57692 | 1, 2, 3 (C terminal) | 11.21 | 4.67 | 4.30 | ||
| Thermoanaerobacterium saccharolyticum | Endo-1,4-β-xylanase A | M97882 | 1, 2 (C terminal) | 10.89 | 4.41 | 4.36 | |
| Bacillus sp. | KSM-635 | Endoglucanase | M27420 | 1, 2, 3 (N terminal) | 10.31 | 4.87 | 4.02 |
pIs of >7 are in boldface (see text).
1/2 1, 2, 3, 1/2 1 means that the first SLH motif is missing its N-terminal part, which is appended to the end of the third SLH motif.
In many previous studies it was demonstrated that the S-layer protein of B. sphaericus CCM 2177 recrystallizes into a regularly structured lattice on a broad spectrum of supports, such as silicon wafers, gold chips, silanized glass, Langmuir films, or liposomes (for a review, see reference 38). Depending on the surface properties of the support, the S-layer subunits attached with either their outer or inner surface, which can be considered a severe disadvantage if S-layer fusion proteins are used for building up highly ordered protein lattices on solid supports. To achieve a predictable oriented binding and recrystallization, the specific interaction between SbpA and the respective type of SCWP will be exploited in future work (Fig. 1).
Thus, one of the goals in the present study was to identify amino acid positions at the C-terminal part of SbpA to which functional sequences can be fused and which after folding of both protein moieties remain located on the outer surface of the S-layer lattice. To generate oriented monolayers and to evaluate the surface accessibility of the introduced functional sequences, all rSbpA forms were recrystallized on peptidoglycan-containing sacculi. Strep-tag I linked to the C-terminal end of rSbpA revealed, even in the soluble form of the full-length S-layer protein, a limited accessibility for macromolecules, such as antibodies or the Strep-Tactin-ALP-conjugate. The accessibility of Strep-tag I could be significantly increased when it was linked to the C-terminally truncated form rSbpA31-1068. Despite the deletion of 200 C-terminal amino acids, rSbpA31-1068/Strep-tag I had retained the ability to self-assemble and to recrystallize on peptidoglycan-containing sacculi into the square S-layer lattice, and Strep-tag I remained accessible on the outer S-layer surface for macromolecule binding.
Due to these properties, the C-terminally truncated SbpA form was selected as the base form for construction of the S-layer fusion protein comprising the Bet v1 sequence, which was produced with the final aim of building up highly ordered Bet v1-specific protein lattices on solid supports. Both features, namely, (i) the ability of rSbpA31-1068/Bet v1 to bind to native peptidoglycan-containing sacculi and to recrystallize into the square lattice structure and (ii) the formation of self-assembly products in suspension, confirmed the functionality of the S-layer protein moiety. Evidence for the location of the fused Bet v1 sequence on the outer surface of the S-layer lattice and proof of its functionality were provided by the high binding density observed for the monoclonal mouse antibody BIP1.
In various other S-layer fusion proteins described so far, only the cell wall-targeting domains such as the three SLH motifs have been exploited. For this purpose, the chimeric genes encoding the sequences of the SLH domains of the S-layer protein Sap or EA1 of Bacillus anthracis and levansucrase of Bacillus subtilis were cloned in B. anthracis (26). The SLH motifs were also incorporated into the fusion protein SLH-Tox, carrying fragment C of the tetanus toxoid of Clostridium tetanii (27). Another example is the green fluorescence-SAC fusion protein, containing the C-terminal cell wall-targeting domain of Lactobacillus acidophilus ATCC 4536 (40). Because of the absence of those domains required for self-assembly, none of these S-layer fusion proteins formed a regularly structured protein lattice.
The example of the rSbpA31-1068/Bet v1 fusion protein demonstrated that functional, regularly structured protein lattices can be built up with S-layer fusion proteins. In such protein lattices, the functional sequences are aligned at a predefined distance in the nanometer range on the outermost surface of the S-layer lattice and therefore remain available for further binding reactions. These specific features imply a considerable application potential for biochip development as required in proteomics or genomics. In the present state of the rSbpA31-1068/Bet v1 monolayer, the density of the fused Bet v1 sequence is determined by the intrinsic properties of the S-layer protein moiety and the resulting center-to-center spacing of the morphological units.
Acknowledgments
This work was supported by the Austrian Science Foundation, projects P12938 and P14689; by the Competence Center “Biomolecular Therapeutics” BMT; and by the Federal Ministry of Education, Science and Culture.
Plasmid pMW175 was kindly provided by BIOMAY, Linz, Austria.
REFERENCES
- 1.Bowditch, R. D., P. Baumann, and A. A. Yousten. 1989. Cloning and sequencing of the gene encoding a 125-kilodalton surface-layer protein from Bacillus sphaericus 2362 and of a related cryptic gene. J. Bacteriol. 171:4178-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brechtel, E., and H. Bahl. 1999. In Thermoanaerobacterium thermosulfurigienes EM1 S-layer homology domains do not attach to peptidoglycan. J. Bacteriol. 181:5017-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitwieser, A., S. Küpcü, S. Howorka, S. Weigert, C. Langer, K. Hoffmann-Sommergruber, O. Scheiner, U. B. Sleytr, and M. Sára. 1996. 2-D protein crystals as an immobilization matrix for producing reaction zones in dipstick-style immunoassays. BioTechniques 21:918-925. [DOI] [PubMed] [Google Scholar]
- 4.Breitwieser, A., C. Mader, I. Schocher, K. Hoffmann-Sommergruber, W. Aberer, O. Scheiner, U. B. Sleytr, and M. Sára. 1998. A novel dipstick developed for rapid Bet v 1-specific IgE detection: recombinant allergen immobilized via a monoclonal antibody to crystalline bacterial cell-surface layers. Allergy 53:786-793. [DOI] [PubMed] [Google Scholar]
- 5.Chauvaux, S., M. Matuschek, and P. Begiun. 1999. Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis. J. Bacteriol. 181:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deblaere, R. January1995. Expression of surface layer proteins. Patent WO 9519371-A.
- 7.Egelseer, E. M., K. Leitner, M. Jarosch, C. Hotzy, S. Zayni, U. B. Sleytr, and M. Sára. 1998. The S-layer proteins of Bacillus stearothermophilus wild-type strains are bound via their N-terminal region to a secondary cell wall polymer of identical chemical composition. J. Bacteriol. 180:1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egelseer, E. M., I. Schocher, U. B. Sleytr, and M. Sára. 1996. Evidence that an N-terminal S-layer protein fragment triggers the release of a cell associated high-molecular-weight amylase from Bacillus stearothermophilus ATCC 12980. J. Bacteriol. 178:5602-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains and surface layer-cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 10.Fujino, T., P. Beguin, and J. P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann-Sommergruber, K., M. Susani, F. Ferreira, P. Jertschin, H. Ahorn, R. Steiner, D. Kraft, O. Scheiner, and H. Breiteneder. 1997. High-level expression and purification of the major birch pollen allergen, Bet v 1. Protein Expr. Purif. 9:33-39. [DOI] [PubMed] [Google Scholar]
- 13.Ilk, N., P. Kosma, M. Puchberger, E. M. Egelseer, H. F. Mayer, U. B. Sleytr, and M. Sára. 1999. Structural and functional analysis of the secondary cell wall polymer of Bacillus sphaericus CCM 2177 serving as an S-layer-specific anchor. J. Bacteriol. 181:7643-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaenicke, R., R. Welsch, M. Sára, and U. B. Sleytr. 1985. Stability and self-assembly of the S-layer protein of the cell wall of Bacillus stearothermophilus. Biol. Chem. Hoppe-Seyler 366:663-670. [DOI] [PubMed] [Google Scholar]
- 15.Jarolim, E., M. Tejkl, M. Rohac, G. Schlerka, O. Scheiner, D. Kraft, M. Breitenbach, and H. Rumpold. 1989. Monoclonal antibodies against birch pollen allergens: characterization by immunoblotting and use for single-step affinity purification of the major allergen Bet vI. Int. Arch. Allergy Appl. Immunol. 90:54-60. [DOI] [PubMed] [Google Scholar]
- 16.Jarosch, M., E. M. Egelseer, C. Huber, D. Moll, D. Mattanovich, U. B. Sleytr, and M. Sára. 2001. Analysis of the structure-function relationship of the S-layer protein SbsC of Bacillus stearothermophilus ATCC 12980 by producing truncated forms. Microbiology 147:1353-1363. [DOI] [PubMed] [Google Scholar]
- 17.Jarosch, M., E. M. Egelseer, D. Mattanovich, U. B. Sleytr, and M. Sára. 2000. S-layer gene of Bacillus stearothermophilus ATCC 12980: molecular characterization and heterologous expression in Escherichia coli. Microbiology 146:273-281. [DOI] [PubMed] [Google Scholar]
- 17a.Korntner, R. 1999. Herstellung und Charakterisierung von Testpräparaten für die Rasterkraftmikroskoie unter Verwendung von zweidimensionalen Proteinkristallen (S-Schichten). Master thesis. University of Agricultural Sciences, Vienna, Austria.
- 18.Kuen, B., A. Koch, E. Asenbauer, M. Sára, and W. Lubitz. 1997. Molecular characterization of the Bacillus stearothermophilus PV72 S-layer gene sbsB induced by oxidative stress. J. Bacteriol. 179:1664-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuen, B., M. Sára, and W. Lubitz. 1995. Heterologous expression and self-assembly of the S-layer protein SbsA of Bacillus stearothermophilus in Escherichia coli. Mol. Microbiol. 19:495-503. [DOI] [PubMed] [Google Scholar]
- 20.Küpcü, S., M. Sára, and U. B. Sleytr. 1995. Liposomes coated with crystalline bacterial cell surface protein (S-layer) as immobilization structures for macromolecules. Biochim. Biophys. Acta 1235:263-269. [DOI] [PubMed] [Google Scholar]
- 21.Lemaire, M., I. Miras, P. Gounon, and P. Beguin. 1998. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology. 144:211-217. [DOI] [PubMed] [Google Scholar]
- 22.Lupas, A. 1996. A circular permutation event in the evolution of the SLH domain? Mol. Microbiol. 20:897-898. [DOI] [PubMed] [Google Scholar]
- 23.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Mader, C., S. Küpcü, U. B. Sleytr, and M. Sára. 2000. S-layer-coated liposomes as a versatile system for entrapping and binding target molecules. Biochim. Biophys. Acta 1463:142-150. [DOI] [PubMed] [Google Scholar]
- 24.Mathews, D. H., T. C. Andre, J. Kim, D. H. Turner, and M. Zuker. 1998. An updated recursive algorithm for RNA secondary structure prediction with improved free energy parameters. Am. Chem. Soc. Symp. Ser. 682:246-257. [Google Scholar]
- 25.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesnage, S., E. Tosi-Couture, and A. Fouet. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927-936. [DOI] [PubMed] [Google Scholar]
- 27.Mesnage, S., M. Weber-Levy, M. Haustant, M. Mock, and A. Fouet. 1999. Cell surface-exposed tetanus toxin fragment C produced by recombinant Bacillus anthracis protects against tetanus toxin. Infect. Immun. 67:4847-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messner, P., F. Hollaus, and U. B. Sleytr. 1984. Paracrystalline cell wall surface layers of different Bacillus stearothermophilus strains. Int. J. Syst. Bacteriol. 34:202-210. [Google Scholar]
- 29.Pum, D., M. Sára, and U. B. Sleytr. 1989. Structure, surface charge, and self-assembly of the S-layer lattice of Bacillus coagulans E38-66. J. Bacteriol. 171:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ries, W., C. Hotzy, I. Schocher, U. B. Sleytr, and M. Sára. 1997. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J. Bacteriol. 179:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rost, B., and C. Sander. 1994. Combining evolutionary information and neutral networks to predict protein secondary structure. Protein 19:55-72. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor, Cold Spring Harbor Laboratory Press, N.Y.
- 33.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci., USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sára, M. 2001. Conserved anchoring mechanism between crystalline cell surface S-layer proteins and secondary cell wall polymers in Gram-positive bacteria? Trends Microbiol. 9:47-50. [DOI] [PubMed] [Google Scholar]
- 35.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, T. G. M., and A. Skerra. 1994. One-step purification of bacterially produced proteins by means of the “ Strep tag” and immobilized recombinant core streptavidin. J. Chromatogr. Sect. A 676:337-345. [DOI] [PubMed] [Google Scholar]
- 37.Sleytr, U. B., and T. J. Beveridge. 1999. Bacterial S-layers. Trends Microbiol. 7:253-260. [DOI] [PubMed] [Google Scholar]
- 38.Sleytr, U. B., P. Messner, D. Pum, and M. Sára. 1999. Crystalline bacterial cell surface layers (S-layers): from supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. 38:1034-1054. [DOI] [PubMed] [Google Scholar]
- 39.Sleytr, U. B., and M. Sára. 1997. Bacterial and archaeal S-layer proteins: structure-function relationship and their biotechnological application. Trends Biotechnol. 15:20-26. [DOI] [PubMed] [Google Scholar]
- 40.Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwels. 2001. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterization of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305:245-257. [DOI] [PubMed] [Google Scholar]
- 41.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuboi, A., R. Uchihi, T. Adachi, T. Sasaki, S. Hayakawa, H. Yamagata, N. Tsukagoshi, and S. Udaka. 1988. Characterization of the genes for the hexagonally arranged surface layer proteins in protein-producing Bacillus brevis 47: complete nucleotide sequence of the middle wall protein gene. J. Bacteriol. 170:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voss, S., and A. Skerra. 1997. Mutagenesis of a flexible loop in streptavidin leads to a higher affinity for the Strep-tag II and improved performance in recombinant protein purification. Protein Eng. 10:975-982. [DOI] [PubMed] [Google Scholar]
- 45.Weiner, C., M. Sára, and U. B. Sleytr. 1994. Novel protein A affinity matrix prepared from two-dimensional protein crystals. Biotechnol. Bioeng. 43:321-330. [DOI] [PubMed] [Google Scholar]