Abstract
As part of its pathogenic life cycle, Phytophthora capsici disperses to plants through a motile zoospore stage. Molecules on the zoospore surface are involved in reception of environmental signals that direct preinfection behavior. We developed a phage display protocol to identify peptides that bind to the surface molecules of P. capsici zoospores in vitro. The selected phage-displayed peptides contained an abundance of polar amino acids and proline but were otherwise not conserved. About half of the selected phage that were tested concomitantly induced zoospore encystment in the absence of other signaling agents. A display phage was shown to bind to the zoospore but not to the cyst form of P. capsici. Two free peptides corresponding to active phage were similarly able to induce encystment of zoospores, indicating that their ability to serve as signaling ligands did not depend on their exact molecular context. Isolation and subsequent expression of peptides that act on pathogens could allow the identification of receptor molecules on the zoospore surface, in addition to forming the basis for a novel plant disease resistance strategy.
Phytophthora capsici is a soilborne pathogenic protist (phylum Oomycota) that infects aerial and subterranean structures of many solanaceous plants. Diseases caused by P. capsici are polycyclic in that multiple cycles of infection and inoculum production occur in a single growing season (20). The pathogen survives unfavorable conditions in soil by forming thick-walled oospores, while dissemination and infection are achieved through the production of motile biflagellate zoospores from oospores. The zoospores swim through water in the soil and are chemotactically attracted to the exudates released by the roots of potential host plants (11, 14). After the zoospores have adhered to the root surface, they encyst and produce a precisely oriented germ tube that grows into adjacent host plant tissue (3). The progression from zoospores to germlings is triggered by environmental signals, some of which are produced by the plant root. Receptors on the surfaces of the zoospores, cysts, and germ tubes detect the environmental signals that trigger or orient each developmental event.
Control of Phytophthora infection remains an ongoing agricultural problem and is most commonly accomplished by the application of biocides, such as methyl bromide or metalaxyl, to the soil. The ability of oospores to persist in soil for long periods obviates the use of crop rotation as an antipest strategy. Alternative, more environmentally benign methods of control will likely have to target host-specific stages of the infectious cycle, since the pathogen is so persistent.
In the present study, we explore the possibility of using specific peptide ligands to interfere with the normal developmental progression of the pathogen. We reasoned that, since zoospores are chemotactic toward the plant surface and since the development of the pathogen involves interaction with the plant surface, surface receptor molecules on the zoospore must exist to signal these events. Therefore, ligands targeting these receptors might provide access points to disrupt pathogen infection and development.
Given that the putative receptors on zoospores have not been characterized at the molecular level, we used a combinatorial approach, specifically phage display, to identify ligands that recognize the zoospores. Since it was first reported (2, 23), phage display technology has become a powerful tool for selecting peptides with affinities for small molecules (22), receptors (1), viruses (8), and whole cells (6). The phage display approach selected peptides that were specific for and induced premature encystment of P. capsici zoospores. Furthermore, we demonstrated specificity at the molecular level by showing that the effects of phage-displayed peptides could be recapitulated by the corresponding peptides in free solution.
MATERIALS AND METHODS
Fungal species and zoospore production.
Isolate ATCC 15399 of P. capsici (4, 5) was maintained on cornmeal agar plates (Difco) at 15°C. To induce sporulation and zoospore release, the isolate was grown on clarified 10% V8 vegetable juice agar (Campbell Soup Company) and incubated for 3 to 6 days at 25°C. Sporangial formation in P. capsici was induced by removing uncolonized agar from around the mycelium and incubating the culture for an additional 1 to 2 days at 25°C under fluorescent lights (12-12 light-dark cycle). Zoospores were released from the sporangia by flooding the plates with sterile deionized water and incubating them at room temperature for 20 to 30 min. The zoospores were filtered through four layers of cheesecloth to remove sporangial cases and mycelial fragments. To determine the concentration of zoospores, an aliquot of the zoospore suspension was vortexed for 30 s to induce encystment, and the number of cysts was determined with a hemocytometer.
Affinity selection of zoospore-binding phage.
Phage display library f8-8mer (18; GenBank accession number AF246447) was used in developing the affinity selection protocol. The library consists of filamentous fd-tet phage displaying random peptide 8-mers fused to all 4,000 copies of the major coat protein pVIII. The library contains 3 × 109 peptide variants.
In the affinity selection procedure, 106 P. capsici zoospores were added to 1011 transducing units (TU) (25) of phage in the presence of 50 mM LiCl, which prevents zoospore encystment (4). The final incubation volume of the mixture was 4 ml. The zoospore-phage mixture was gently agitated on an orbital shaker at room temperature in a petri dish (60 by 15 mm; Fisher Scientific Company). After 30 min, phage that bound to zoospores were separated from the nonbound phage in suspension by centrifugation at 1,000 × g for 45 s in a swinging-bucket rotor (microcentrifuge model 59A; Fisher Scientific). The pellet was resuspended in 150 μl of 50 mM LiCl, and the phage-zoospore complexes were repelleted. After 10 washes in LiCl, the bound phage were eluted from the pelleted zoospores by the addition of 200 μl of elution buffer (0.1 N HCl [pH 2.2] with glycine, 1 g of bovine serum albumin/liter).
The eluted phage suspension was neutralized with 1 M Tris-Cl, pH 9.0 (typically 40 μl was added) and titered as TU, and the residual population was amplified by infection of Escherichia coli K91BluKan cells (28). The amplified phage were twice purified by precipitation with polyethylene glycol (PEG) and resuspended in TBS buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl) (25). The phage were titered in E. coli K91BluKan cells as TU before being reapplied to freshly released zoospores. Phage isolation was continued for a total of three affinity purification and two amplification steps. After each round of affinity selection, the yield of zoospore-binding phage was expressed as phage TU eluted from the zoospores (25).
Phage from the final round of affinity selection were used to infect E. coli K91BluKan cells. These cells were plated on Luria-Bertani plates containing 40 mg of tetracycline/liter to select for growth of cells containing phage. Bacterial colonies, each containing a single phage clone, were selected for analyses of peptide diversity and bioactivity.
DNA sequencing and peptide analysis.
The diversity of affinity-selected, phage-displayed peptides was assessed by sequencing the region of the phage genome that coded for the inserted 8-mer peptide and translating to the amino acid sequence. Phage clones were randomly selected after the final affinity selection step, and single-stranded DNA was isolated from the recombinant phage particles of each clone (25). The DNA was sequenced from the 3′ end on a Prism 377 automated sequencer (Applied Biosystems, Foster City, Calif.) at the DNA Core Laboratory, University of Missouri—Columbia.
The DNA sequences were translated using the translate program on the ExPASy molecular biology server (http://www.expasy.ch/). The diversity of translated peptide sequences was assessed with GeneDoc (15) and with ClustalW (27) using a PAM250 weight table.
Assessment of zoospore encystment induced by phage-displayed peptides.
Representative affinity-selected, phage-displayed peptides were assessed for the ability to induce premature zoospore encystment. Phage clones were selected to represent the diversity of phage-displayed peptides. Each clone was amplified by E. coli infection, twice purified using PEG precipitation, and resuspended in sterile deionized water. For encystment assays, phage were extensively dialyzed versus 0.1 mM K(2-N-morpholino)ethanesulfonate (MES), pH 6.2, to reduce potential background encystment caused by residual PEG. Phage concentrations were calculated from UV absorbance measurements as virions (virus particles) per microliter (26).
Replicate 20-μl droplets containing ∼400 newly released zoospores in MES buffer were incubated in the bottom of a plastic petri dish in the presence of various concentrations of phage. Phage were present in 106- to 107-fold excess in each experiment. The number of encysted zoospores in a droplet was determined by visual counts under a compound microscope or by analysis of time-lapse micrographs. Control experiments to quantify spontaneous encystment in the zoospore population included zoospores incubated in the absence of phage, in the presence of fuse5 vector phage (23), or with an equivalent number of virions from the unfractionated library. Kinetics of encystment were measured by time-lapse photography. Digital micrographs (1- to 2-s exposure time) were scored for the presence of nonmotile cysts and motile zoospores.
Specificity of phage-displayed peptide binding to zoospores and cysts.
Phage-displayed peptides with high and low encystment induction abilities were compared for the ability to bind to P. capsici zoospores and cysts. Phage clones Pc87 and Pc45 were selected as the representative clones that induced high and low levels of encystment, respectively. Phage vector was included as a control treatment. Phage clones were amplified by E. coli infections and purified as described above. For each binding reaction, 5 × 1010 TU of phage was incubated with 200,000 P. capsici zoospores. The binding reaction and washes were performed as described above for the library screening. Phage eluted from the zoospore population were titered in E. coli K91BluKan cells and expressed as total TU. Although it was consistent within a single experiment, this value varied from experiment to experiment because the infectivity of the starved host cells varies from preparation to preparation (25). A similar procedure was used to determine whether the selected phage bound to P. capsici cysts.
RESULTS
Affinity selection of display phage that bind to P. capsici zoospores.
There are potentially a large number of receptors and other surface molecules on the zoospore. Since there was no prior information about which of these molecular species would be involved in signal recognition or zoospore development, we decided to carry out affinity selection for only a limited number of generations. We reasoned that more extensive affinity selection would ultimately lead to one or a few phage-displayed peptides which might or might not target a developmentally significant event (13). The affinity selection protocol effectively recovered phage-displayed peptides that bound to P. capsici zoospores. The zoospores remained intact throughout the affinity selection procedure; no encystment or cell lysis was observed. After each round of affinity selection, the number of phage eluted from the zoospores (yield) was determined prior to reamplification in E. coli. The phage yield in each round of selection typically varied between 10−5 and 10−6 input phage; this was 3 to 4 orders of magnitude greater than the yield obtained in the absence of zoospores (25). The overall yield of phage did not increase with each round of selection. This is commonly observed in whole-cell screenings and likely reflects the large molar ratio of input phage to receptors (17).
The diversity of phage-displayed peptides was evaluated in a collection of 21 phage clones randomly selected from the final eluted phage population. Nineteen of these clones encoded different peptide sequences. Application of the Chao1 sampling diversity function (12) resulted in an estimate that the overall set contained approximately 120 peptide sequences. Given that the input library contained approximately 109 members (18), this suggests that each round of the selection procedure enriched the population of binding phage about 200-fold. Among four similarity groups defined by ClustalW analysis (Table 1), group 1 contained a single clone, Pc56. Group 2 contained nine members, of which Pc78 and Pc87 differed by two residues. Similarity group 3 contained eight members, of which clones Pc43 and Pc64 displayed the same peptide. Group 4 contained three members, of which Pc12 and Pc42 were identical.
TABLE 1.
Sequences of phage-displayed peptidesa
| Phage | Peptide |
|---|---|
| Group 1 | |
| Pc56 | AAPDLQDAMb |
| Group 2 | |
| Pc19 | ADRLNSDAG |
| Pc36 | ADRPSTTSL |
| Pc78 | ADPPRTVST |
| Pc87 | ADRPSMSPTc |
| Pc11 | ADRTSNAST |
| Pc76 | ADKSYIPSS |
| Pc65 | AVRNPSHHS |
| Pc44 | ADPTPRGHS |
| Pc58 | ADPTRQPHS |
| Group 3 | |
| Pc45 | AEHQNSAGP |
| Pc14 | ADARSAGAIS |
| Pc39 | ADSKNAGPM |
| Pc53 | AETKFSGSA |
| Pc15A | ADPKGSGVT |
| Pc15B | AGLTSPNDM |
| Pc43 | ADITDPMGA |
| Pc64 | ADITDPMGA |
| Group 4 | |
| Pc29B | AVGTHTPDS |
| Pc12 | AVSPNVHDG |
| Pc42 | AVSPNVHDGc |
Species shown in boldface were selected for further characterization. All insert sequences in the library begin with A; the second residue can be one of five amino acids: D, E, A, V, or G.
Corresponding free peptide (200 μM) was ineffective at inducing encystment of P. capsici zoospores.
Corresponding free peptide (200 μM) was effective at inducing encystment.
Preferential selection for specific amino acid residues occurred during the affinity selection procedure, although no single sequence motif was highly represented. For example, the majority of the sequenced peptides contained either an aspartic acid or a glutamic acid residue at the second (first randomized) position, although valine and alanine are also encoded in the library at this position with equal frequency (18). Similarly, proline was present at about twice its expected frequency within the random inserted sequences. In the abundant groups 2 and 3, the most common amino acid residues were hydrophilic or polar, while aromatic and hydrophobic amino acid residues were underrepresented.
Influence of phage-displayed peptides on zoospore encystment.
We used time-lapse photography or visual counts to distinguish between P. capsici zoospores and cysts (Fig. 1). In a typical time-lapse image, motile zoospores leave elongated tracks as they continue swimming during the period of exposure. Cysts, on the other hand, remain stationary and appear as dots in the image. Nonmotile zoospores appear as elongated stationary images in the cultures at higher magnification; they did not appear in these experiments.
FIG. 1.
Micrograph (negative of original dark-field image) showing a mixture of nonmotile and motile zoospores. Encysted zoospores do not move during the period of exposure (1 to 2 s), and their images are period shaped, while the tracks of the motile zoospores are elongated. The large spherical objects are sporangia and are disregarded in the measurements.
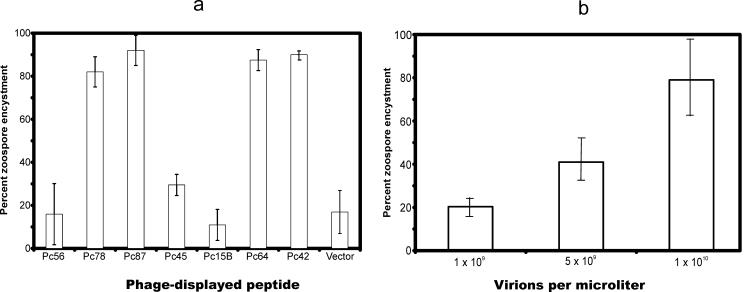
The abilities of phage clones to induce encystment varied (Fig. 2a). For example, at a concentration of 2.5 × 109 virions/μl, clones Pc78, Pc87, Pc64, and Pc42 induced >80% encystment within 20 min. Conversely, only about 20% of zoospores exposed to Pc56, Pc45, Pc15B, or the fuse5 phage vector were encysted during this period. In a separate experiment, we compared the abilities of identical numbers of vector phage and the unselected library to induce encystment. In separate experiments, the vector and the unselected library induced similar amounts of encystment (7.5 and 8%, respectively, after 20 min of incubation). The extent of encystment induced by phage Pc87 was concentration dependent (Fig. 2b).
FIG. 2.
Premature encystment of P. capsici zoospores induced by phage-displayed peptides. (a) Effectiveness of different selected phage. A 20-μl droplet containing 400 zoospores was incubated with phage-displayed peptides (∼5 × 1010 particles) for 20 min at room temperature. The number of zoospores encysted was counted and expressed as a percentage. The percentages represent the means of two experiments. The percent encystment for the control zoospore population that contained no phage varied between 0 and 10%. (b) Concentration dependence of encystment. Phage (in water) were mixed with zoospores, and the amount of encystment was determined visually after 5 min at room temperature. The error bars indicate standard deviations.
Specificity of phage-displayed peptide binding to zoospores and cysts.
In several cases, the ability of phage to cause encystment of P. capsici zoospores reflected their ability to bind to the zoospores. When phage were mixed with zoospores under the conditions used for selection, >107 TU of cyst-inducing phage Pc87 (>50 per zoospore) was recovered after 30 min of incubation, while only about 50,000 TU (∼0.25 per zoospore) of noninducing phage Pc45 or phage vector was recovered under the same conditions. Moreover, binding was specific to zoospores: <104 Pc87 TU (∼0.05 per cyst) was recovered from cysts. This level of binding was similar to the background binding of control vector phage (Table 2).
TABLE 2.
Stage specificity of display phage binding to P. capsicia
| Source of recovered phage | Binding (TU)
|
|
|---|---|---|
| Vector | Pc87 | |
| Cysts | 3,000 ± 800 | 4 × 104 ± 3 × 104 |
| Zoospores | 4 × 104 ± 3 × 104 | 1.0 × 108 ± 3 × 107 |
Bound phage (of 5 × 1010 input phage) were eluted and titrated as TU. The values represent mean binding in three experiments.
Zoospore encystment induced by free peptides in solution.
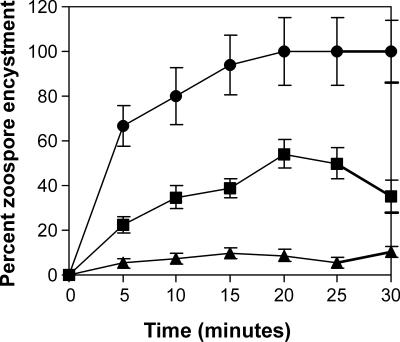
We synthesized peptides corresponding to several of the sequences identified by phage display and tested these species for the ability to encyst P. capsici zoospores. The results (Fig. 3) correlated qualitatively with the behavior of the peptide sequences in their phage display context. Thus, 200 μM peptide 87, corresponding to phage Pc87, induced complete encystment of zoospores. In contrast, peptide 56, corresponding to the ineffectual phage Pc56, did not induce significant encystment at the same concentration. Peptide 42, corresponding to phage Pc42, induced an intermediate level of encystment.
FIG. 3.
Zoospore encystment induced by free peptides. The percentages of encystment induced by peptide 87 (•), peptide 42 (▪), and peptide 56 (▴) are shown. The error bars represent the calculated Poisson standard deviation as determined from the number of cysts and zoospores counted in each determination. These varied, since zoospores were able to swim in and out of the field during the experiment while cysts were necessarily fixed.
DISCUSSION
Control of Phytophthora infection is currently achieved biocidally, e.g., by fumigation with methyl bromide or treatment with metalaxyl. This strategy has significant drawbacks, including the danger of occupational exposure, as well as concomitant loss of microbial diversity and soil tilth. An alternative, environmentally more benign strategy could result from engineering plants to be less susceptible to infection. In this context, the expression of antimicrobial peptides would seem to be an attractive approach to plant protection. Antimicrobial peptides are expressed by a number of metazoan species (29), including humans, in which they function in primary antimicrobial defense. Given that they are biomolecules, peptides are nonpersistent and potentially pathogen specific. Expression of these peptides can also be restricted to regions of the plant where infection is most likely to occur (e.g., the growing root tip), which would make them unlikely to enter the human food chain. All these features would be a significant advantage over existing measures to control Phytophthora parasitism.
A successful peptide-based anti-infection strategy requires the identification of specific peptides as a first step. Since limited genetic or genomic information is available for most plant pathogens, the design of molecular species that target pathogens is time-consuming and expensive. We chose, therefore, to use a combinatorial strategy for the identification of peptides that bound to the pathogens and then to screen these binding peptides for the ability to interfere with pathogen development. Phytophthora zoospores exhibit chemotaxis and orient their development with respect to the plant surface. Consequently, it seems likely that molecular receptors exist on the zoospore surface to control these events.
Combinatorial methods offer a powerful means of obtaining ligands that bind to receptors or other molecules. In many ways, combinatorial strategies mimic the immune system, and it is not surprising that phage display methods have been used to identify antibodies for virus diagnosis (10, 26). An antibody-phage display strategy has been used to similar effect against surface-exposed epitopes on germlings and spores of Phytophthora infestans (9, 21). The isolated antibodies cross-reacted with Phytophthora cactorum, Phytophthora citricola, and Phytophthora megasperma, indicating that these species contain similar epitopes; however, the abilities of the antibodies to affect development were not assessed.
We used a whole-cell-based screening procedure to identify phage-displayed peptides rather than antibodies. Selections of phage-displayed peptides against whole cells have been less common than selection against purified molecules (24). In several cases, however, tumor- and tissue-specific phage have been isolated by whole-animal screening (17), pointing out the power of phage display methods to identify specific effectors of biological events in the absence of detailed information about the actual receptors involved. Together with the present work, these studies point to the utility of phage display as a means of identifying molecules that disrupt pathogenic processes.
It was possible that the putative developmental receptors we wished to target would be only a minority of the molecular species on the zoospore surface. In this case, it was possible that all the phage we isolated would bind to other, more numerous components of the cell surface which were uninvolved in development. If this were so, only a relatively few phage-displayed peptide sequences would be isolated, and these would not be capable of interfering with pathogen motility or development. In order to minimize this possibility, we carried out the selection procedure for only three rounds of binding-elution-amplification, and at constant stringency, before testing specific clones. We hypothesized that this strategy would allow the identification of diverse peptide species, some of which would be relevant to development (13).
The results shown in Tables 1 and 2 and in Fig. 2 indicate that this strategy was successful. The selected phage displayed diverse but related sequences, with only two clones represented twice. This yielded an estimate that the selected population consisted of approximately 120 molecular species as determined by the Chao1 sampling diversity function (12). Perhaps remarkably, an appreciable fraction of the clones we tested were effective in inducing encystment, even though some of the phage, e.g., Pc45, apparently escaped the selection process and did not bind strongly to zoospores.
Phage-displayed peptide Pc87 bound to the zoospore stage of P. capsici, against which it was selected, and its ability to induce encystment was concentration dependent. This phage, therefore, fulfills the definition of a pharmacological agonist, i.e., it is a molecular species that induces a physiological response in a concentration-dependent manner.
The limited selection strategy employed in this work, while selecting for affinity only, nonetheless enriched for phage species that were able to induce encystment. Approximately half of the phage species selected only for the ability to bind to the zoospore surface concomitantly induced encystment. This result could mean that the receptors targeted by these phage are relatively abundant on the zoospore surface or that these receptors present surface characteristics that are especially effective in binding other molecules. Obviously both situations could exist simultaneously.
How effective are the selected peptides in inducing encystment on a molecular basis? Given that the landscape library we used exhibits approximately 4,000 copies of the peptide on the phage surface (18), it is possible to compute a total peptide concentration from the number of virus particles in the assay. At a total peptide concentration equivalent to 16 μM, several phage-displayed peptides induced >80% encystment within 20 min, whereas zoospores mixed with nonselected phage remained motile for 4 h or more. The above concentration estimate assumes that all peptides on the phage surface are simultaneously accessible to the zoospores. This assumption is conservative, since only a few of the inserted peptides on a single phage particle could be involved in any single zoospore-binding event; the rest would be physically removed from the zoospore surface. It is therefore likely that the phage-displayed peptide sequences are active at low to submicromolar concentrations. This activity compares favorably with the affinities of other pharmacologic agents for their targets and suggests that these peptides could be protective against P. capsici when produced by a plant.
Free peptides were approximately 10-fold less effective on a molar basis at inducing encystment than were their corresponding phage-displayed sequences. The greater activity of the phage-borne peptides could be due to two possible causes. First, multivalency could enhance the local concentration of adjacent phage-displayed peptides at a single surface receptor molecule. Second, the phage-displayed peptides in the landscape library we used are conformationally restricted (18) while small peptides adopt a large number of conformations. A plant defense strategy involving expression of these peptides from the plant will likely require an investigation of the effects of either of these two variables on the overall effectiveness of the peptides.
In conclusion, we have described the utility of whole cells of a plant pathogen as targets to select bioactive peptides from phage display libraries without molecular characterization of the receptors. The selected peptides are potentially protective against infection when produced by the plant. If successful, this approach may lead to useful disease resistance, complementary to the strategy of engineering plants to express defensins or cecropins (7, 15, 16, 19). In addition, these peptides provide reagents for the identification of receptors involved in developmental progression.
Acknowledgments
We thank George Smith, Valery Petrenko, and the University of Missouri Combinatorial Biotechnology Group for libraries and helpful discussions; J. H. Im and F. Gallazzi for peptide synthesis; Monique Johnston for scoring micrographs; and Matt Martin for technical assistance.
This research was sponsored by grants from the Monsanto Corp. and the Illinois-Missouri Biotechnology Alliance.
REFERENCES
- 1.Balass, M., Y. Heldman, S. Cabilly, D. Givol, E. Katchalski-Katzir, and S. Fuchs. 1993. Identification of a hexapeptide that mimics a conformation-dependent binding site of acetylcholine receptor by use of a phage-epitope library. Proc. Natl. Acad. Sci. USA 90:10638-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cwirla, S. E., E. A. Peters, R. W. Barrett, and W. J. Dower. 1990. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. USA 87:6378-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deacon, J. W., and S. P. Donaldson. 1993. Molecular recognition in the homing responses of zoosporic fungi, with special reference to Pythium and Phytophthora. Mycol. Res. 97:1153-1171. [Google Scholar]
- 4.Érsek, T., J. T. English, and J. E. Schoelz. 1995. Creation of species hybrids of Phytophthora with modified host ranges by zoospore fusion. Phytopathology 85:1343-1347. [Google Scholar]
- 5.Érsek, T., J. E. Schoelz, and J. T. English. 1994. PCR amplification of species-specific DNA sequences can distinguish among Phytophthora species. Appl. Environ. Microbiol. 60:2616-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong, S., L. V. Doyle, J. J. Devlin, and M. V. Doyle. 1994. Scanning whole cells with phage-display libraries—identification of peptide ligands that modulate cell function. Drug Dev. Res. 33:64-70. [Google Scholar]
- 7.Gao, A.-G., S. M. Hakimi, C. A. Mittanck, Y. Wu, B. M. Woerner, D. M. Stark, D. M. Shah, J. Liang, and C. M. T. Rommens. 2000. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18:1307-1310. [DOI] [PubMed] [Google Scholar]
- 8.Gough, K. C., W. Cockburn, and G. C. Whitelam. 1999. Selection of phage display peptides that bind to cucumber mosaic virus coat protein. J. Virol. Methods 79:169-180. [DOI] [PubMed] [Google Scholar]
- 9.Gough, K. C., Y. Li, T. J. Vaughan, A. J. Williams, W. Cockburn, and G. C. Whitelam. 1999. Selection of antibodies to surface epitopes of Phytophthora infestans. J. Immunol. Methods 228:97-108. [DOI] [PubMed] [Google Scholar]
- 10.Griep, R. A., C. van Twisk, H. J. H. G. Keller, R. J. Kerschbaumer, R. Kormelink, R. W. Goldbach, and A. Schots. 2000. Applications of phage display in selecting tomato spotted wilt virus-specific single-chain antibodies (scFvs) for sensitive diagnostic in ELISA. Phytopathology 90:183-190. [DOI] [PubMed] [Google Scholar]
- 11.Hardham, A. R., and E. Suzaki. 1986. Encystment of zoospores of the fungus Phytophthora cinnamomi is induced by specific lectin and monoclonal antibody binding to the cell surface. Protoplasma 133:165-173. [Google Scholar]
- 12.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levitan, B. 1998. Stochastic modeling and optimization of phage display. J. Mol. Biol. 277:893-916. [DOI] [PubMed] [Google Scholar]
- 14.Morris, P. F., E. Bone, and B. M. Tyler. 1998. Chemotropic and contact responses of Phytophthora sojae hyphae to soybean isoflavonoids and artificial substrates. Plant Physiol. 117:1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas, K. B., H. B. Nicholas, Jr, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. http://www.psc.edu/biomed/dissem/genedoc/index.html.
- 16.Osusky, M., G. Zhou, L. Osuska, R. E. Hancock, W. W. Kay, and M. Mishra. 2000. Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat. Biotechnol. 18:1162-1166. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualini, R., and E. Ruoslahti. 1996. Organ targeting in vivo using phage display peptide libraries. Nature 380:364-366. [DOI] [PubMed] [Google Scholar]
- 18.Petrenko, V. A., G. P. Smith, X. Gong, and T. Quinn. 1996. A library of organic landscapes on filamentous phage. Protein Eng. 9:797-801. [DOI] [PubMed] [Google Scholar]
- 19.Rajasekaran, K., K. D. Stromberg, J. W. Cary, and T. W. Cleveland. 2001. Broad-spectrum antimicrobial activity in vitro of the synthetic peptide D4E1. J. Agric. Food Chem. 49:2799-2803. [DOI] [PubMed] [Google Scholar]
- 20.Ristaino, J. B. 1991. Influence of rainfall drip irrigation and inoculum density on the development of Phytophthora root and crown rot epidemics and yield in bell pepper. Phytopathology 81:922-929. [Google Scholar]
- 21.Robold, A. V., and A. R. Hardham. 1998. Production of species-specific monoclonal antibodies that react with surface components on zoospores and cysts of Phytophthora nicotianae. Can. J. Microbiol. 44:1161-1170. [Google Scholar]
- 22.Saggio, I., and R. Laufer. 1993. Biotin binders selected from a random peptide library expressed on phage. Biochem. J. 293:613-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott, J. K., and G. P. Smith. 1990. Searching for peptide ligands with an epitope library. Science 249:386-390. [DOI] [PubMed] [Google Scholar]
- 24.Smith, G. P. 1991. Surface presentation of protein epitopes using bacteriophage expression systems. Curr. Opin. Biotechnol. 2:668-673. [DOI] [PubMed] [Google Scholar]
- 25.Smith, G. P., and J. K. Scott. 1993. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217:228-257. [DOI] [PubMed] [Google Scholar]
- 26.Susi, P., A. Ziegler, and L. Torrance. 1998. Selection of single chain variable fragment antibodies to black currant reversion associated virus from a synthetic phage display library. Phytopathology 88:230-233. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, J., and G. P. Smith. 1996. Affinity maturation of phage-displayed peptide ligands. Methods Enzymol. 267:3-27. [DOI] [PubMed] [Google Scholar]
- 29.Zasloff, M. 1992. Antibiotic peptides as mediators of innate immunity. Curr. Opin. Immunol. 4:3-7. [DOI] [PubMed] [Google Scholar]