Abstract
Nematophagous fungi are soil-living fungi that are used as biological control agents of plant and animal parasitic nematodes. Their potential could be improved by genetic engineering, but the lack of information about the molecular background of the infection has precluded this development. In this paper we report that a subtilisin-like extracellular serine protease designated PII is an important pathogenicity factor in the common nematode-trapping fungus Arthrobotrys oligospora. The transcript of PII was not detected during the early stages of infection (adhesion and penetration), but high levels were expressed concurrent with the killing and colonization of the nematode. Disruption of the PII gene by homologous recombination had a limited effect on the pathogenicity of the fungus. However, mutants containing additional copies of the PII gene developed a higher number of infection structures and had an increased speed of capturing and killing nematodes compared to the wild type. The paralyzing activity of PII was verified by demonstrating that a heterologous-produced PII (in Aspergillus niger) had a nematotoxic activity when added to free-living nematodes. The toxic activity of PII was significantly higher than that of other commercially available serine proteases. This is the first report showing that genetic engineering can be used to improve the pathogenicity of a nematode-trapping fungus. In the future it should be possible to express recombinant subtilisins with nematicidal activity in other organisms that are present in the habitat of parasitic nematodes (e.g., host plant).
Many nematodes are parasites on plants or animals and can cause serious crop losses and severe diseases. The development of new methods to control such nematodes is of major importance because the traditional methods that are based on the use of nematicides and antihelminthic drugs are associated with major environmental and health concerns. In addition, resistance development in target organisms requires new nematicides to be developed continuously. An alternative strategy is to control parasitic nematodes by using their natural enemies (23). Among them, the nematophagous fungi, which comprise a group of mainly soil-living fungi that are infectious to nematodes, have drawn the largest interest. A common strategy has been to mass-produce the fungi on solid substrates followed by application to the environment. Although there are several reports of successful biological control of both plant and animal parasitic nematodes by using nematophagous fungi (e.g., references 24 and 31), they are commonly perceived to perform poorly compared to traditional nematicides and antihelminthics.
One way to improve the control potential of nematophagous fungi would be to use genetic engineering to increase the pathogenicity and survival of the introduced fungus. Until recently, however, such a development has been precluded due to the lack of information about the molecular background of fungal infections of nematodes (11). We have identified two putative pathogenicity factors in the common nematophagous fungus Arthrobotrys oligospora, a carbohydrate binding protein (lectin) and an extracellular serine protease designated PII (1, 20, 26). PII belongs to the subtilisin family of serine proteases (27). Extracellular subtilisins have also been isolated from other species of nematophagous fungi (3, 13, 21). The function of these proteases is not yet clear, but it has been assumed that they have a role during the fungal penetration of the nematode cuticle and/or during the digestion of the internal tissues of the host.
In this report, the role of PII has been studied in more detail by generating several PII mutants, using a recently developed transformation system for A. oligospora (28). Notably, mutants containing additional copies of the PII gene demonstrated an increased pathogenicity. This is the first report showing that genetic engineering can be used to improve the virulence of a nematode-trapping fungus. Furthermore, PII was expressed in a heterologous system (Aspergillus niger), and it was demonstrated that the recombinant enzyme has a nematotoxic activity in vitro.
MATERIALS AND METHODS
Organisms and plasmids.
A. oligospora Fres. (ATCC 24927) was maintained on CMA7-containing cornmeal agar (Difco) supplemented with 2 g of KH2HPO4 per liter. Trap-containing mycelium was grown in a liquid medium containing 0.01% soya peptone (neutralized; Oxoid) supplemented with 0.05 g of phenylalanine and 0.05 g of valine per liter (27). Two auxotrophic pyrG mutant strains of A. niger were used for heterologous expression of PII: AB1.13pcl3FOA17 (designated AB1.13pclA), which is deficient in both aspergillopepsin A and aspergillopepsin B (due to a regulatory mutation, prt-13) (14) and with the pclA gene deleted (processing enzyme) (P. J. Punt, unpublished data), and A. niger AB1.18, which is mutated in the pepA gene encoding aspergillopepsin A (14). The nematode Panagrellus redivivus L. (Goodey) was grown axenically in a soya peptone-liver extract medium (15). The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description/use | Source |
|---|---|---|
| pUBH | Genomic clone of PII | 1 |
| pAotubF2 | Gene fragment of tubA (from A. oligospora) | This study |
| pBHE | Gene disruption vector for PII | This study |
| pCB1003 | Transformation vector containing hph | 5 |
| pAN7-1 | Transformation vector containing hph | 19 |
| pCB1003-PII | Transformation vector containing hph and PII | This study |
| pAFR | PCR clone of PII | This study |
| pAN52-7Not | glaA promoter (from A. niger) | 30 |
| pASP | gluA promoter linked to PII | This study |
| pAMDSPYRG#1 | amdS/pyrG selection marker | (P. J. Punt, unpublished data) |
Bioassays.
An infection assay using a dialysis membrane technique was employed (20). Briefly, conidia of A. oligospora were inoculated onto pieces of dialysis membrane and incubated on agar plates containing a low nutrient mineral salt (LNM) medium. The formation of infection structures (traps) was induced by adding a few specimens of the nematode P. redivivus to the hyphae growing on the dialysis membranes. After another 7 to 10 d, the infection experiments were started by adding 20 to 30 nematodes cm−2. The number of adhered (captured) and killed (captured and not moving) nematodes was counted by using a microscope. The toxic effects of proteases on free-living P. redivivus were tested using an assay in microtiter wells (27). The numbers of mobile and immobile (i.e., with arrested movements) nematodes were determined by using a light microscope. The effect of PII was compared with that of proteinase K and trypsin (bovine pancreas) (Sigma). The enzymes were dissolved in water or phosphate-buffered saline (50 mM sodium phosphate buffer [pH 6.5], 0.15 M NaCl). The nematotoxic activities of linoleic acid and ivermectin (from Sigma) were also examined. The compounds were dissolved in 5% methanol (in water). The bioassays were performed with 10 to 15 parallels and repeated at least twice.
Competitive reverse transcription-PCR.
Total RNA was isolated from the mycelia growing on the dialysis tubing by acid-guanidinium thiocyanate-phenol-chloroform extraction (20), dissolved in diethyl pyrocarbonate-H2O, and then stored at −20°C. Traces of genomic DNA were digested by RQ1 DNase (Promega). First-strand synthesis was performed in 10-μl reaction mixtures by using oligo(dT)12-18 primer (Gibco/BRL), 2 μl of total RNA, 30 U of RNasin RNase inhibitor (Promega), and 100 U of Superscript II RNaseH reverse transcriptase (Gibco/BRL) with other conditions according to the manufacturer. After termination, the first-strand product was diluted by adding 20 μl of 1× first-strand buffer (Gibco/BRL). As competitive PCR templates, plasmids containing the A. oligospora PII gene (pUBH) and a fragment of the partially characterized A. oligospora tubulin gene (tubA) were used. Here, the tubA template was used as a positive control and for normalizing the data between different measurements and experiments. For cloning of the tubA gene fragment, standard PCR was conducted on A. oligospora genomic DNA by using the primers Bt2a (5′-GGTAACCAAATCGGTGCTGCTTTC) and Bt2b (5′-ACCCTCAGTGTAGTGACCCTTGGC) (10). A PCR product of the expected size (567 bp) was ligated into the pGEM-T Easy vector (Promega), generating the plasmid pAotubF2. The insert was sequenced and verified as a β-tubulin gene fragment by homology to other fungal tubulin genes (GenBank accession number AY028375).
The primers used for competitive PCR were designed to flank intron regions, resulting in a size difference of the products amplified from genomic DNA (gDNA) and the corresponding cDNA template. For PII (GenBank accession number X94121), the primers P106 (5′-TGAGGTCGACTACGTTGAACAAG) and P107 (5′-GGAATCAGTCTTGTCAACAGAGTT) were used, giving PCR products of 320 bp (gDNA) and 259 bp (cDNA), respectively, whereas for tubA the primers were P114 (5′-CTCGACGGCTCCGGTGTTTA) and P115 (5′-CAGGAAACAGCTATGAC), yielding products of 320 bp (gDNA) and 179 bp (cDNA), respectively. PCR products were separated on a 1.5% agarose gel in 0.5× Tris-borate-EDTA and visualized with EtBr and UV light. Equivalence points were calculated by plotting molar ratios of gDNA to cDNA band intensities, and the molar amount of cDNA was determined from the point at the regression line where the ratio between gDNA and cDNA was equal to 1.
Construction of PII mutants.
The genomic PII clone pUBH was PCR amplified (Expand High-Fidelity PCR system; Roche Molecular Biochemicals) with two primers designed from the PII sequence: a1 (5′-TGGGAGATGCGAATACCGGT; position 854 to 873; GenBank accession no. X94121) and a2 (5′-ACCGTGAATGGTTCATCCCG; position 948 to 967). The hph gene under the control of the trpC promoter from Aspergillus nidulans was excised from the plasmid pCB1003 using EcoRI (5). End regions of both the PCR fragment and the excised hph gene were filled in with Klenow before ligation, producing the gene disruption vector pBHE. This vector contains the 5′ and 3′ ends (approximately 1.5 and 1.6 kb, respectively) of the PII gene with a displacement of 74 bp (a 61-bp intron and a 13-bp exon region) and a 1.4-kb hph insertion, as well as an intact plasmid backbone (see Fig. 2A).
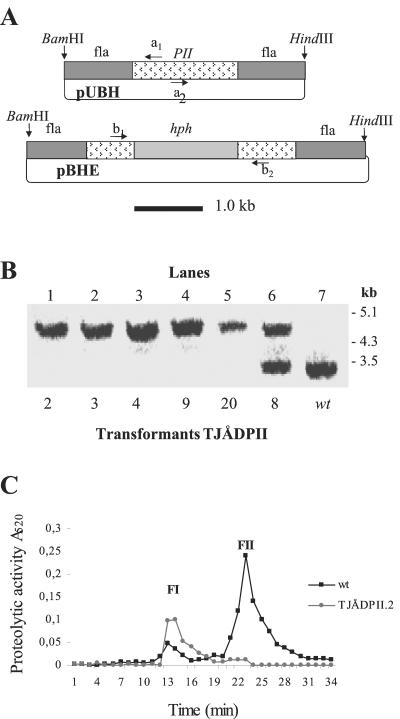
FIG. 2.
Construction of ΔPII mutants. (A) Gene disruption vector pBHE constructed by inserting the hph gene (Escherichia coli hygromycin B resistance gene, under the control of the trpC promoter from A. nidulans) in the open reading frame of PII present in the vector pUBH. fla, flanking region. Arrows indicate PCR primers used for cloning (a1 and a2) and for screening (b1 and b2). (B) Southern analysis of gene disruption mutants obtained using the vector pBHE. Genomic DNA was digested with BamHI/HindIII and probed with a 32P-labeled fragment of the PII gene (BamHI/HindIII fragment excised from pUBH) (30). Lanes 1 to 5 are mutants (designated TJÅDPII.2, -3, -4, -9, and -20) for which the mutated PII gene has been homologously integrated into the genome. Lane 6 shows a mutant (TJÅDPII.8) containing nonhomologous integration of the mutated PII gene. Lane 7 is the wild type. (C) Partial purification of the extracellular serine proteases produced by the wild type and the deletion mutant TJÅDPII.2 of A. oligospora when grown in liquid medium that stimulated the formation of infection structures (traps). The extracts were applied on a Mono Q column equilibrated with a Tris-HCl buffer (pH 7.5) and eluted with a gradient of NaCl using a flow of 1 ml/min (27). One-milliliter fractions were collected and assayed for protease activity by using the chromogenic substrate Azocoll.
For generating mutants containing additional copies of the PII gene, the pUBH vector was cotransformed with the pAN7-1 vector at a molar ratio of 1:1. Multicopy mutants were also generated by transforming the fungus with a vector containing both the PII and hph genes. The vector designated pCB1003-PII was constructed by excising a BamHI/HindIII fragment containing the entire PII gene from the plasmid pUBH. The fragment was inserted into the BamHI/HindIII sites of pCB1003.
Transformation of A. oligospora was performed according to the methods described by Tunlid et al. (28). To confirm the stability of the transformants, they were transferred several times between media with and without selection pressure (hygromycin). Transformants were analyzed by Southern blotting as described below. The pBHE gene disruption transformants were also analyzed by PCR amplification. Two primers positioned in the PII gene adjacent to the inserted hph gene were used: b1 (5′-TTTGACAAGGCAACTCTCCAGG; position 650 to 671; GenBank/EMBL accession no. X94121) and b2 (5′-CCACCAGCGCTAAGAACCTTAAC; position 1092 to 1114) (see Fig 2A). Transformants were screened for extracellular serine protease activity by growing the fungus for 2 to 3 days (130 rpm; 20°C) in a liquid medium containing 0.5% (wt/vol) soya peptone. Transformants were also grown under nitrogen-limited conditions and transferred to an inducing medium containing bovine serum albumin (BSA; 1 mg/ml) as the nitrogen and carbon source (1).
Heterologous expression of PII.
A gene fragment covering the entire PII gene (position 358 to 1682; GenBank accession no. X94121), including regions encoding the signal peptide, propeptide, and the mature enzyme, was obtained by PCR amplifying the genomic clone pUBH using two primers: Aspfw (5′-TCCATCATGATTACGAACGG) and Asprw (5′-CTTGGTCATGAGTCTTTGGG). Both primers contained RcaI sites (underlined). The product was inserted into a pGEM-T vector (Promega), generating the plasmid pAFR. The expression vector pAN52-7Not, containing the glaA promoter (30), was cut with NcoI and the RcaI fragment from pAFR was inserted to produce the expression vector pASP. Junction regions of pASP were sequenced to verify the orientation and cloning sites of the insert. The two auxotrophic pyrG mutant strains of A. niger (AB1.13pclA and AB1.18) were cotransformed with pASP and the amdS/pyrG selection marker (isolated as a NotI fragment from pAMDSPYRG#1 [P. J. Punt, unpublished data]) (18). Transformants with strong growth on plates with acetamide-acrylamide as sole nitrogen (and carbon) source were selected (29). To analyze the production of recombinant PII, the transformants of A. niger were grown for 48 h at 30°C (200 rpm) in minimal growth medium (2) supplemented with 5% maltodextrin.
Peptide sequencing and protease assays.
Proteases were purified from the culture filtrates of A. oligospora and A. niger by high-performance liquid chromatography (HPLC) (27). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a MiniProtean II electrophoresis cell (Bio-Rad). Gels were stained with Coomassie brilliant blue. Before peptide sequencing, the protease activity was inhibited by adding phenylmethylsulfonyl fluoride (PMSF; 1 mM). The N-terminal region was analyzed by Edman degradation using an ABI 476A protein sequenator. The hydrolytic activity of PII was measured using the peptide substrate benzoyl (Bz)-Phe-Val-Arg-nitroanilide (NA) and the chromogenic protein substrate Azocoll (Sigma) (27). The substrate specificity, the effects of inhibitors, and the pH dependency of the recombinant PII were examined as previously described (27).
Hydrolysis of nematode cuticle and tissues.
The cuticle of P. redivivus was isolated according to the protocol of Cox et al. (7). Briefly, nematodes were sonicated in a Tris buffer (10 mM Tris-HCl, pH 7.4), the samples were centrifuged, and the supernatant was collected (intracellular fraction). The pellet was treated with an SDS-containing buffer (7) and, following centrifugation, the supernatant was collected (cuticle-SDS fraction). The pellet was washed with the Tris buffer, lyophilized, and resuspended to give a concentration of 1.0 mg/ml (cuticle-fragment fraction). Before being used in the assays, the intracellular fraction was filtered (0.2-μm pore size), and the cuticle-SDS fraction was desalted using a PD-10 column (Pharmacia) equilibrated with the Tris buffer. Ten microliters of recombinant PII, proteinase K, or trypsin (1.0 × 10−3 proteolytic units [PU]; 0.3 to 0.8 μg of protein) was mixed with 300 μl of the intracellular fraction (total protein content, 18 mg/ml), cuticle-SDS fraction (0.12 mg/ml), and the cuticle-fragments fraction (1.0 mg/ml), and incubated at 37°C for 2 h. Samples were centrifuged and the products of protease hydrolysis were assayed by HPLC using a size-exclusion column (Superdex PC 3.2/30) connected to a SMART system (Pharmacia). The buffer was 10 mM Tris (pH 7.4) containing 0.15 M NaCl. Following the subtraction of a chromatogram (280 nm) obtained from a sample incubated in buffer only, the areas of low-molecular-weight peaks (less than ca. 13 kDa) were integrated using the SMART software. The proteolytic activity is given as area counts per minute.
Southern analysis and RNA dot blots.
Genomic DNA was isolated (28), digested, separated on a 0.8% agarose gel, and transferred to a Hybond-N+ membrane (Amersham).
RNA was isolated from liquid-grown mycelia using the RNeasy Plant Mini Kit (Qiagen) and quantified by measuring the absorbance at 260 nm. The RNA samples were blotted on a Hybond-N membrane (Amersham) using a Bio-Dot microfiltration apparatus (Bio-Rad). The amount of applied RNA was approximately 1.0 μg as determined by absorbance at 260 nm.
The probes were labeled with [α-32P]dCTP (≈3,000 Ci mmol−1) using the random-primed DNA method (rediPrime DNA labeling kit; Amersham).
RESULTS
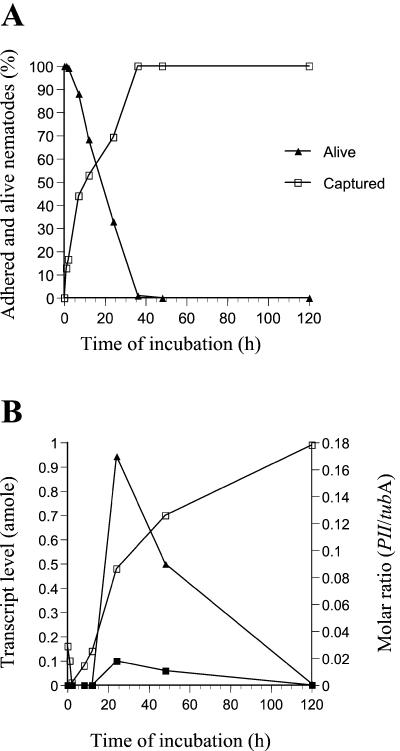
PII is expressed during the colonization and digestion of the nematode. The PII transcript was not detected (<0.001 amole) during the early stages of the infection, including the phase of adhesion (ca. 0 to 12 h of incubation) (Fig. 1). However, the PII mRNA was detected 24 h after adding the nematodes. At this time, the fungus had penetrated and killed ca. 70% of the added nematodes, and fungal hyphae were growing inside the infected nematodes. No PII transcript was detected after 5 days of incubation, by which time all of the added nematodes had been killed and completely degraded.
FIG. 1.
Expression of the subtilisin PII during the infection of nematodes. (A) Adhesion and killing of the nematode P. redivivus during infection by the fungus A. oligospora. The numbers of adhered (captured) and killed (captured and not moving) nematodes were counted using a microscope. (B) Results of analysis of transcript levels of PII and tubA, using competitive reverse transcription-PCR. Tubulin was used as a positive control gene fragment and for normalizing the data. ▪, PII transcript level; □, tubA transcript level; ▴, PII-tubA molar ratio.
Construction of ΔPII mutants.
Twenty mutants of A. oligospora transformed with the gene disruption vector pBHE were examined for proteolytic activity when grown in soya peptone medium. Five of them had significantly reduced proteolytic activity. In Southern analysis, these mutants contained one hybridizing fragment of ca. 4.5 kb (Fig. 2B, lanes 1 to 5) but not the band of 3.2 kb present in the wild type (Fig. 2B, lane 7). This pattern of hybridization is expected after homologous integration of the hph-disrupted PII gene (Fig. 2A). The other 15 mutants were shown to contain both the 3.2-kb as well as the 4.5-kb fragment (Fig. 2B, lane 6), which show the presence of both the native PII gene as well as the disrupted PII gene.
The proteolytic activities of two of the replacement transformants (TJÅDPII.2 and -3) were further examined and compared with the activity of the wild-type strain by using BSA as the inducing substrate (Fig. 2C and 3A). Under these conditions, the proteolytic activities in TJÅDPII.2 and -3 were ca. 70% lower than in the wild-type strain (the maximum activity in the wild type corresponding to 100% was 4.2 pmol of NA ml−1 min−1 mg−1). The serine protease inhibitor PMSF (1 mM) completely inhibited the residual activity against Bz-Phe-Val-Arg-NA in both TJÅDPII.2 and -3. That the reduction of proteolytic activity in the deletion mutants was due to the lack of PII activity was further verified by partly purifying extracellular proteases expressed by TJÅDPII.2 during growth in liquid medium. Under these conditions, the wild-type strain produced two fractions of proteolytic activity designated FI and FII, with the latter containing PII (27). In TJÅDPII.2, the proteolytic activity in FII was almost completely absent (Fig. 2C). That A. oligospora contains several genes encoding serine proteases of the subtilisin family has been indicated by Southern analyses (1).
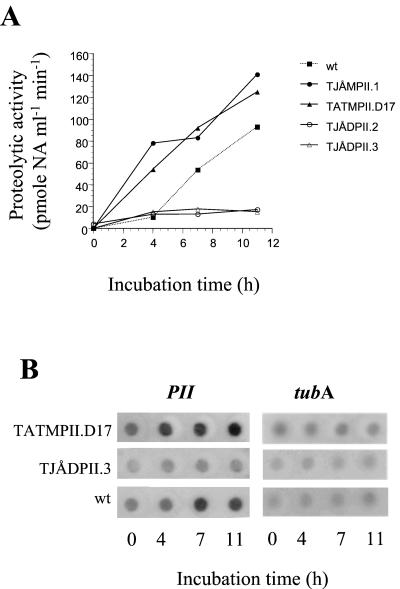
FIG. 3.
Proteolytic activity and expression of mRNA in PII mutants of A. oligospora. Mycelia were grown for 2 days in a nitrogen-limited medium before being transferred to a medium containing albumin as nitrogen and carbon source. (A) Proteolytic activity (in the culture filtrates) as estimated by measuring hydrolysis of the peptide substrate Bz-Phe-Val-Arg-NA. (B) Dot blot of total RNA hybridized with a 32P-labeled fragment of the PII gene (BamHI/HindIII fragment excised from pUBH) (1) and tubA (EcoRI fragment excised from pAotubF2) (cf. Table 1).
Construction of overexpression mutants.
In an initial screen, 100 of the mutants obtained after transformation using the pUBH and pAN7-1 vectors and 50 mutants obtained using the pCB1003-PII vector were analyzed for protease activity when grown in soya peptone medium for 2 to 3 days. None of them showed elevated levels of protease activity. However, Southern analysis of randomly selected mutants indicated that several of them contained additional copies of the PII gene. The induction of proteases in 45 of these mutants was analyzed in more detail using BSA as carbon and nitrogen source. Notably, two of the mutants (TJÅMPII.1 and TATMPII.D17) started to secrete a protease activity more rapidly than the wild-type strain (Fig. 3A). The integration of DNA in TJÅMPII.1 (transformed with the pUBH and pAN7-1 vectors) was analyzed by Southern blotting. Genomic DNA was digested with BamHI and HindIII, and the membranes were hybridized to a BamHI/HindIII gene fragment of the PII gene excised from pUBH. Two fragments were visualized, one the size of the PII band in the wild-type strain and one that was larger (data not shown). As judged from the intensities of these bands, the TJÅMPII.1 mutant contained one additional copy of the PII gene. However, the fact that multiple fragments were observed in the Southern blots indicates that there were gene rearrangements of the inserted DNA. Southern analysis of the TATMPII.D17 mutant (transformed with the pCB1003-PII vector) by HindIII digestion and hybridization, using the pCB1003 plasmid as probe, showed three DNA fragments, with one the size of the pCB1003-PII vector (8 kb). This suggests integration of a tandemly repeated array of at least two copies of the pCB1003-PII vector (data not shown).
The variations in the level of PII mRNA followed approximately the changes in the activity of PII in the wild type, the deletion mutant TJÅDPII.3, and the overexpression mutant TATMPII.D17 (Fig. 3B).
Pathogenicity of PII mutants.
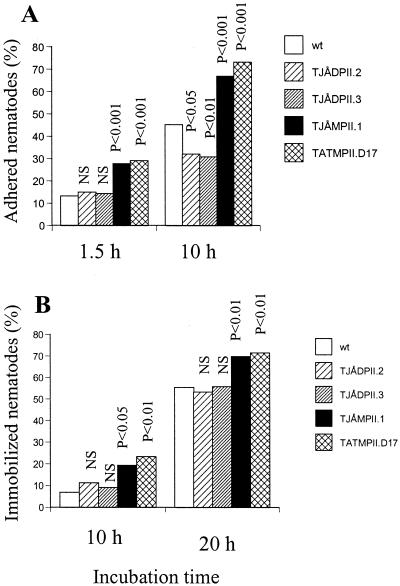
The pathogenicities of two ΔPII mutants (TJÅDPII.2 and -3) and two overexpressing mutants (TJÅMPII.1 and TATMPII.D17) were examined using a bioassay with the nematode P. redivivus. The ΔPII mutants had a lower number of infection structures (traps) than the wild-type strain (Table 2). However, disruption of PII had a limited effect on the capture of nematodes, as significant differences between the ΔPII mutants and the wild type in the capture of nematodes were only observed after 10 h of infection (Fig. 4A). The overexpression mutants had a significantly higher number of traps than the wild-type strain and had an increased speed of capturing nematodes than the wild type (Table 2 and Fig. 4A). The effect of the gene-disrupted and the overexpressing mutants on the immobilization of captured nematodes was also investigated. No significant difference in the immobilization of nematodes was observed between the two ΔPII mutants and the wild-type strain (Fig. 4B). However, the percentage of immobilized nematodes was higher in the overexpressing strains than in the wild type; thus, the overexpressing mutants showed a more rapid killing of nematodes than the wild-type strain.
TABLE 2.
Development of infection structures in various mutants of A. oligosporaa
| Strain | Traps (cm−2) (mean [SD]) |
|---|---|
| Wild type | 81 (36) |
| TJÅDPIL2 | 51 (20) |
| TJÅDPII.3 | 41 (19) |
| TJÅMPII.1 | 202 (67) |
| TATMPII.D17 | 229 (72) |
The fungus was grown on dialysis membranes for 3 d, whereupon nematodes were added to induce trap formation. After another 7 to 10 days, the numbers of traps were counted using a microscope. n = 10.
FIG. 4.
Infection of nematodes by various mutants of A. oligospora. (A) Adhesion of nematodes; (B) immobilization of captured nematodes. Indicated significance levels are from χ2 tests (df = 1) comparing the frequencies of captured and free nematodes (A) or mobile and immobile nematodes (B) in the mutant strains versus the wild type. NS, not significant.
Heterologous expression of PII.
The expression vector pASP was constructed by cloning the open reading frame of PII in front of the glaA (glucoamylase) promoter in A. niger. Twenty-eight transformants (19 from strain AB1.13pclA and 9 from AB1.18) of A. niger that showed extensive growth on the selective acetamide-acrylamide medium were isolated. When grown in liquid medium after induction of the glaA promoter with dextrin, all of these transformants had an increased protease activity compared to the nontransformed control. Southern analysis of genomic DNA from nine different transformants digested with BamHI/HindIII showed three or more bands hybridizing to a probe of the PII gene (data not shown). Since these enzymes do not cut in the inserted PII gene fragment, these data indicate that the transformants contained three or more copies of the PII gene.
The proteolytic activities of the 10 AB1.13pclA and 7 AB1.18 transformants showing the highest levels of extracellular protease expression were purified by HPLC. The protease activity of all AB1.13pclA transformants was eluted in one peak by using a size-exclusion column (Fig. 5). Furthermore, the AB1.18 transformants had an additional peak, with activity eluting in a fraction corresponding to protein(s) with a higher molecular weight than the protease expressed in the AB1.13pclA transformants. The AB1.13pclA transformant showing the highest proteolytic activity (designated AB1.13pclA:3) was chosen for further studies. SDS-PAGE analysis of the HPLC fractions with the highest activity against Bz-Phe-Val-Arg-NA revealed a polypeptide of approximately 48 kDa in mass (Fig. 5, insert). The N-terminal sequence of this polypeptide was XEQTDSTWGLDRISHEDYSA, with X denoting an unidentified residue. Previous peptide sequencing of PII from A. oligospora indicated a blocked N terminus (27). However, based on the alignment of the deduced amino acid sequence of PII with other subtilisin-related fungal serine proteases, it was predicted that the mature PII protease has an N-terminal sequence of TYAEQTDSTWGL (27), which contains two additional residues compared to that of the recombinant PII. The difference observed between the molecular mass of the recombinant PII (48 kDa when using SDS-PAGE) and the native enzyme (40 kDa) (27) could be explained by differences in glycosylation. An alteration in the pattern of glycosylation has been observed for many heterologously produced proteins in Aspergillus, including a Rhizomucor miehi aspartic proteinase (6).
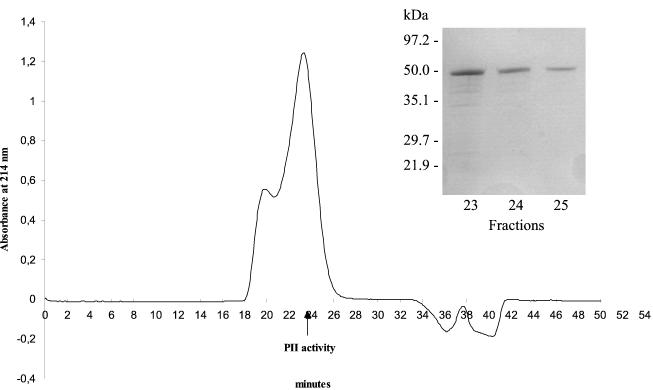
FIG. 5.
Heterologous expression of PII and purification of heterologous PII from A. niger strain AB1.13pclA:3. Extracellular proteases were purified by anion-exchange, hydrophobic-interaction, and size-exclusion chromatography (shown). Protease activities in the different fractions (every minute) were analyzed using the peptide substrate Bz-Phe-Val-Arg-NA. The arrow indicates major PII activity. Inserted is an SDS-PAGE of fraction 23-25.
The yield of PII obtained from AB1.13pclA:3 was 120 μg/liter, which is significantly higher than the production by A. oligospora (0.7 μg/liter) (27). The specific activity of the heterologous PII was 0.8 PU/mg of protein, which is lower than that of PII from A. oligospora (1.9 PU/mg) (27). There were no significant differences between the recombinant and native PII with regard to the effects of various inhibitors, pH optimum, and hydrolysis of synthetic peptide substrates (data not shown). Thus, both enzymes were inhibited by PMSF, chymostatin, and antipain, both had a broad pH optimum in the alkaline range (approximately 7 to 9), and their preferred peptide substrate was Bz-Phe-Val-Arg-NA (27).
Hydrolytic and nematotoxic activity of recombinant PII.
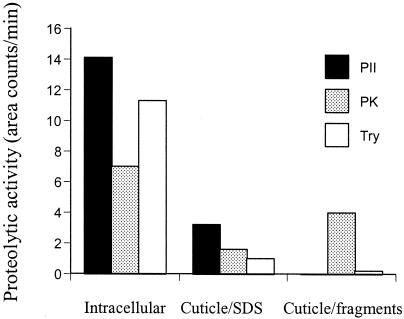
Recombinant PII showed high hydrolytic activity against an intracellular fraction of proteins isolated from the nematode P. redivivus (Fig. 6). Some activity was observed against proteins present in the cuticle-SDS fraction, which is supposed to contain muscle fragments and the tissue remains of the cuticle wall (7). The activity against the purified cuticle fragments was very low. Trypsin had a pattern of hydrolytic activity against the various cellular fractions similar to that of PII. Proteinase K degraded proteins present in all three fractions, including the cuticle fragments.
FIG. 6.
Hydrolysis of nematode cuticle and tissues by various serine proteases. The nematode P. redivivus was sonicated and separated into three fractions. Heterologous-produced PII, proteinase K (PK), or trypsin (Try) were added to the fractions and incubated at 37°C for 2 h. The amount of released peptides was assayed by using HPLC.
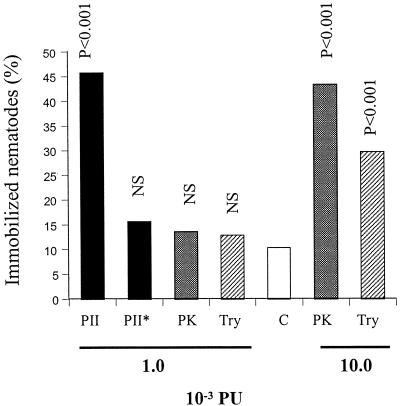
When added to free-living nematodes, the heterologous-produced PII had a nematotoxic activity, i.e., the nematodes were arrested in their movements (Fig. 7). The effect was not observed after boiling the preparation, indicating that it was due to the presence of protease activity in PII. No toxic effect was observed after adding a similar level of proteolytic activity of two other commercially available serine proteases, the fungal subtilisin proteinase K and trypsin. At least 10 times higher proteolytic activity was needed for these enzymes to immobilize nematodes at a level corresponding to that of PII (Fig. 7).
FIG. 7.
Nematotoxic activity of heterologous PII. The proteases were incubated with the nematode P. redivivus in microtiter wells. After 20 to 22 h the number of mobile and immobile (i.e., with arrested movements) nematodes were counted in a light microscope. Indicated significance levels are from χ2 tests (df = 1) comparing the frequencies of mobilized and immobilized nematodes in treated versus control samples (without enzymes). NS, not significant. PII, PII produced in A. niger; PII∗, boiled PII (10 min); PK, proteinase K; Try, trypsin; C, control (buffer). The proteolytic activities of the added enzymes, as measured by using the substrate Azocoll, are indicated.
The concentration of heterologous PII that immobilized ca. 50% of the nematodes in the microwell assay (Fig. 7) was calculated to be 13 μg/ml (2.7 × 10−8 M). Using the same microwell plate assay, linoleic acid and ivermectin immobilized ca. 50% of the nematodes at 100 μg/ml (3.6 × 10−4 M) and 0.5 μg/ml (5.7 × 10−7 M), respectively. Linoleic acid is the only nematotoxic compound that so far has been isolated from A. oligospora (22). Ivermectin is a macrocyclic lactone and belongs to the avermectin group of antihelminthics and insecticides (4).
DISCUSSION
A deletion of the PII gene in A. oligospora by homologous recombination had only a limited effect on the capture and infection of nematodes. This could be explained by the presence of a significant residual proteolytic activity in the ΔPII mutants. However, the role of PII in virulence was demonstrated by analyzing mutants containing additional copies of PII. Two of these mutants showed an increased speed in capturing nematodes compared to the wild type.
That the activity of PII has a role in the initiation or development of traps in A. oligospora was indicated by the fact that the deletion mutants had a lower number and the multicopy mutants a larger number of traps compared to the wild type. It is well documented that the formation of traps in A. oligospora and several other nematode-trapping fungi is stimulated by peptides released from the nematode host (8). Proteases secreted by the fungi could have a role in the generation of such peptides by degrading and solubilizing proteins of the host cuticle. The chymoelastase serine protease Pr1 in Metarhizium anisopliae is induced by peptides from the insect cuticle produced by the basal activity of Pr1 and Pr2 (trypsin-like serine protease) (17). An alternative explanation for the stimulation of trap formation by the PII mutants is that they more rapidly digested the tissues of the infected nematodes, thus making nutrients available for the development of infection structures. Whether the activity of PII could affect the adhesion of nematodes to the traps of A. oligospora is difficult to evaluate, since the ratio of added nematodes to trap numbers varied between the various strains tested. However, that the PII transcript was not detected during adhesion (Fig. 1) and that treatment of A. oligospora with inhibitors against serine proteases did not affect the capture of nematodes (26) make it unlikely that PII has a role in adhesion.
The factor(s) that contributes to the comparable rapid killing of nematodes captured by nematophagous fungi has intrigued scientists for a long time. Early studies indicated that toxic fungal metabolites might be involved in the immobilization of nematodes by A. oligospora (16). Based on experiments demonstrating that only extracts from nematodes infected by the fungus, but not extracts from the fungus or nematodes alone, were toxic to nematodes, it was proposed that A. oligospora secretes a nematotoxic substance which paralyzes nematodes following the capture and penetration of them (16). This earlier paper stimulated a number of studies attempting to isolate and identify nematotoxic compounds from nematophagous fungi. Despite extensive efforts from a number of different laboratories, the results have been confusing (11). The only nematotoxic compounds identified so far from nematophagous fungi are fatty acids, including linoleic acid (12, 22). The observations that recombinant PII immobilized free-living nematodes in vitro indicate that PII can function as a nematotoxic compound. In agreement with the observations by Olthof and Estey (16), this toxic activity is produced after the nematodes are caught by the adhesive trapping nets (Fig. 1). Notably, the toxic activity of PII was significantly higher than that of two other commercially available serine proteases. The difference in nematotoxic activity between the enzymes was apparently not related to their hydrolytic activity on purified cuticle fragments, since proteinase K had a significantly higher enzymatic activity on cuticle fragments but lower nematicidal activity than PII. In combination with the observation that PII is mainly degrading proteins in the intracellular fraction, we propose that the added recombinant PII may be transported inside the nematode and display an intracellular, not-yet-characterized cytotoxic effect.
This is the first report demonstrating that genetic engineering can be used to improve the virulence of a nematophagous fungus. In contrast to some other fungi, including the mycoparasite Trichoderma harzianum (9), it appeared to be difficult to insert a large number of copies of PII into the genome of A. oligospora. Previous studies of integration patterns of transformants generated by using the pAN7-1 vector also indicated a low frequency of multiple and tandem-repeated integrations in A. oligospora (28). Furthermore, the Southern analysis of generated mutants indicated a high frequency of rearrangements of the inserted DNA. Alternatively, the expression levels of PII could be modified or elevated by using a constitutive promoter. This approach was recently successfully used in the entomopathogen M. anisopliae (25). In the future it should be possible to express recombinant subtilisins with nematicidal activity in other organisms that are present in the habitat of parasitic nematodes (e.g., host plant).
Acknowledgments
This study was supported by grants from the Swedish Natural Science Research Council, the Swedish Council for Forestry and Agricultural Research Council, and an EMBO short fellowship (to J.Å.).
We thank Bo Ek for peptide sequencing.
REFERENCES
- 1.Åhman, J., B. Ek, L. Rask, and A. Tunlid. 1996. Sequence analysis and regulation of a gene encoding a cuticle-degrading serine protease from the nematophagous fungus Arthrobotrys oligospora. Microbiology 142:1605-1616. [DOI] [PubMed] [Google Scholar]
- 2.Bennet, J. W., and L. L. Lasure. 1991. Growth media, p. 444-445. In J. W. Bennet and L. L. Lasure (ed.), More gene manipulations in fungi. Academic Press, San Diego, Calif.
- 3.Bonants, P. J. M., P. F. L. Fitters, H. Thijs, E. Den-Belder, C. Waalwijk, and J. W. D. M. Henfling. 1995. A basic serine protease from Paecilomyces lilacinus with biological activity against Meloidogyne hapla eggs. Microbiology 141:775-784. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, W. C. 1982. Progress and prospects in the chemotherapy of nematode infections of man and other animals. J. Nematol. 15:608-615. [PMC free article] [PubMed] [Google Scholar]
- 5.Carrol, A. M., J. A. Sweigard, and B. Valent. 1995. Improved vectors for selecting resistance to hygromycin. Fungal Gen. News 41:22. [Google Scholar]
- 6.Christensen, T., H. Woeldike, E. Boel, S. B. Mortensen, L. T. Hjortshoej, and M. T. Hansen. 1988. High level expression of recombinant genes in Aspergillus oryzae. Bio/Technology 6:1419-1422. [Google Scholar]
- 7.Cox, G. N., M. Kusch, and R. S. Edgar. 1981. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J. Cell Biol. 90:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijksterhuis, J., M. Veenhuis, W. Harder, and B. Nordbring-Hertz. 1994. Nematophagous fungi: physiological aspects and structure-function relationships. Adv. Microb. Physiol. 36:111-143. [PubMed] [Google Scholar]
- 9.Flores, A., I. Chet, and A. Herrera-Estrella. 1997. Improved biocontrol activity of Trichoderma harzianum by overexpression of the proteinase-encoding gene prb1. Curr. Genet. 31:30-37. [DOI] [PubMed] [Google Scholar]
- 10.Glass, N. L., and C. G. Donaldson. 1995. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansson, H.-B., A. Tunlid, and B. Nordbring-Hertz. 1997. Biological control of nematodes, p. 38-50. In T. Anke (ed.), Fungal bio/technology. Chapman and Hall, Weinheim, Germany.
- 12.Kwok, O. C. H., R. Plattner, D. Weisleder, and D. T. Wicklow. 1992. A nematicidal toxin from Pleurotus ostreatus NRRL 3526. J. Chem. Ecol. 18:127-136. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Llorca, L. V., and W. M. Robertson. 1992. Immunocytochemical localization of a 32-kDa protease from the nematophagous fungus Verticillium suchlasporium in infected nematode eggs. Exp. Mycol. 16:261-267. [Google Scholar]
- 14.Mattern, I. E., J. M. Van Noort, P. Van den Berg, D. B. Archer, I. N. Roberts, and C. A. M. J. J. Van den Hondel. 1992. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol. Gen. Genet. 234:332-336. [DOI] [PubMed] [Google Scholar]
- 15.Nordbring-Hertz, B. 1977. Nematode induced morphogenesis in the predacious fungus Arthrobotrys oligospora. Nematologica 23:443-451. [Google Scholar]
- 16.Olthof, T. H. A., and R. H. Estey. 1963. A nematotoxin produced by the nematophagous fungus Arthrobotrys oligospora. Nature 197:514-525. [Google Scholar]
- 17.Paterson, I. C., A. K. Charnley, R. M. Cooper, and J. M. Clarkson. 1994. Partial characterization of specific inducers of a cuticle-degrading protease from the insect pathogenic fungus Metarhizium anisopliae. Microbiology 140:3153-3159. [DOI] [PubMed] [Google Scholar]
- 18.Punt, P. J., and C. A. M. J. J. Van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance marker. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 19.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. M. J. J. Van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 20.Rosén, S., K. Sjollema, M. Veenhuis, and A. Tunlid. 1997. A cytoplasmic lectin produced by the fungus Arthrobotrys oligospora functions as a storage protein during saprophytic and parasitic growth. Microbiology 143:2593-2604. [DOI] [PubMed] [Google Scholar]
- 21.Segers, R., T. M. Butt, B. R. Kerry, and J. F. Peberdy. 1994. The nematophagous fungus Verticillium chlamydosporium Goddard produces a chymoelastase-like protease which hydrolyses host nematode proteins in situ. Microbiology 140:2715-2723. [DOI] [PubMed] [Google Scholar]
- 22.Stadler, M., H. Anke, and O. Sterner. 1993. Linoleic acid—the nematicidal principle of several nematophagous fungi and its production in trap-forming submerged cultures. Arch. Microbiol. 160:401-405. [Google Scholar]
- 23.Stirling, G. R. 1991. Biological control of plant parasitic nematodes: progress, problems and prospects. CAB International, Wallington, United Kingdom.
- 24.Stirling, G. R., K. A Licastro, L. M. West, and L. J. Smith. 1998. Development of commercially acceptable formulations of the nematophagous fungus Verticillum chlamydosporium. Biol. Control 11:217-223. [Google Scholar]
- 25.St. Leger, R. J., L. Joshi, M. J. Bidochka, and D. W. Roberts. 1996. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. USA 93:6349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tunlid, A., and S. Jansson. 1991. Proteases and their involvement in the infection and immobilization of nematodes by the nematophagous fungus Arthrobotrys oligospora. Appl. Environ. Microbiol. 57:2868-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tunlid, A., S. Rosén, B. Ek, and L. Rask. 1994. Purification and characterization of an extracellular serine protease from the nematode-trapping fungus Arthrobotrys oligospora. Microbiology 140:1687-1695. [DOI] [PubMed] [Google Scholar]
- 28.Tunlid, A., J. Åhman, and R. P. Oliver. 1999. Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 173:111-116. [DOI] [PubMed] [Google Scholar]
- 29.Verdoes, J. C., P. J. Punt, J. M. Schrickx, H. W. Van-Verseveld, A. H. Stouthamer, and C. A. M. J. J. Van den Hondel. 1993. Glucoamylase overexpression in Aspergillus niger: molecular genetic analysis of strains containing multiple copies of the glaA gene. Transgen. Res. 2:84-92. [DOI] [PubMed] [Google Scholar]
- 30.Verdoes, J. C., P. J. Punt, A. H. Stouthamer, and C. A. M. J. J. Van den Hondel. 1994. The effect of multiple copies of the upstream region on expression of the Aspergillus niger glucoamylase-encoding gene. Gene 145:179-187. [DOI] [PubMed] [Google Scholar]
- 31.Wolstrup, J., P. Nansen, J. Grönvold, S. A. Henriksen, and A. Zorn. 1996. Toward practical biological control of parasitic nematodes in domestic animals. J. Nematol. 28:129-132. [PMC free article] [PubMed] [Google Scholar]