Abstract
Cyclic lipopeptides (CLPs) with antibiotic and biosurfactant properties are produced by a number of soil bacteria, including fluorescent Pseudomonas spp. To provide new and efficient strains for the biological control of root-pathogenic fungi in agricultural crops, we isolated approximately 600 fluorescent Pseudomonas spp. from two different agricultural soils by using three different growth media. CLP production was observed in a large proportion of the strains (approximately 60%) inhabiting the sandy soil, compared to a low proportion (approximately 6%) in the loamy soil. Chemical structure analysis revealed that all CLPs could be clustered into two major groups, each consisting of four subgroups. The two major groups varied primarily in the number of amino acids in the cyclic peptide moiety, while each of the subgroups could be differentiated by substitutions of specific amino acids in the peptide moiety. Production of specific CLPs could be affiliated with Pseudomonas fluorescens strain groups belonging to biotype I, V, or VI. In vitro analysis using both purified CLPs and whole-cell P. fluorescens preparations demonstrated that all CLPs exhibited strong biosurfactant properties and that some also had antibiotic properties towards root-pathogenic microfungi. The CLP-producing P. fluorescens strains provide a useful resource for selection of biological control agents, whether a single strain or a consortium of strains was used to maximize the synergistic effect of multiple antagonistic traits in the inoculum.
Cyclic lipopeptides (CLPs) are produced by distinctively different groups of bacteria, both gram-positive (20) and gram-negative (28). The high diversity of CLP-producing microorganisms (28) and differences in chemical structure suggest that the CLP compounds may serve different, and possibly multiple, purposes. This may explain why the specific role of CLP production is often unclear (28, 40). For a limited number of CLPs (28), the reported functions include promotion of bacterial swarming (12, 26) and biosurfactant properties (19, 24, 41). In many cases, CLP compounds are also known to exert a role in antagonistic interactions with other organisms (28), e.g., plant pathogenicity (5) and antifungal (19, 30, 31, 38, 44), antibacterial (11), antiviral (49), or cytotoxic (16) activity.
Synthesis of CLPs is nonribosomal and catalyzed by large peptide synthetase complexes (27). Various environmental stimuli may affect CLP production, i.e., carbon substrate (36), limitation by C, N, or P (15, 37), Fe limitation (15), growth phase conditions (15), and interaction with interfaces (32). Little information is available on production rates and regulating factors for the compounds in natural environments. Asaka and Shoda (2) detected surfactin and iturine production by Bacillus subtilis RB14 in a sterilized vermiculite-soil system, and Nakayama et al. (31) detected xanthobaccin A production by a Stenotrophomonas sp. strain, SB-K88, in a hydroponic sugar beet rhizosphere system, but documentation for in situ production of CLPs in natural soils has been lacking.
Despite the fact that several new CLP-producing Pseudomonas spp. strains have recently been isolated from soil (37, 38), an overview of the different types (chemical structure) and properties (surface tension, antibiosis) of CLP compounds, together with frequencies of the producing Pseudomonas spp. strains, has never been made. In this study, we present an extensive survey on lipopeptide production among approximately 600 fluorescent Pseudomonas spp. isolated from the sugar beet rhizosphere. Several new CLP compounds are reported. A total of eight subgroups of strains producing CLPs of different molecular weights (MWs) (amino acid number) and compositions (amino acid substitution) were identified among the Pseudomonas spp. isolates. The strain groups and hence the production of specific CLPs could be affiliated to a limited number of Pseudomonas fluorescens biovars (I, V, and VI). Important differences in biosurfactant and antagonistic properties of the CLPs were documented.
MATERIALS AND METHODS
Isolation of surfactant-producing Pseudomonas spp. strains.
Soil was collected from a fallow field (loamy sand; Højbakkegård field station, Tåstrup [near Copenhagen], Denmark) and a sugar beet field (sandy loam; Danisco Seed, Holeby, Lolland, Denmark) and kept at 5°C until use. The loamy sand and sandy loam contained 46.3 and 21.1% coarse sand, 39.1 and 39.0% fine sand, 5.9 and 17.8% silt, and 7.2 and 19.5% clay, respectively. Total organic C in the Højbakkegård and Danisco soils was 9 and 17 g kg of dry soil−1 and total N was 0.6 and 0.9 g kg of dry soil−1, respectively. The pH values of the two soils in CaCl2 were 6.3 and 7.3, respectively. In all experiments, the soils were air dried to 10% (wt/wt) before sieving through a 2-mm-diameter mesh screen. The water content was then adjusted to 13% (wt/dry weight) by spraying with tap water and gently mixing in a polyethylene bag.
Fifty grams of soil (wet weight) was weighed into 50-ml polyethylene vials and compressed to a bulk density of 1.1 g cm−3. Three sugar beet seeds (Madison variety) were sown in each vial before samples were incubated at 15°C under a 16-h light and 8-h dark cycle. After 7 days, the plants were excavated and five roots plus adhering soil were transferred to 10 ml of sterile 0.9% NaCl to comprise one combined sample. The sample was vortexed for 1 min and sonicated for 0.5 min before the liquid phase was diluted and plated onto solid media for isolation of fluorescent Pseudomonas spp. colonies.
To obtain a high diversity among the isolated Pseudomonas spp., three different isolation media were used. (i) Gould's S1 medium, containing (per liter) 10 g of sucrose, 10 ml of glycerol, 5 g of Casamino Acids, 1 g of NaHCO3, 1 g of MgSO4 · 7H2O, 2.3 g of K2HPO4, 1.2 g of Na-lauryl sarcosin, and 15 g of agar, was autoclaved, and then 5 ml of solution containing 100 mg of trimethoprim (Sigma T-7883), 8.5 ml of methanol, and 16.5 ml of Milli-Q water was added to 1 liter of the medium. Due to the selectivity of Gould's S1 medium for Pseudomonas spp. (13), all colonies appearing on this medium were eligible for random picking. (ii) On King's B medium (Pseudomonas F; Difco catalog no. 0448-17-1), fluorescent Pseudomonas spp. could be detected by illuminating the agar plates with UV light (254 nm) and randomly picking the fluorescent colonies. (iii) To isolate Pseudomonas spp. from plates covered with R2A medium (Difco catalog no. 1826-15-3), a polyclonal antibody targeting outer membrane protein in genuine Pseudomonas spp. (rRNA homology group I) was used in a colony blot procedure (22). For all media, the R2A plates were incubated at 15°C and colonies were picked randomly from plates with the highest dilution (containing more than 10 Pseudomonas spp. colonies). Eventually, all isolates from the three media were streaked again onto Gould's S1 agar and checked for fluorescence before culturing in 3 ml of Luria-Bertani medium containing (per liter) 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, and 1 g of glucose (pH 7.2) for subsequent preservation at −80°C.
The first screening step for surfactant production among a total of 581 fluorescent Pseudomonas spp. isolates was a test for each isolate to lower the interfacial tension between an oil-covered surface and a drop of spent growth medium, as described by Bodour and Miller-Maier (6). All isolates were cultured in 3 ml of Luria-Bertani medium (room temperature, overnight), and 5 μl of culture was transferred into the 10W-40 Pennzoil-coated wells of a microtiter lid. Surfactant production was determined up to 3 min after incubation, and a comparison to a surfactant-producing P. fluorescens strain, DR54 (37), and a nonproducing P. fluorescens strain, DF57 (43), was made.
Structural diversity of Pseudomonas spp. surfactants.
The first characterization of the Pseudomonas spp. surfactants by high-pressure liquid chromatography (HPLC) analysis was performed after culturing of all isolates at 20°C for 2 days in 25-ml glass tubes with 3 ml of Davis minimal broth (DMB) medium. DMB contained 30 mM K2HPO4, 14 mM KH2PO4, 0.4 mM MgSO4, 7.6 mM (NH4)2SO4, 120 mM carbon-C (glucose), and 1 ml of trace element solution per liter (pH 7.3); the trace element solution contained (per liter) 20 mg of CoCl2 · 6H2O, 30 mg of H3BO3, 10 mg of ZnSO4 · 7H2O, 1 mg of CuCl2 · 2H2O, 2 mg of NiCl2 · 6H2O, 3 mg of NaMoO4 · 2H2O, 10 mg of FeSO4 · 7H2O, and 2.6 mg of MnSO4 · H2O. Samples were obtained by extraction for 1 h with 5 ml of ethyl acetate containing 1% formic acid. HPLC analysis of surfactant compounds was performed by using a Hypersil BDS C18 column (100 by 4.6 mm; 3-μm particle diameter) held at 40°C, and UV detection (200 to 400 nM) was performed on a Hewlett-Packard model 1100 HPLC diode array detector. The samples were analyzed in a gradient of 85% eluent A (0.1% o-phosphoric acid) and 15% eluent B (acetonitrile) at 0 min, increasing eluent B to 100% after 40 min. Eluent flow rate was 1 ml per min. Chromatograms were analyzed using the Hewlett-Packard ChemStation software package. Surfactants were considered identical when retention times in HPLC chromatograms varied by less than 0.1 min. The retention times of one (occasionally two) major surfactant peak were used to cluster the isolates, hereafter referred to as Pseudomonas spp. strain groups.
Final purification of surfactants from representative Pseudomonas strains was conducted by preparative HPLC (Waters Delta-Pak C18 column; 15-μm inner diameter, 100 Å, 300 by 19 mm) using gradients of acetonitrile in water with 0.1% trifluoroacetic acid at a flow rate of 20 ml min−1. Structural analyses of the surfactants were based on one-dimensional and two-dimensional nuclear magnetic resonance (NMR) spectra recorded on a Varian 400 MHz FT-NMR spectrometer with deuterated dimethyl sulfoxide (DMSO-d6) as solvent. Fast atom bombardment-mass spectrometry (MS) data were obtained on a JMS-HX/HX110A tandem mass spectrometer (JEOL, USA Inc.) (positive ion mode), and stereochemical data were obtained by Chiral gas chromatography. In the case of tensin and amphisin, the complete structure was established by X-ray crystallographic studies (17, 42).
Diversity of surfactant-producing Pseudomonas spp. strains.
Pseudomonas strains representing the production of all surfactant groups were tentatively identified to the species and biovar levels described by Palleroni (39). The simplified scheme suggested by Sørensen et al. (43) and Nielsen et al. (34) based on the occurrence of denitrification, levan production, and growth in mannitol, meso-inositol, or sorbitol was used. Strains representing all the surfactant groups were also compared by their C utilization patterns by using Biolog GN plates as described by the manufacturer (Biolog Inc., Hayward, Calif.).
Additional phenotypic characteristics tested included extracellular enzymes and secondary metabolites often associated with antagonism by the fluorescent Pseudomonas spp. Endochitinase activity (hydrolytic splitting of chitin polymer) was tested on potato dextrose broth-Bacto agar (PDA; Difco catalog no. 0013-17) medium containing 1.5 mg of chromogenic substrate (carboxymethylcellulose-chitin-RBV; Loewe, Sauerlach, Germany) per ml. Protease activity (casein degradation) was tested as described by Nielsen and Sørensen (35), except using PDA instead of one-fifth-strength tryptic soy agar. Both endochitinase and protease were determined semiquantitatively by the diameters of clearing zones around colonies on the agar plates. Hydrogen cyanide (HCN) production was determined as described by Bakker and Schippers (4) by using the protocol of Nielsen et al. (34). All antagonistic traits were classified visually into categories of no, weak, moderate, or strong activity. Other metabolites tested included diacetyl phloroglucinol, phenazine-1-carboxylic acid, pyrrolnitrin, and pyoluteorin by following the HPLC protocol of Nielsen et al. (37). Absorption spectra (200 to 400 nm) were compared to library spectra by the Hewlett-Packard three-dimensional ChemStation software.
Surface-active properties of Pseudomonas spp. surfactants.
Biosurfactant properties (surface tension) were tested using the spent growth medium of Pseudomonas spp. cultures. One representative strain from each strain group producing a specific CLP profile was cultured in DMB with 20 mM glucose in triplicate for 2 days at 15°C. Cells were removed by centrifugation (10,000 × g) for 10 min at 4°C, and the supernatant was frozen for later analysis. Reduction of surface tension was measured on a Wilhelmy plate mounted in a Sigma model 703 instrument (KSV Instruments Ltd., Helsinki, Finland) (1).
To test the effect of surfactant production on the swarming abilities of the bacteria, randomly selected isolates from each strain group were dotted on soft-agar medium composed of potato dextrose broth (Difco) with 0.6% Bacto agar (Difco) or ABTG (AB medium of Clark and Maaløe [7] containing [per liter] 2.5 mg of thiamine, 4 g of glucose, and 4 g of Casamino Acids [Difco]) with 0.6% Bacto agar (Difco). After inoculation for 1 or 2 days at 20°C, swarming was recorded by the radial growth of the colonies, measured in millimeters per hour.
Antagonistic activity of Pseudomonas spp. surfactants.
Antagonistic properties of purified surfactants were tested by growing the pathogenic fungi Pythium ultimum and Rhizoctonia solani in the vicinity of the compounds on PDA medium. Purified surfactant (0.1 mg) in a 5-mm Whatman GF/C filter was placed 10 mm from the edge of the fungal inoculum. Triplicate plates were then incubated at 25°C, and the antagonistic activity was registered after 1 and 2 days by measuring the inhibition zone between the filter and the growing mycelium (37). In parallel, whole-cell antagonistic activity by representative cultures from each of the strain groups was tested by dotting the cultures 20 mm from the edge of an agar plug with mycelium on PDA or ABTG medium. Triplicate plates were incubated at 25°C, and inhibition zones were recorded after 1 and 2 days.
RESULTS
Abundance of surfactant-producing Pseudomonas spp. isolates in the sugar beet rhizosphere.
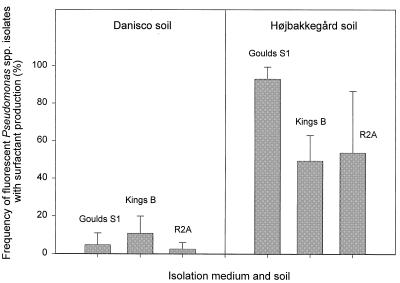
The abundance of fluorescent Pseudomonas spp. was approximately 5 × 106 colonies per g of rhizosphere soil sample (Danisco sandy loam) and approximately 1.5 × 106 colonies per g of rhizosphere soil sample (Højbakkegård loamy sand) when tested on the three different media (data not shown). When a total of 353 and 228 fluorescent Pseudomonas spp. isolates from Danisco and Højbakkegård soils, respectively, were tested, their frequencies of surfactant production by the drop assay (6) were highly variable in the two soils. As shown in Fig. 1, approximately 6% of the fluorescent Pseudomonas spp. isolated on the three media from the Danisco soil were able to produce surfactants while the average value was approximately 60% for isolates from the Højbakkegård soil. For both soils, the frequencies of surfactant producers among the Pseudomonas spp. isolates retrieved on the three different media were quite similar. Finally, when calculating the total number of surfactant-producing fluorescent Pseudomonas spp. per gram of rhizosphere soil sample, their abundance was approximately three times higher in the Højbakkegård soil than in the Danisco soil.
FIG. 1.
Frequency of surfactant-producing fluorescent Pseudomonas spp. isolates from the sugar beet rhizosphere in Danisco and Højbakkegård soils. Error bars indicate standard deviations.
CLP surfactants produced in Pseudomonas spp. isolates.
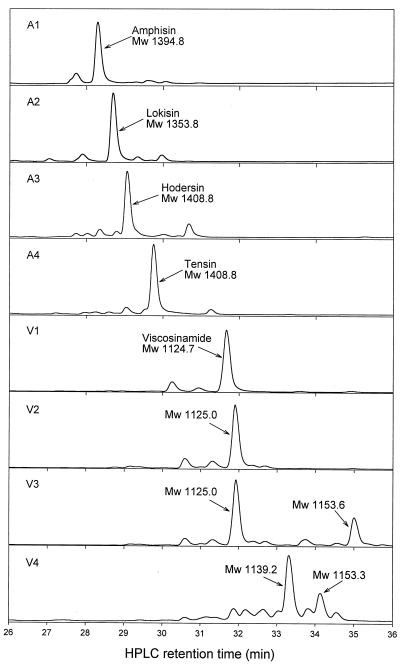
The initial screening step for surfactant-producing Pseudomonas spp. strains was based on the drop collapse assay, but CLP production was subsequently verified by HPLC analysis. By determining the retention time of all major peaks (retention times between 27 and 36 min), we found that 155 out of 169 surfactant-producing isolates from the Danisco and Højbakkegård soils were clustered into eight distinct strain groups. Figure 2 shows representative chromatograms from each of these groups. One major surfactant peak was present in most strain groups, while two peaks representing different surfactants appeared in groups V3 and V4. The smaller peaks surrounding the major surfactant peak in all chromatograms are derivates of the major compounds (D. Sørensen, unpublished data). No major peaks with retention times between 27 and 36 min were observed for strains testing negative in the drop collapse assay. The absorption spectra recorded by the HPLC diode array detector showed that all surfactants had a maximum absorption at approximately 200 nm (endpoint absorption).
FIG. 2.
HPLC chromatograms showing major surfactants (CLPs) produced in batch culture (spent growth medium) by fluorescent Pseudomonas spp. strain groups isolated from the sugar beet rhizosphere.
Further analysis of purified surfactants by MS and NMR demonstrated two major groupings of the eight strain groups producing CLPs. In Fig. 2 are included MW data for all the major CLPs and their chemical names, where available (37, 38, 42). Four of the strain groups (A1 through A4) produced CLPs with MW values of approximately 1,350 to 1,430. This A group (so called because it is associated with the production of amphisin-like compounds) produced amphisin (A1), lokisin (A2), hodersin (A3), and tensin (A4), all of which contained 11 amino acids in the cyclic peptide structure and a 3-hydroxydecanoyl moiety. The group A1 surfactant amphisin, produced by a single isolate, P. fluorescens DSS73 (42), contains leucine (l-Leu, positions 7 and 9, and d-Leu, positions 1, 4, and 5), isoleucine (l-Ile, position 10), aspartic acid (d-Asp, position 2, and l-Asp, position 11), glutamine (d-Gln, position 8), allo-threonine (d-allo-Thr, position 3), and serine (d-Ser, position 6) (42). By comparison, the group A2 surfactant lokisin, produced by 6% of the CLP-positive isolates, differs from the amphisin compound by replacement of the glutamine (d-Gln, position 8) with a serine (d-Ser, position 8) (Sørensen, unpublished). The group A3 surfactant hodersin, produced by 12% of the CLP-positive isolates, is currently being investigated to determine the chemical structure. Finally, the group A4 compound tensin, produced by 29% of the surfactant-producing isolates, differs from amphisin by replacement of the aspartic acid (l-Asp, position 11) with glutamic acid (l-Glu, position 11) (17).
By comparison, the V group (so called because it is associated with the production of viscosinamide-like compounds) produced CLPs with MW values of approximately 1,120 to 1,150. The four strain groups produced viscosinamide (V1), an unnamed 1,125.0-MW compound (V2), two unnamed 1,125.0-MW and 1,153.6-MW compounds (V3), and two unnamed 1,139.2-MW and 1,153.3-MW compounds (V4). Although the chemical structures of all the viscosinamide-like compounds were not fully investigated, HPLC and MS analyses so far indicate a close similarity between them (Sørensen, unpublished). The V1 group surfactant viscosinamide, produced by less than 1% of the surfactant-producing isolates, thus contains only 9 amino acid residues in the peptide moiety but has the same 3-hydroxydecanoyl residue as the amphisin-like compounds described above. The surfactants from groups V2 and V3, representing 12 and 28% of the surfactant-producing isolates, respectively, have not been chemically characterized, but the two groups share one of the surfactants (MW, 1125.0) produced. Finally, the group V4 surfactants, produced by 8% of the surfactant-producing isolates, are also under further study but have shown high similarity to viscosinamide based on preliminary NMR data (Sørensen, unpublished), indicating amino acid replacement(s) in the peptide moiety.
Affiliation of CLP surfactant production to Pseudomonas species and biovars.
Table 1 shows a comparison of selected phenotypic characters among the biosurfactant-producing Pseudomonas spp. strains. By using the limited number of growth characteristics (43), it was possible to tentatively assign each surfactant-producing strain group to a specific P. fluorescens biovar. The groups producing the relatively large amphisin-like lipopeptides (A1 through A4) could all be assigned to biovar VI, while most of the groups producing the relatively small viscosinamide-like lipopeptides (V1, V2, and V4) could be assigned to biovar I. One exception was group V3, which differed from the V1, V2, and V4 groups only by the absence of levan production on sucrose medium; this group was tentatively assigned to biovar V (Table 1).
TABLE 1.
Phenotypic variation among CLP-producing P. fluorescens strain groups isolated from the sugar beet rhizosphere
| P. fluorescens | Biolog GN carbon utilization activity ofb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain group | Biovara | Ino | Sor | Ado | Gla | Glu | Ita | Hyd | Ery | Put |
| A1 | VI | − | − | − | − | − | − | − | − | + |
| A2 | VI | − | − | − | − | − | − | − | − | + |
| A3 | VI | − | − | − | − | − | − | − | − | + |
| A4 | VI | − | − | − | − | − | − | − | − | + |
| V1 | I | + | + | + | + | + | + | + | + | − |
| V2 | I | + | + | + | + | + | + | − | − | − |
| V3 | V | + | + | + | + | + | + | − | − | − |
| V4 | I | + | + | + | + | + | + | + | + | − |
Biovar affiliation was based on levan production, denitrification activity, and growth on mannitol, meso-inositol, and sorbitol.
Utilization of meso-inositol (Ino), d-sorbitol (Sor), adonitol (Ado), d-galacturonic acid (Gla), d-glucuronic acid (Glu), itaconic acid (Ita), p-hydroxy phenyl acetic acid (Hyd), i-erythritol (Ery), and putrescine (Put).
Also included in Table 1 is a number of selected items from the Biolog GN carbon utilization pattern, including the presence of glucuronidase and galacturonidase activity and the utilization of adonitol, i-erythritol, p-hydroxy phenylacetic acid, and putrescine. These selected substances primarily affiliated the two major A and V groups with biovars VI and I, respectively. Even if levan production affiliated group V3 with biovar V, the additional test substances in Table 1 suggested that group V3 was closely related to group V2, representing biovar I.
Role of CLP surfactants in surface motility and fungal antagonism.
The reduction in surface tension in spent growth medium was indicative of surfactant activity. In Table 2, representative strains from the A and V groups are shown to lower the surface tension of DMB from 71 to 27 mN m−1 in the A groups and to 25 through 27 mN m−1 in the V groups. For control strains, those with no CLP production, and for a mutant impaired in the peptide synthetase responsible for amphisin production (B. Koch et al., submitted for publication), surface tension reductions were 63 and 48 mN m−1, respectively.
TABLE 2.
Surface tension (in spent DMB medium) and swarming ability (on PDA and ABTG soft-agar media) recorded in CLP-producing P. fluorescens strain groups isolated from the sugar beet rhizosphere
| P. fluorescens strain group | Surface tension (mN m−1) in spent DMB mediuma | Swarming motility (mm h−1) onb:
|
|
|---|---|---|---|
| PDA | ABTG | ||
| A1 | 27.4 ± 0.7 | 1 | 1 |
| A2 | 27.4 ± 0.4 | 1.0 ± 0.3 | 1.6 ± 0.5 |
| A3 | 26.9 ± 1.2 | 1.2 ± 0.6 | 1.7 ± 0.5 |
| A4 | 26.7 ± 0.0 | 1.3 ± 0.5 | 1.2 ± 0.4 |
| V1 | 26.8 ± 0.2 | 0 | 0.3 ± 0.1 |
| V2 | 26.7 ± 0.2 | 0 | 0.4 ± 0.3 |
| V3 | 24.7 ± 0.3 | 0 | 0.7 ± 0.3 |
| V4 | 24.5 ± 0.1 | 0.5 ± 0.5 | 0.7 ± 0.3 |
| Control 1c | 63.3 ± 6.6 | 0 | 0 |
| Control 2d | 48.3 ± 5.7 | 0 | 0 |
Surface tension in spent DMB medium after 2 days of incubation at 20°C is given as the mean of three replicates ± the standard deviation, except for control 1, which is given as the mean of six strains ± the standard deviation. Surface tension in uninoculated DMB medium was 71 mN m−1.
Swarming velocity on PDA and ABTG media after 24 and 48 h of incubation at 20°C is given as the mean of 3 to 22 strains ± the standard deviation, except for those for groups A1 and control 2, which represent one strain.
Strain group without CLP production.
A1 mutant impaired in the peptide synthetase responsible for amphisin production (Koch et al., submitted).
Surface motility may be greatly stimulated by surfactants, as is known for the CLP serrawettin produced by Serratia spp. (26). Inoculation of the P. fluorescens strains on soft-agar medium (Table 2) revealed that strains producing the relatively large, amphisin-like CLPs (group A) had a higher swarming motility on both PDA and ABTG media than the strains in group V. In contrast, all strains producing the relatively small, viscosinamide-like CLPs (group V) all exhibited surface motility when propagated on soft-agar ABTG medium whereas only strains belonging to group V4 demonstrated swarming motility on the soft-agar PDA medium. Finally, control strains, including the A1 mutant that did not produce CLPs, had no swarming motility on either of the two media.
CLP compounds may also play a role in antagonism, which was the basis for the selection of fluorescent Pseudomonas spp. strains in the biological control of root-pathogenic microfungi (37, 38, 45). In the present study, whole-cell incubation of representative strains from all eight surfactant-producing strain groups was performed to test their effects against P. ultimum and R. solani on PDA and ABTG media. As shown in Table 3, the strain groups A1 through A4, producing the relatively large amphisin-like CLPs, generally demonstrated the highest level of antagonism against the microfungi. One exception was the V1 group, which was already known to be very active against the two fungi on PDA medium (37). Interestingly, the highly antagonistic strain groups A1 through A4 also had the most complete set of antagonistic traits tested (protease, chitinase, and HCN production) while one or two of these substances were always absent in the strain groups V1 through V4.
TABLE 3.
Whole-cell antagonistic activity towards P. ultimum and R. solani (on PDA and ABTG media) and selected antagonistic traits recorded in CLP-producing P. fluorescens strain groups isolated from the sugar beet rhizosphere
| P. fluorescens strain group | Antagonism to P. ultimum and R. solania
|
Production ofb:
|
|||
|---|---|---|---|---|---|
| PDA | ABTG | Protease | Chitinase | HCN | |
| A1 | 4.0, 6.3 | 1.6, 2.3 | ++ | ++ | +++ |
| A2 | 1.9 ± 1.4, 0.9 ± 0.7 | 0.9 ± 0.7, 1.0 ± 0.6 | ++ | ++ | +++ |
| A3 | 2.3 ± 0.9, 1.4 ± 0.6 | 0.6 ± 0.6, 0.7 ± 0.5 | ++ | ++ | +++ |
| A4 | 0.6 ± 0.3, 0.7 ± 0.4 | 0.6 ± 0.4, 1.2 ± 0.5 | ++ | ++ | +++ |
| V1 | 1.6 ± 0.5, 2.6 ± 0.2 | 0.2 ± 0.4, 0.3 ± 0.4 | +++ | +++ | − |
| V2 | 0, 0.2 ± 0.3 | 0, 0 | − | − | − |
| V3 | 0, 0 | 0, 0 | − | − | − |
| V4 | 0, 0.4 ± 0.2 | 0.1 ± 0.3, 0.4 ± 0.8 | +++ | +++ | − |
| Controlc | 0, 0 | 0.1 ± 0.1, 0.2 ± 0.3 | +/− | − | +/− |
Inhibition zone (in millimeters) between fungal mycelium (P. ultimum or R. solani) and a bacterial colony after 1 or 2 days of incubation at 25°C on PDA or ABTG medium. Values are means of 3 to 22 strains ± standard deviations, except for those for group A1, which are given as data from one strain.
These antagonistic traits were visually classified into categories of no (−), weak (+), moderate (++), or strong (+++) activity. Values are means of 3 to 22 strains ± standard deviations, except for those for group A1, which are data from one strain. +/−, variable activity.
Strain group without CLP production. Values are means of 14 strains ± standard deviations.
As shown in Table 4, the purified CLPs from the most antagonistic P. fluorescens groups, A1 through A4 and V1, were also tested against P. ultimum and R. solani on PDA medium. Except for the completely inactive lipopeptide (hodersin) from strain group A3, the relatively large lipopeptides (amphisin, lokisin, and tensin) generally appeared to be more antagonistic towards the fungi than the smaller viscosinamide from P. fluorescens DR54 (37).
TABLE 4.
In vitro antagonistic activity to P. ultimum and R. solani (on PDA medium) by purified CLPs produced in P. fluorescens strain groups isolated from the sugar beet rhizosphere
| P. fluorescens strain group | CLP compound | Antagonism to P. ultimum and R. solania |
|---|---|---|
| A1 | Amphisin | 2.3 ± 0.6, 1.7 ± 0.6 |
| A2 | Lokisin | 2.7 ± 0.6, 3.3 ± 0.6 |
| A3 | Hodersin | 0.0 ± 0.0, 0.0 ± 0.0 |
| A4 | Tensin | 1.7 ± 0.6, 2.3 ± 0.8 |
| V1 | Viscosinamide | 1.0 ± 0.9, 0.7 ± 0.3 |
Inhibition zone (in millimeters) between fungal mycelium (P. ultimum or R. solani) and a GF/C filter with purified CLP compound (0.1 mg) after 1 or 2 days of incubation at 25°C on PDA medium. Values are means of three replicates ± standard deviations.
DISCUSSION
Surfactant-producing Pseudomonas spp. in two Danish agricultural soils.
It was surprising to find that approximately 60% or more of the fluorescent Pseudomonas spp. isolates inhabiting the sugar beet rhizosphere from loamy sand soil (Højbakkegård field station) were able to produce CLP. The high frequency of CLP-producing isolates in this soil was confirmed by other isolations from the sugar beet rhizosphere, showing approximately 40% CLP producers within the fluorescent Pseudomonas spp. population (data not shown). This suggests that the trait may be of ecological importance in the soil environment. The data further indicate that the soil type may be important for the frequency of CLP-producing strains, since only approximately 6% of the fluorescent Pseudomonas spp. isolated from the sandy loam (Danisco soil) were found to be CLP producers.
The total number of surfactant-producing Pseudomonas spp. isolates retrieved from the sandy loam (Danisco soil) was only 17 compared to 152 from the loamy sand (Højbakkegård soil), and the surfactant groups identified in the Danisco soil were already represented in the Højbakkegård soil samples (data not shown). Most noteworthy in the whole strain collection was that some surfactants appeared to be rare whereas others appeared to be very common. For instance, amphisin (group A1) was found in only one strain (P. fluorescens DSS73) while tensin (group A4) was observed in as much as 29% of the surfactant-producing Pseudomonas spp. population in the Højbakkegård soil. The high abundance of isolates producing tensin was confirmed in a different isolation, showing that 56% of all surfactant-producing Pseudomonas spp. strains retrieved from this soil actually produced this surfactant (data not shown). Even if tensin production among the fluorescent Pseudomonas spp. population appears to be common in the Højbakkegård soil, the reason will remain obscure until a functional role, including a possible selective value of the CLP production, has been identified.
The possible advantage of surfactant production among Pseudomonas spp. in the rhizosphere could involve such traits as facilitated surface motility, adhesion, nutrient availability, competition, or antagonism. Still, the task of identifying the actual role of CLP production may be difficult, and predicting the selective value (if there is one) may be just too difficult, since many factors influence the diversity and activity of soil Pseudomonas spp. populations. Hence, when looking at Pseudomonas spp. populations in tomato and flax rhizospheres in different soil types, Latour et al. (23) observed that diversity was most influenced by soil type and less by crop type. Bachmann and Kinzel (3) also suggested that soil type was the most important factor determining the overall diversity of bacterial populations in the rhizosphere. Höper et al. (18) suggested that basic soil characteristics such as pH and texture may influence the density of fluorescent pseudomonads in soil. Indeed, the two soils at Danisco and Højbakkegård differed significantly in both pH value and texture, but at the moment, we are unable to identify a particular role or selective value of CLP productions that would explain the very different frequencies of surfactant-producing Pseudomonas spp. strains in the two soils.
An attempt to link CLP production to the abundance of specific Pseudomonas spp. was made by affiliating the CLPs produced with specific P. fluorescens biovars. This resulted in the almost exclusive assignment of CLP production to P. fluorescens biovars I, V, and VI. In a study on CLP production in Pseudomonas spp. isolates from the sugar beet rhizosphere (34), the exclusive assignment of viscosinamide production (group V1 [this study]) to P. fluorescens biovar I was reported. For the viscosinamide-like compounds, Laycock et al. (24) affiliated the viscosin-producing strain SH10-3B with P. fluorescens biovar II. The extensive survey of the present study strongly supports the predominance of a few selected P. fluorescens biotypes (I, V, and VI) as CLP producers in the sugar beet rhizosphere. This observation does not contradict the larger, overall diversity of fluorescent Pseudomonas spp. populations in the rhizosphere, which may commonly include the other P. fluorescens biovars II, III, and IV as well (23, 25, 34). Hence, CLP production could well be associated with only a smaller, exclusive subpopulation of the total fluorescent Pseudomonas spp. population in soil.
Structural differences among CLP surfactants.
Although an increasing number of CLPs with surfactant properties have been described in Pseudomonas spp., the present survey is the first to demonstrate the predominance in a soil environment of two major groups of compounds, sharing the same 3-hydroxydecanoic acid fatty acid moiety but with either 9 or 11 amino acids in the peptide ring structure. We suggest that the group of larger compounds is termed “amphisin-like,” based on a recent discovery of the highly active amphisin (42). Similarly, we suggest that the group of smaller compounds is termed “viscosinamide-like,” referring to the highly active viscosinamide (37).
The structural differences in both amphisin-like and viscosinamide-like CLPs are all caused by single amino acid replacements in the peptide moiety. Compared to the amino acid sequence of the amphisin molecule (d-Leu-d-Asp-d-allo-Thr-d-Leu-d-Leu-d-Ser-l-Leu-d-Gln-l-Leu-l-Ile-l-Asp), tensinhas l-Glu (position 11) (38) and lokisin has d-Ser (position 8) according to our investigations (Sørensen, unpublished). Lokisin has the same sequence as pholipeptin isolated from P. fluorescens strain BMJ279-76F1 (48), but pholipeptin features the l-isomer of threonine in position 3, all leucines are d-isomers, and the lactone ring is slightly different, since it connects the side chain of C-terminal d-Asp to l-Thr. Similarly, the amino acid sequence of viscosinamide (l-Leu-d-Glu-d-allo-Thr-d-Val-l-Leu-d-Ser-l-Leu-d-Ser-l-Ile) is differentfrom viscosin isolated from Pseudomonas viscosa (21) by only the d-Glu substitution (position 2). In turn, the WLIP compound from Pseudomonas reactans (30) varies only stereochemically from viscosin by the d-Leu in position 5. Finally, the massetolides A through H from several marine Pseudomonas spp. strains (11) vary by amino acid substitutions (Val or Ile in position 4; Val, Leu, or Ile in position 9) as well as by the length of the fatty acid chain (10 to 12 C atoms). The MWs of massetolide F (1,125.6 kDa), massetolides A, D, and G (1,139.7 kDa), and massetolides B and H (1,153.7 kDa) are all very similar to those found for V2, V3, and V4, suggesting that these unresolved structures may be very similar to the massetolide compounds.
A possible structure-function relationship may be depicted from the amino acid sequences of CLPs. Hydrophobic amino acids (leucine, isoleucine, and valine) thus occur abundantly (positions 1, 4, 5, 7, and 9) between hydrophilic ones (aspartic acid, glutamic acid, glutamine, serine, and allo-threonine in positions 2, 3, 6, and 8, respectively). This provides an amphiphilic ring structure, which may facilitate molecular binding such as the formation of ligands of metal ions (siderophore function) and other cations (14) of importance for cellular (membrane) transport, e.g., Ca2+. Both the topological configuration of the amino acid residues and the existence of carboxylic groups in the peptide moiety are important for water solubility and surfactant properties of CLPs (29). Three-dimensional structures depicted from crystalline amphisin (42) and tensin (17) clearly show how the localization and stereochemistry of amino acids make the ring structure hydrophobic or hydrophilic, respectively, at the two sides of the molecule. The hydrophobic fatty acid tail, together with the amphiphilic property of the peptide, structure may finally play an important role in penetration and binding of CLPs within biological membranes. This in turn may support their role as surfactants and as antibiotics, e.g., disrupting membrane functions leading to excess Ca2+ influx into target cells (46).
Functional roles of CLP surfactants in P. fluorescens.
A unifying trait of the CLP compounds is the high capability to lower the surface tension in a medium. Surface tension measured in spent growth medium (DMB) was thus very low (approximately 25 to 27 mN m−1), indicating that the compounds were very powerful surfactants. The recorded values are as low as the ones (approximately 27 mN m−1) reported for other CLPs, surfactin from B. subtilis (8) and viscosin from P. viscosa (33). One likely role of the CLP compounds, therefore, is an enhancement of surface motility, as has previously been observed with induction of the swarming phenotype associated with serrawettin production in Serratia spp. (26). The strong correlation observed in this study between CLP production and swarming motility in fluorescent Pseudomonas spp. was further supported by results obtained by using the P. fluorescens DSS73-15C2 mutant impaired in the peptide synthetase responsible for amphisin production (Koch et al., submitted). This mutant lowered the surface tension to 48 mN m−1 in spent DMB medium, which was only slightly lower than the values obtained for strains without CLP production. The mutant, further, had no swarming motility (J. B. Andersen et al., submitted for publication). Whether CLP-enhanced surface motility actually plays a role in the rhizosphere, e.g., for seed or root colonization, needs further investigation.
The antifungal activity of purified CLPs seemed somewhat higher for at least three of the larger amphisin-like compounds (amphisin, lokisin, and tensin) than that for the smaller viscosinamide and viscosinamide-like compounds (data not shown), but the difference could not be directly linked to the amino acid number or sequence of the peptide moiety. Most interestingly, the hodersin compound of the amphisin-like type (group A3) was completely inactive in antagonizing the two plant-pathogenic fungi, P. ultimum and R. solani. This was quite surprising, as the whole-cell assay with producing strains indeed showed inhibition of both fungi. One explanation may be that this compound is inactive because of a structural difference. Preliminary NMR results suggest that hodersin and tensin are structural isomers (Sørensen, unpublished) and that stereochemical changes may be responsible for the differences in antagonistic activity. Changes of single amino acids may indeed cause drastic changes in antagonistic activity (9, 47) and surfactant properties (14). Alternatively, compounds other than CLPs may have been produced and exerted the antagonism in the whole-cell assay.
While most of the purified CLPs showed antifungal properties, the compounds were likely to act in synergism with other antifungal elements, such as antibiotic compounds or cell wall-degrading enzymes. Hence, the differences between antagonistic activities in the strain groups tested in the whole-cell assays may be attributed to differences in lipopeptide type (amphisin-like or viscosinamide-like), though the synergistic enzyme or antibiotic production may also be important. In the present study, we observed that the P. fluorescens strains (groups A1 through A4) demonstrating the largest inhibition towards the fungi also had the most complete array of cell wall-degrading enzymes (protease and chitinase) and HCN production. In contrast, none of the surfactant-producing Pseudomonas spp. strains tested here had the often-reported antifungal traits of diacetyl phloroglucinol, phenazine-1-carboxylic acid, pyrrolnitrin, or pyoluteorin production (10). In future studies, the well-characterized collection of CLP-producing P. fluorescens strains and their purified compound will be used to study the functional role of CLP production on surfaces (seed, root, fungal mycelium) in soil microcosms and to identify further structure-function relationships for the different CLP compounds.
Acknowledgments
This study was supported by the Danish Agricultural and Veterinary Research Council (J. no. 9702796).
We thank Dorte Rasmussen, Ann Dorrit Steffensen, and Malene F. Lihme for excellent technical assistance, Ole Nybroe for providing protocols for immunodetection using the OprF antibody, and Martin Lund and Anders Johansen for assistance in surface tension measurements.
REFERENCES
- 1.Adamson, A. W. 1990. Physical chemistry of surfaces, 5th ed. Wiley Interscience, New York, N.Y.
- 2.Asaka, O., and M. Shoda. 1996. Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol. 62:4081-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann, G., and H. Kinzel. 2001. Physiological and ecological aspects of the interactions between plant roots and rhizosphere. Soil Biol. Biochem. 24:543-552. [Google Scholar]
- 4.Bakker, A. W., and B. Schippers. 1987. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp.-mediated plant growth-stimulation. Soil Biol. Biochem. 19:451-457. [Google Scholar]
- 5.Bender, C. L., F. Alarcón-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodour, A. A., and R. M. Miller-Maier. 1998. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 32:273-280. [Google Scholar]
- 7.Clark, D. J., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 8.Cooper, D. J., and E. Zajic. 1980. Surface-active compounds from microorganisms. Adv. Appl. Microbiol. 26:229-253. [Google Scholar]
- 9.Danders, W., A. Marahiel, M. Krause, N. Kosui, T. Kato, N. Izumiya, and H. Kleinkauf. 1982. Antibacterial action of gramicidin S and tyrocidines in relation to active transport, in vitro transcription, and spore outgrowth. Antimicrob. Agents Chemother. 22:785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowling, D. N., and F. O'Gara. 1994. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 12:133-141. [Google Scholar]
- 11.Gerard, J., R. Lloyd, T. Barsby, P. Haden, M. T. Kelly, and R. J. Andersen. 1997. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 60:223-229. [DOI] [PubMed] [Google Scholar]
- 12.Givskov, M., J. Östling, L. Eberl, P. Lindum, A. B. Christensen, G. Christiansen, S. Molin, and S. Kjelleberg. 1998. Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:742-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould, W. D., C. Hagedorn, T. R. Bardinelli, and R. M. Zablotowicz. 1985. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl. Environ. Microbiol. 49:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grangemard, I., J. Wallach, R. Marget-Dana, and F. Peypoux. 2001. Licenysin: a more efficient cation chelator than surfactin. Appl. Biochem. Biotechnol. 90:199-210. [DOI] [PubMed] [Google Scholar]
- 15.Gross, D. C. 1985. Regulation of syringomycin synthesis in Pseudomonas syringae pv. syringae and defined conditions for its production. J. Appl. Bacteriol. 58:167-174. [DOI] [PubMed] [Google Scholar]
- 16.Harrigan, G. G., B. L. Harrigan, and B. S. Davidson. 1997. Kailuins A-D, new cyclic acyldepsipeptides from cultures of a marine-derived bacterium. Tetrahedron 53:1577-1582. [Google Scholar]
- 17.Henriksen, A., U. Anthoni, T. H. Nielsen, J. Sørensen, C. Christophersen, and M. Gajhede. 2000. Cyclic lipoundecapeptide tensin from Pseudomonas fluorescens strain 96.578. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 56:113-115. [DOI] [PubMed] [Google Scholar]
- 18.Höper, H., C. Steinberg, and C. Alabouvette. 1995. Involvment of clay type and pH in the mechanisms of soil suppressiveness to fusarium wilt of flax. Soil Biol. Biochem. 27:955-967. [Google Scholar]
- 19.Hutchison, M. L., and K. Johnstone. 1993. Evidence for the involvement of the surface active properties of the extracellular toxin tolaasin in the manifestation of brown blotch disease symptoms by Pseudomonas tolaasii on Agaricus bisporus. Physiol. Mol. Plant Pathol. 42:373-384. [Google Scholar]
- 20.Katz, E., and A. L. Demain. 1977. The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol. Rev. 41:449-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kochi, M., D. W. Weiss, L. H. Pugh, and V. Groupé. 1951. Viscosin, a new antibiotic. Bacteriol. Proc. 29-30.
- 22.Kragelund, L., K. Leopold, and O. Nybroe. 1996. Outer membrane protein heterogeneity within Pseudomonas fluorescens and P. putida and use of an OprF antibody as a probe for rRNA homology group I pseudomonads. Appl. Environ. Microbiol. 62:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latour, X., T. Corberand, G. Laguerre, F. Allard, and P. Lemanceau. 1996. The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl. Environ. Microbiol. 62:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laycock, M. V., P. D. Hildebrand, P. Thibault, J. A. Walter, and J.-L. C. Wright. 1991. Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of Pseudomonas fluorescens. J. Agric. Food Chem. 39:483-489. [Google Scholar]
- 25.Lemanceau, P., T. Corberand, L. Gardan, X. Latour, G. Laguerre, J.-M. Boeufgras, and C. Alabouvette. 1995. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl. Environ. Microbiol. 61:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindum, P., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marahiel, M. A., T. Stacelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 28.Moffitt, M. C., and B. A. Neilan. 2000. The expansion of mechanistic and organismic diversity associated with non-ribosomal peptides. FEMS Microbiol. Lett. 191:159-167. [DOI] [PubMed] [Google Scholar]
- 29.Morikawa, M., Y. Hirata, and T. Imanaka. 2000. A study on the structure-function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta 1488:211-218. [DOI] [PubMed] [Google Scholar]
- 30.Mortishire-Smith, R. J., J. C. Nutkins, L. C. Packman, C. L. Brodey, P. B. Rainey, K. Johnson, and D. H. Williams. 1991. Determination of the structure of an extracellular peptide produced by the mushroom saprotroph Pseudomonas reactans. Tetrahedron 47:3645-3654. [Google Scholar]
- 31.Nakayama, T., Y. Homma, Y. Hashidoko, J. Mizutani, and S. Tahara. 1999. Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl. Environ. Microbiol. 65:4334-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neu, T. R. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neu, T. R., T. Härtner, and K. Poralla. 1990. Surface active properties of viscosin: a peptidolipid antibiotic. Appl. Microbiol. Biotechnol. 32:518-520. [Google Scholar]
- 34.Nielsen, M. N., J. Sørensen, J. Fels, and H. C. Pedersen. 1998. Secondary metabolite- and endochitinase-dependent antagonism toward plant-pathogenic microfungi of Pseudomonas fluorescens isolates from sugar beet rhizosphere. Appl. Environ. Microbiol. 64:3563-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, P., and J. Sørensen. 1997. Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere. FEMS Microbiol. Ecol. 22:183-192. [Google Scholar]
- 36.Nielsen, T. H., C. Christophersen, U. Anthoni, and J. Sørensen. 1998. A promising new metabolite from Pseudomonas fluorescens for biocontrol of Pythium ultimum and Rhizoctonia solani, p. 51-55. In B. K. Duffy, U. Rosenberger, and C. Douglas (ed.), Molecular approaches in biological control. IOBC/WPRS Bulletin, vol. 21, no. 9. International Organization for Biological and Integrated Control of Noxious Animals and Plants, West Palaearctic Regional Section, INRA-Centre de Recherches de Dijon, Dijon, France.
- 37.Nielsen, T. H., C. Christophersen, U. Anthoni, and J. Sørensen. 1999. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 86:80-90. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, T. H., C. Thrane, C. Christophersen, U. Anthoni, and J. Sørensen. 2000. Structure, production characteristics and fungal antagonism of tensin—a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J. Appl. Microbiol. 89:992-1001. [DOI] [PubMed] [Google Scholar]
- 39.Palleroni, N. J. 1984. Genus I. Pseudomonas, p. 141-199. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 40.Ron, E. Z., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg, E., and E. Z. Ron. 1999. High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 52:154-162. [DOI] [PubMed] [Google Scholar]
- 42.Sørensen, D., T. H. Nielsen, C. Christophersen, J. Sørensen, and M. Gajhede. 2001. Cyclic lipoundecapeptide amphisin from Pseudomonas sp. strain DSS73. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 57:1123-1124. [DOI] [PubMed] [Google Scholar]
- 43.Sørensen, J., J. Skouv, A. Jørgensen, and O. Nybroe. 1992. Rapid identification of environmental isolates of Pseudomonas aeruginosa, P. fluorescens and P. putida by SDS-PAGE analysis of whole-cell protein patterns. FEMS Microbiol. Ecol. 101:41-50. [Google Scholar]
- 44.Takesako, K., H. Kuroda, T. Inoue, F. Haruna, Y. Yoshikawa, I. Kato, K. Uchida, T. Hiratani, and H. Yamaguchi. 1993. Biological properties of aureobasidin A, a cyclic depsipeptide antifungal antibiotic. J. Antibiot. 46:1414-1420. [DOI] [PubMed] [Google Scholar]
- 45.Thrane, C., T. H. Nielsen, M. N. Nielsen, S. Olsson, and J. Sørensen. 2000. Viscosinamide-producing Pseudomonas fluorescens DR54 exerts biocontrol effect on Pythium ultimum in sugar beet rhizosphere. FEMS Microbiol. Ecol. 33:139-146. [DOI] [PubMed] [Google Scholar]
- 46.Thrane, C., S. Olson, T. H. Nielsen, and J. Sørensen. 1999. The use of vital fluorescent probes for the detection of stress in the fungi Pythium ultimum and Rhizoctonia solani challenged with viscosinamide from Pseudomonas fluorescens DR54. FEMS Microbiol. Ecol. 30:11-23. [Google Scholar]
- 47.Trischman, J. A., P. R. Jensen, and W. Fenical. 1994. Halobacillin: a cytotoxic cyclic acylpeptide of the iturin class produced by a marine Bacillus. Tetrahedron Lett. 35:5571-5574. [Google Scholar]
- 48.Ui, H., T. Miyake, H. Linuma, S. Hattori, M. Hamada, T. Takeuchi, S. Umezawa, and K. Umezawa. 1995. A novel cyclic lipoundecapeptide, pholipeptin, isolated from Pseudomonas sp. Tetrahedron Lett. 36:7479-7480. [Google Scholar]
- 49.Vollenbroich, D., M. Özel, J. Vater, R. M. Kamp, and G. Pauli. 1997. Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from Bacillus subtilis. Biologicals 25:289-297. [DOI] [PubMed] [Google Scholar]