Abstract
The history, taxonomy, geographic distribution, clinical disease, and therapy of the pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria (RGM) are reviewed. Community-acquired disease and health care-associated disease are highlighted for each species. The latter grouping includes health care-associated outbreaks and pseudo-outbreaks as well as sporadic disease cases. Treatment recommendations for each species and type of disease are also described. Special emphasis is on the Mycobacterium fortuitum group, including M. fortuitum, M. peregrinum, and the unnamed third biovariant complex with its recent taxonomic changes and newly recognized species (including M. septicum, M. mageritense, and proposed species M. houstonense and M. bonickei). The clinical and taxonomic status of M. chelonae, M. abscessus, and M. mucogenicum is also detailed, along with that of the closely related new species, M. immunogenum. Additionally, newly recognized species, M. wolinskyi and M. goodii, as well as M. smegmatis sensu stricto, are included in a discussion of the M. smegmatis group. Laboratory diagnosis of RGM using phenotypic methods such as biochemical testing and high-performance liquid chromatography and molecular methods of diagnosis are also discussed. The latter includes PCR-restriction fragment length polymorphism analysis, hybridization, ribotyping, and sequence analysis. Susceptibility testing and antibiotic susceptibility patterns of the RGM are also annotated, along with the current recommendations from the National Committee for Clinical Laboratory Standards (NCCLS) for mycobacterial susceptibility testing.
INTRODUCTION
The species of rapidly growing mycobacteria (RGM) capable of producing disease in humans consist primarily of the Mycobacterium fortuitum group, the M. chelonae/abscessus group, and the M. smegmatis group. Key features for identification of these groups are the presence of typical long-chain fatty acids known as mycolic acids, growth of readily visible colonies on primary isolation within 7 days on multiple types of solid media, arylsulfatase activity within 3 days or 2 weeks, and the absence or slow appearance of any pigmentation (144).
Historically, the M. fortuitum group has been composed of two known species and a taxon which has been reported to include more than one species (82). The species include M. fortuitum (formerly M. fortuitum biovar fortuitum), M. peregrinum (formerly M. fortuitum biovar peregrinum) and the taxon known as the unnamed third biovariant complex (96). Several additional taxa to be discussed later are candidates for inclusion in this group.
M. chelonae (formerly M. chelonae subsp. chelonae) and M. abscessus (formerly M. chelonae subsp. abscessus) (89), along with the newly recognized species M. immunogenum (212), are members of a group known collectively as the M. chelonae-abscessus group.
Finally, the M. smegmatis group contains M. smegmatis sensu stricto and two newly described species: M. goodii, and M. wolinskyi (17, 199).
Prior to recent molecular reevaluations, the taxa in the M. chelonae-abscessus and the M. fortuitum group were considered “subspecies” or “biovariants,” respectively. However, the introduction and evolution of 16S ribosomal gene (rDNA) sequencing provided strong evidence that these biovars and subspecies were in fact separate species. Gene sequencing also permitted much easier recognition of new taxa, since investigators could use data banks for strain comparisons rather than performing the much more technically difficult genomic DNA-DNA pairing experiments with all potentially related taxa. The genomic DNA-DNA pairing experiments that show <70% homology to other species still remain the “gold standard” for recognition of new species, but such studies are performed infrequently and many new species of mycobacteria are based genetically on 16S rDNA comparisons only.
In this review, we update the taxonomy of these nonpigmented RGM and point out many of the changes brought about by such newer technologies as 16S rRNA gene sequencing; high-performance liquid chromatography (HPLC) of mycolic acid esters, including fluorescence-HPLC; and PCR restriction fragment length polymorphism (RFLP) analysis (PRA) of a 439-bp fragment (referred to as the Telenti fragment) of the 65-kDa heat shock protein-encoding gene (hsp65) (174). Clinical disease caused by these groups, together with their drug susceptibility, and most effective drug treatment are also addressed. The disease syndromes caused by these organisms are listed in Table 1.
TABLE 1.
Species or taxonomic group and their most frequently recognized clinical disease syndromes
| Species or group | Disease syndromes in:
|
|
|---|---|---|
| Normal hosts | Immunosuppressed persons | |
| M. fortuitum group | Localized post-traumatic wound infections | |
| Catheter infections | Catheter infections | |
| Surgical wound infections, especially following augmentation mammaplasty cardiac surgery | ||
| M. chelonae | Localized post-traumatic wound infections Post-traumatic or postsurgical corneal infections | Disseminated skin infections in patients receiving corticosteroids and in organ transplant recipients |
| Catheter infections | Catheter infections | |
| M. abscessus | Chronic lung infections | |
| Localized post-traumatic wound infections | ||
| Surgical wound infections Chronic otitis media | Disseminated skin infections in patients receiving corticosteroids and in organ transplant recipients | |
| Catheter infections | Catheter infections | |
| M. smegmatis group | Localized post-traumatic wound infections | |
| Surgical wound infections | ||
| Osteomyelitis following open fractures | ||
| Lung infection complicating lipoid pneumonia or achalasia | ||
| M. mucogenicum | Sputum contaminant | Catheter infections |
| M. fortuitum third biovariant complex, sorbitol positive and sorbitol negative (proposed M. houstonense and M. bonickei) | Localized post-traumatic wound infections Osteomyelitis following open fractures Surgical wound infections | |
EPIDEMIOLOGY
The nonpigmented RGM are extremely hardy and thrive in even the most hostile of environments (208, 209, 213-215). Some of the taxa—such as a subgroup of M. peregrinum, some members of the unnamed third biovariant complex of M. fortuitum, and most isolates in the M. smegmatis group—are able to grow at 45°C (17, 194, 199). Additionally, some species, such as the M. chelonae/abscessus group and M. mucogenicum, resist the activity of disinfectants and biocides such as organomercurials, chlorine, and alkaline glutaraldehyde (166, 198, 210).
These hardy species of RGM are commonly seen in municipal tap water (31). One study by Carson et al. (31) showed that 55% of the incoming city water in hemodialysis centers throughout the United States contained RGM. Some outbreaks of human infection related to these organisms have involved hospital water systems as the microbial reservoirs. Recently, the presence of acid-fast mycobacteria in up to 90% of biofilms (the slime layer present at water-solid interfaces) taken from piped water systems has been described (153). The presence of acid-fast mycobacteria in these biofilms probably serves as a major environmental reservoir for organisms such as M. kansasii, M. mucogenicum, M. simiae, M. xenopi, and M. gordonae.
Because of the ubiquity of the RGM, human infections have been reported from most geographic areas in the world (213) and species of RGM have been recovered from 30 to 78% of soil samples throughout the United States (117). Most nosocomial (hereafter referred to as health care-associated) outbreaks and pseudo-outbreaks have occurred in the United States and seem to be concentrated mainly in the South. M. senegalense, originally found in Africa, has never been described elsewhere. Such a localized distribution among the RGM seems to be rare, however.
Health care-associated outbreaks and pseudo-outbreaks commonly involve exposure to tap water or water sources such as ice, ice water, and water-based solutions (29, 39, 80, 146, 189, 198). Contaminated ice machines are a relatively important hospital reservoir for the RGM, especially M. fortuitum. Reported disease outbreaks have included sternal wound infections (189, 198), surgical wound infections following plastic surgery (192, 194), and postinjection abscesses (57, 183). Catheter infections also have been associated with the RGM, including M. mucogenicum (189, 200, 205). Pseudo-outbreaks of disease, defined as clusters of false infections or artifactual clustering of real infections, have been associated with contaminated bronchoscopes and automated endoscopic cleaning machines with tap water as the source of the organism (54, 55).
Localized infection in sporadic community-acquired disease usually occurs after a traumatic injury followed by potential soil or water contamination (9, 203, 205). Such injuries as stepping on a nail, motor vehicle accidents, compound fractures, etc., are typical of the clinical histories seen in patients with RGM disease (203, 205).
TAXONOMY AND CLINICAL SIGNIFICANCE
M. fortuitum Group
The M. fortuitum group has historically included three taxa: M. fortuitum, M. peregrinum, and the unnamed third biovariant complex. Here, we propose the addition of six more species to the group: M. mucogenicum (for reasons given below), M. senegalense, M. mageritense, and three recently described (M. F. Schinsky, M. P. Douglas, A. G. Steigerwalt, R. W. Wilson, M. M. Floyd, M. I. Daneshvar, B. A. Brown-Elliott, R. J. Wallace, Jr., M. M. McNeil, D. J. Brenner, and J. M. Brown, Abstr. 12th Int. Symp. Biol. Actino., abstr. P-117, 2001) species: M. septicum, M. houstonense, and M. bonickei. Few studies have accurately separated these eight taxa, and they were often referred to simply as “M. fortuitum,” as if they were a single species. For purposes of this review, “M. fortuitum group” is used when no subgrouping was performed or the data include all subgroups. Names of the specific taxa are used when the isolates were so identified by current methods of carbohydrate utilization (156) or PRA of the hsp65 gene (165, 174). Common features within this group are a positive 3-day arylsulfatase, the absence of pigmentation, a positive nitrate reductase, a positive iron uptake, and susceptibility to multiple drugs including polymyxin B, sulfonamides, and the newer fluoroquinolones. Most of these taxa grow better at 30 than 35°C. Their 16S rDNA sequences generally differ by 15 bp or less.
Historical perspective.
(i) M. fortuitum.
M. fortuitum was the designation given by da Costa Cruz (42) to a strain of RGM (ATCC 6841T) isolated from a human postinjection abscess in 1938. Subsequently, Stanford and Gunthorpe (162) determined that the isolate was identical to an established species known as M. ranae, with the type strain isolated from a frog by Küster in 1905 (88). Runyon (144) challenged the name M. ranae, in part because many isolates identified as M. ranae were subsequently found to be M. smegmatis and in part because the name M. fortuitum was more widely recognized and established in the medical literature. Hence, in 1972, the Judicial Commission of the International Committee of Systematic Bacteriology of the International Association of Microbiological Societies ruled in favor of Runyon's recommendation to maintain the species designation M. fortuitum, which has remained to the present (144).
(ii) M. peregrinum.
M. peregrinum was first proposed in 1962 when Bojalil and colleagues published the first Adansonian or numerical classification of mycobacteria (10, 11). This analysis separated mycobacteria into 12 different categories or branches based on physiological characteristics. Branch 1 consisted of M. smegmatis, M. phlei, and a group of other strains that showed the greatest metabolic capacity of all the strains analyzed. The name M. peregrinum sp. nov. was proposed for this latter group (Latin adjective meaning strange or foreign) because “they were the only non-pigmented strains in Branch 1” (10, 11). The type strain is ATCC 14467T.
(iii) M. fortuitum third biovariant complex.
In 1966 Bönicke divided isolates of the M. fortuitum group into three subgroups on the basis of differences in acid production from carbohydrates and designated them biotypes A, B, and C (13). Biotype A had no unique carbohydrates, biotype B was mannitol positive, while biotype C was mannitol and inositol positive. Subsequently, Pattyn et al. (124) renamed these three Bönicke biotypes M. fortuitum biovariant fortuitum, M. fortuitum biovariant peregrinum, and an unnamed third biovariant.
In 1984, Tsang et al. (176) used a combination of chemical analysis, seroagglutination, and enzyme-linked immunosorbent assay to compare the glycolipids of the RGM and provided evidence that M. fortuitum biovar peregrinum was a separate species (176). M. fortuitum biovar peregrinum was later confirmed as an independent species, as M. peregrinum, based on genomic DNA-DNA relatedness studies by Levy-Frébault et al. (96) and Kusonoki and Ezaki (89) that showed <70% genomic DNA-DNA homology of M. peregrinum (ATCC 14467T) to other RGM species. By 16S rDNA sequencing, M. peregrinum (ATCC 14467T) is unique, with a Hamming distance of 9 to 15 bp from other members of the M. fortuitum group. Wallace et al. noted two subgroups within M. peregrinum. One group contained the ATCC type strain (ATCC 14467T), which was pipemidic acid susceptible; while the other, represented by ATCC 35755, was pipemidic acid resistant. The two groups have different PRA patterns with the hsp65 gene (165). An analysis of the taxonomic status of this second group using phenotypic and genetic analysis is ongoing (M. F. Schinsky, R. E. Morey, M. P. Douglas, A. G. Steigerwalt, R. W. Wilson, M. M. Floyd, M. I. Daneshvar, B. A. Brown-Elliott, R. J. Wallace, Jr., M. M. McNeil, D. J. Brenner, and J. M. Brown, unpublished).
M. fortuitum biovar fortuitum was also elevated to species status (M. fortuitum) based on DNA-DNA pairing studies which showed <70% homology of M. fortuitum ATCC 6841T to other taxa (89,96). By 16S rDNA sequencing, M. fortuitum differs by only 6 bp from the unnamed third biovariant sorbitol-negative group, by 8 bp from M. senegalense, and by 15 bp from M. peregrinum (ATCC 14467T).
The unnamed third biovariant, first described by Bönicke in 1966 (13), was later characterized and then subdivided into two groups by Wallace et al. (194) on the basis of a number of characteristics including sorbitol utilization. The two unnamed third biovariant groups were known as M. fortuitum third biovariant, sorbitol positive, and M. fortuitum third biovariant, sorbitol negative (194). By 16S rDNA sequencing, these two groups had a Hamming distance of 9 (160), with proposed representative strains being ATCC 49403 (sorbitol positive) and ATCC 49404 (sorbitol negative) (194). They differed by up to 20 bp from other M. fortuitum group members. Both sorbitol-positive and negative groups appeared heterogeneous, especially when studied for β-lactamase alleles (225) but also when compared by PRA of the hsp65 gene (165).
(a) M. houstonense and M. bonickeii (proposed species). Investigations currently under way may better delineate the multiple taxa or species within the third biovariant complex. Currently, at least six new species have been delineated. The majority of the sorbitol-positive group have been renamed M. houstonense, while the majority of the sorbitol-negative group have been renamed M. bonickei (Schinsky et al., Abstr. 12th Int. Symp. Biol. Actino.). Additional details of these species await publication of the entire study.
(b) M. septicum.M. septicum is one of the new species within the third biovariant complex. The type strain (ATCC 700731T) was the causative agent of central- line sepsis in a child with metastatic hepatoblastoma (74, 152). By HPLC analysis of mycolic acids, the isolate was distinctive but closely related to other members of the M. fortuitum group (156, 194), including M. senegalense. Standard biochemical testing showed the isolate to be similar to members of the unnamed third biovariant complex sorbitol-negative group (i.e., mannitol positive, inositol positive, sorbitol negative). Although initial testing showed the isolate to be arylsulfatase negative, it has been found to be positive in other laboratories (K. Jost, unpublished data). Analysis of the 16S rDNA showed a sequence related to but not identical to M. fortuitum, M. peregrinum, and M. senegalense (Schinsky et al., Abstr. 12th Int. Symp. Biol. Actino.).
(c) M. mageritense.The first five reported organisms in the new species M. mageritense were isolated from human sputa in two hospitals in Spain but were not considered clinically significant (46). Recently, six clinical isolates of this species were recovered in the United States, four of which were associated with clinical disease (190). Phenotypically, by antibiotic susceptibility patterns and biochemical tests, isolates of M. mageritense resemble sorbitol-positive members of the M. fortuitum third biovariant complex (positive for mannitol, inositol, and sorbitol) (190). By sequencing of its 16S rDNA, however, genetically, M. mageritense is more closely aligned with members of the M. smegmatis group (17). M. mageritense differs by only 9 bp from the type strain of M. wolinskyi (ATCC 700010T), by 16 bp from the type strain of M. goodii (ATCC 700504T), and by 18 bp from the type strain of M. smegmatis sensu stricto (ATCC 19420T). It generally differs by 23 to 28 bp from members of the M. fortuitum group. Future phenotypic and molecular studies of M. mageritense that include larger numbers of isolates may provide a more complete and accurate taxonomic placement of this species.
(iv) M. mucogenicum.
M. mucogenicum has been recognized as a species since 1995. The organism was first called M. chelonae-like organism (MCLO) in 1982, when it was reported as the etiologic agent in a peritonitis outbreak involving two peritoneal dialysis units (5). It was given the designation MCLO because the outbreak strain, just as M. chelonae, was nitrate negative and growth was inhibited on 5% NaCl. In 1993, a large number of sporadic clinical isolates were evaluated by Wallace and colleagues (200) by using biochemical reactions, HPLC of mycolic esters, and antibiotic susceptibility patterns. Subsequently, in 1995, Springer et al. proposed the name M. mucogenicum for this organism group, reflecting the highly mucoid character of the isolates (160). This species has always been grouped with the M. chelonae-abscessus group, but is unlike that group in that approximately 50% of isolates are nitrate positive, have a weak but positive iron uptake, and are much more susceptible to antibiotics including the fluoroquinolones, amoxicillin-clavulanic acid, polymyxin B, and cephalothin. In addition, by 16S rDNA sequencing M. mucogenicum is more closely related to the M. fortuitum group than to the M. chelonae-abscessus group (160). The type strain (ATCC 49650T) of M. mucogenicum differs by 11 to 18 bp from members of the M. fortuitum group but differs by 35 bp from M. abscessus and by 38 bp from M. chelonae. Hence, we propose to add this species to other members within the M. fortuitum group.
(v) M. senegalense.
M. senegalense was originally described by Chamoiseau in 1973 as a subspecies of M. farcinogenes (34). Later, however, it was recognized as a different species closely related to M. fortuitum (137). Like M. peregrinum, M. senegalense is positive only on mannitol, when tested on common sugars, but has a unique PRA pattern with the hsp65 Telenti fragment (165, 174). Its 16S rDNA sequence differs by 4 bp from the unnamed M. fortuitum third biovariant complex (sorbitol-positive) strain ATCC 49403; by 5 bp from the unnamed third biovariant complex (sorbitol-negative) strain ATCC 49404; and by only 8 bp from M. fortuitum (ATCC 6841T). By DNA comparison studies, however, it is a species apart from these other organism groups.
Type of disease.
(i) Community-acquired disease.
The M. fortuitum group accounts for 60% of cases of localized cutaneous infections caused by RGM but is a rare cause of chronic mycobacterial pulmonary disease (1, 3, 203, 205). Localized cutaneous disease generally occurs in previously healthy hosts, and drug-induced immune suppression appears to result in minimal increase in this risk. Wallace et al. (203) studied 123 patients with extrapulmonary disease caused by RGM and reported that 76 (63%) of these infections were due to the M. fortuitum group. Griffith et al. (63) studied 154 patients with pulmonary disease due to RGM and reported that only 16% of the infections encountered during the 15-year study were due to the M. fortuitum group (63). The M. fortuitum group was a common lung pathogen (50% of cases) only in the setting of chronic aspiration secondary to underlying gastroesophageal diseases such as achalasia. Approximately 25% of M. fortuitum group infections based on one study (205) have been associated with a variety of diseases other than skin or soft tissue infections including cervical lymphadenitis, mastoiditis, and meningitis. (91, 202, 205). Species in the M. fortuitum group are relatively rare causes of disseminated disease compared to other pathogenic RGM species, especially M. chelonae and M. abscessus.
(ii) Health care-associated disease.
The M. fortuitum group is responsible for the majority (60 to 80%) of cases of postsurgical wound infections and catheter infections caused by the RGM (77, 134, 198). Most of the responsible organisms are M. fortuitum and are detailed below in the specific section on M. fortuitum (38, 198).
Geography.
Cutaneous disease caused by the M. fortuitum group, although reported from all over the United States and worldwide, has been recognized most commonly in the southeastern United States (9, 198, 220). A report in 1989 (198) indicated that about 80% of wound isolates related to cardiac surgery were from seven southern coastal states: Texas, Louisiana, Georgia, Maryland, Alabama, Florida, and South Carolina. A second report published in the same year found that 92% of 37 identified cases of surgical wound infections following augmentation mammaplasty were in patients from southern coastal states, with the majority being from Texas, Florida, and North Carolina (201). Approximately 80% of isolates in both studies combined belonged to the M. fortuitum group.
Individual taxa.
(i) M. fortuitum.
(a) Community-acquired disease. In essentially all series of community-acquired or health care-associated disease attributed to the M. fortuitum group, most or all of the cases are due to M. fortuitum. In a study of 154 patients with RGM pulmonary disease, Griffith et al. (63) reported 13% of infections were due to M. fortuitum and only 3% were due to other members of the M. fortuitum group. In a series of cases of extrapulmonary disease cased by the M. fortuitum group, Wallace et al. (205) reported that almost 80% of the infections were due to M. fortuitum. Disseminated infections with M. fortuitum are rare. The first case report appeared in 1990, when Sack (145) described a patient with a history of intravenous IV drug abuse and AIDS, who had cutaneous lesions from which M. fortuitum was isolated. Cultures of specimens from lymph nodes, urine, pleural effusions, and feces all yielded M. fortuitum.
(b) Health care-associated disease. M. fortuitum has been implicated in numerous outbreaks of hospital or health care-associated infections (70, 77, 189). These include sternal wound infections; postinjection abscesses related to electromyography needles (72, 115); and a respiratory disease outbreak in Washington, D.C. (24). M. fortuitum has also been recovered from sporadic cases of surgical wound infections and catheter-related infections and is the most common RGM species in women with surgical wound infections following augmentation mammaplasty (189, 201). Details of some of the outbreaks are listed below.
Cardiac disease outbreaks. Isolates of M. fortuitum have been found in sternal wound infections in Hong Kong, Colorado, Nebraska, and Texas (189). Interestingly, three of the four involved multiple RGM species or multiple strains of the same species. The first outbreak occurred in 1976 in a hospital in Colorado. Of nine patients who underwent cardiovascular surgery within a 2-week period, four became infected with a single genetic strain (70) of M. fortuitum. Despite intense infection control efforts to recover the organism from environmental sources and subsequent molecular analysis of the isolates recovered, no environmental source was identified (70, 198).
In 1981, Preheim and colleagues (L. C. Preheim, M. J. Bittner, D. K. Giger, and W. E. Sanders Jr., Program Abstr. 22nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 564, 1982) reported an outbreak in a Nebraska hospital that involved five patients who developed sternotomy infections following coronary bypass surgery. All had multiple debridements. One patient died (Preheim et al., 22nd ICAAC). Later analysis by plasmid profiles (198), multilocus enzyme electrophoresis (MEE), and pulsed-field gel electrophoresis (70) showed the presence of two strains of M. fortuitum.
Another outbreak in 1981 involving M. abscessus and M. fortuitum, was reported in a hospital in Corpus Christi, Tex. (87). Of 51 patients undergoing surgery over a 6-month period, 6 (11.3%) developed infections with one of these RGM. One patient with sternal wound infection due to M. fortuitum died as a result of complications from sternectomy and antimicrobial therapy. A subsequent study was performed in which culture media were inoculated with samples taken from environmental sites including the municipal water system, water from the cold water tap in the operating room, ice machines, swabs of lamps, oxygen tanks, suction apparatus, and commercial bone wax. Multiple sites were culture positive for M. fortuitum strains (87) which, by MEE and subsequent pulsed-field gel electrophoresis, were identical to the disease strains (70, 189, 198). This is the first and only cardiac surgery outbreak in which the piped-water system has been implicated as the source of the pathogenic RGM. In this case, the cardioplegia solution was cooled in ice made from tap water contaminated with the outbreak strains.
The fourth and latest outbreak of M. fortuitum infection following cardiac surgery came from Hong Kong (220). Both M. fortuitum and M. peregrinum were implicated. The outbreak spanned the years 1987 to 1989, and 7 (33%) of 21 wound infections were due to M. fortuitum. (Details of the M. peregrinum strains are presented in the M. peregrinum section, below.) Subsequent ribotyping and susceptibility testing suggested that multiple strains were involved (220, 221). No source for the prolonged Hong Kong outbreak was identified.
Postinjection abscess disease. Most outbreaks of postinjection abscesses caused by RGM have involved M. abscessus. However, Nolan et al. (115) reported an outbreak of M. fortuitum in five (83%) of six patients undergoing electromyography at a facility in Washington state. The outbreak was traced to a break in a manual procedure used for sterilizing the reusable needle electrodes. When subsequent sterilizations were performed using an autoclave, no further cases were reported (115).
Plastic surgery-related disease. No large outbreaks of wound infection following plastic surgery due to M. fortuitum have been reported, but sporadic wound infections following plastic surgery have been well documented (205). Many of these infections, involving M. fortuitum, occurred after augmentation mammaplasty (201). A study by Wallace et al. (198) identified 37 cases of surgical wound infection with RGM following augmentation mammaplasty. Of these isolates, 26 (70%) were identified as M. fortuitum. Several surgeons had more than one case, suggesting that local environmental factors were important to disease development. Six cases of M. fortuitum disease associated with spontaneous breast abscesses with no history of surgery or trauma were also reported.
Pseudo-outbreaks. One pseudo-outbreak involving M. fortuitum occurred in 1987 in Houston, Tex., and involved ice contaminated with M. fortuitum. Although M. fortuitum was recovered from specimens from four patients who underwent bone marrow aspiration, none of the patients had evidence of disease due to M. fortuitum. It was later noted that each of the patients whose aspirates yielded M. fortuitum on culture also had viral cultures performed, and this required that the syringe containing the aspirate be plunged into ice for transport. Samples taken from the ice machine located on the same floor as the patients with positive aspirate samples were also positive for M. fortuitum. Samples from ice machines on other floors did not grow M. fortuitum (76). No molecular typing was performed.
(ii) Unnamed third biovariant complex including M. septicum, M. mageritense, M. houstonense (proposed), and M. bonickei (proposed).
Pattyn et al. (124) published one of the earliest studies of the unnamed third biovariant complex approximately 30 years ago. The authors commented that most of the isolates they studied were environmental strains and none had a definite disease association.
(a) Community-acquired disease. In 1991, Wallace et al. (192), characterized 85 clinical isolates of unnamed M. fortuitum third biovariant complex, all of which were disease associated. These represented 16% of 410 isolates of the M. fortuitum group submitted to a Texas laboratory and 22% of 45 isolates submitted to the Queensland, Australia, state laboratory. Over 75% of the infections involved skin, soft tissue, or bone. Clinical histories were available for 52 patients with skin and soft tissue infections, and the type of injury responsible for infection was reported for 42 patients. Of these 42 patients, 29% had osteomyelitis confirmed by bone biopsy. Most infections occurred following puncture wounds or compound (open) fractures. Only two patients (children) had no history of trauma leading to their infection. Metal puncture wounds (48%) or motor vehicle accidents (26%) were the most common histories given, and approximately 40% of the injuries involved the foot or leg. Stepping on a nail was the classic scenario (194). No cases of disseminated disease were observed in this study, and, to date, none have been reported by other authors. This study also identified 26 isolates of the third biovariant complex from pulmonary sources. Clinical significance was not determined, but almost certainly some of these were disease producing. None of these isolates were studied by molecular methods that would identify them as one of the recently described species within the complex (i.e., M. houstonense [proposed], M. bonickei [proposed], M. septicum, or M. mageritense) (190; Schinsky et al., submitted).
(b) Health care-associated disease. A small number of cases have been reported in association with hospital-acquired disease in other studies, including wound infections following cardiac surgery (198) and augmentation mammaplasty (201).
(c) Geography. The original description by Pattyn et al. (124) of this group was based on isolates from Europe and Africa. Later in 1983, Levy-Frébault et al. characterized 23 additional isolates of M. fortuitum from France and found 6 environmental isolates that were identified subsequently as M. fortuitum unnamed third biovariant complex (95). A later study by Wallace et al. included 10 isolates from Australia. The remaining 70 isolates were from the United States, of which approximately 70% were from Texas, 6% were from Florida, and 5% were from Alabama and Georgia (194). Four percent or less of the third biovariant isolates identified in this study were from other states including Tennessee, Louisiana, New Hampshire, South Carolina, Arkansas, and Connecticut (our unpublished data).
(iii) M. peregrinum.
Currently, there is no published series or review evaluating the clinical significance of M. peregrinum. No case of disseminated infection due to M. peregrinum has been reported. However, a small number of cases of sporadic infections have been reported and have been associated with diseases similar to other members of the M. fortuitum group. These include chronic lung disease (63, 64), sternal wound infections (198), and cutaneous disease (205). In general, they represent only 1 to 2% of sporadic community-acquired or health care-associated infections due to RGM. M. peregrinum has been reported as a cause of a pseudo-outbreak of respiratory disease due to a contaminated ice machine (93) and as the etiologic agent of 67% of cases in an outbreak of sternal wound infections from Hong Kong that occurred from 1987 to 1989 (220, 221).
(iv) M. mucogenicum.
M. mucogenicum (160), formerly MCLO, was originally discovered in two outbreaks of peritonitis in 1976 and 1978, in 5 (18%) of 22 and 5 (63%) of 8 patients, respectively, undergoing intermittent chronic peritoneal dialysis (5). Infections were traced to the use of contaminated automated chronic peritoneal dialysis machines in two dialysis centers. Seven sporadic cases of peritonitis due to M. mucogenicum from the same two centers were also diagnosed. The investigators' findings (5) suggested that ineffective disinfection of the equipment followed by colonization of the machines by this newly described species was responsible for the outbreaks. Three studies showed that M. mucogenicum, like other pathogenic species of RGM, is relatively resistant to formaldehyde and glutaraldehyde disinfectants, which are typically used to disinfect dialysis equipment (5, 31, 69). In 1985, Bolan et al. (12) described a mycobacterial disease outbreak in a hemodialysis center in Louisiana. Of 26 identified isolates, 25 were M. abscessus and 1 was M. mucogenicum. The single factor common to all patients was their exposure to hemodialyzers (artificial kidneys) that had been ineffectively treated with concentrations of disinfectant (formaldehyde) that were below effective levels for these RGM species (12). Although M. mucogenicum is often recovered as a laboratory contaminant, these three outbreaks alert us to the potential significance of this waterborne organism.
In 1993, Wallace et al. (200) evaluated 87 sporadic isolates of M. mucogenicum. Of these isolates, 54 (62%) were respiratory, and only 2 (4%) of them (both from patients with AIDS) were clinically significant. For the remaining 33 nonrespiratory isolates, significant clinical diseases included posttraumatic wound infections and catheter-related sepsis. Recovery of M. mucogenicum from skin, wound, or blood cultures was most often associated with clinical disease. In contrast, a single positive sputum culture is almost never clinically significant. Goldblatt and Ribas (61) recently reported the first case of a patient with granulomatous hepatitis caused by M. mucogenicum. No cases of disseminated cutaneous disease due to M. mucogenicum have been reported. The frequent presence of this organism in tap water, including ice machines (39), may contribute to the transient colonization or contamination of sputum samples (200). In a study of 113 mycobacterial isolates from tap water samples from various geographic sites, the most frequently occurring nontuberculous mycobacterium (41%) was M. mucogenicum (39). This study underscores the potential health risks of these ubiquitous organisms.
M. mucogenicum was first described by Band et al. (5) in the peritonitis outbreak in Washington state. Since that time, isolates have been recovered from Texas, Arizona, Maryland, Delaware, Illinois, Pennsylvania, Missouri, Florida, Washington D.C. and California (R. J. Wallace, Jr., unpublished data).
(v) M. senegalense.
M. senegalense is an etiologic agent of farcy, a disease of skin and superficial lymphatics in African bovines (208). It has not been reported from environmental or clinical cultures in the United States or Europe.
(vi) M. septicum.
The type strain of M. septicum (ATCC 700731T) was recovered in Australia from three separate blood cultures and a central venous catheter tip after its removal (74, 152). No other isolates have been reported.
M. chelonae-abscessus Group
The M. chelonae-abscessus group contains three species: M. chelonae, M. abscessus, and M. immunogenum. When researching the literature, it is important to recognize the difficulty in establishing which species is responsible for cases of disease associated with this group. This is particularly cogent for diseases during the period from 1972 to 1992 (generally prior to 1990), when M. chelonae and M. abscessus were considered to belong to the same species (“M. chelonei” or “M. chelonae”). Although they have been recognized as subspecies since an international collaborative study published by Kubica et al. (86) in 1972, little effort was made to separate them. Furthermore, interpretation of the literature after 1992 from investigators unaware that these organisms are, in fact, two species continues to be a problem. For purposes of this review, the description “M. chelonae-abscessus group” is used for cases when no subgrouping was performed or the data include all subgroups; the specific species names are used when an isolate was characterized by its ability to utilize citrate (156) or by genetic methods (eg., PRA of the hsp65 Telenti gene fragment) (165, 174). Common features within this group are a positive 3-day arylsulfatase, the absence of pigmentation, better growth at 30 than 35°C, a negative nitrate reductase, a negative iron uptake, and resistance to polymyxin B and most other antimicrobial drugs except amikacin and clarithromycin.
Historical perspective.
In 1953, Moore and Frerichs recovered an RGM (now ATCC 19977T) from a knee abscess. The authors thought the isolate was distinctive biochemically and morphologically from other RGM and identified it as a new species, M. abscessus. This name was selected because of the ability of the organism to produce deep subcutaneous abscesses (111).
Stanford et al. (163) first reported studies on clinical isolates of what was then known as M. borstelense (125). Among the isolates they studied were isolates from postinjection abscess outbreaks in Holland and England, human strains from other parts of Europe and Africa, and environmental strains. This study resulted in the official adoption of the name “M. chelonei” for these isolates, a name which was later changed to the more correct Latin, M. chelonae.
Many early investigators, however, believed M. chelonae and M. abscessus to be the same organism because they showed almost identical biochemical features. A cooperative numerical phenotypic study by the International Working Group on Mycobacterial Taxonomy (IWGMT) published in 1972, however, demonstrated that the two taxa were sufficiently different to be classified as subspecies and renamed them M. chelonae subspecies chelonae and M. chelonae subspecies abscessus (86).
Using genomic DNA-DNA hybridization studies, M. chelonae subspecies abscessus (ATCC 19977T) was later shown to be a separate species on the basis of <70% genomic homology with other RGM taxa, including M. chelonae subspecies chelonae (ATCC 35752T) (96). In 1992, Kusunoki and Ezaki elevated M. abscessus comb. nov. to species status, and M. chelonae subsp. chelonae once again became M. chelonae (89). Interestingly, by 16S rDNA sequencing, M. chelonae and M. abscessus differ by only 4 bp and are examples of the few different nontuberculous mycobacterial species that have identical 16S rDNA hypervariable region A sequences (89).
M. immunogenum, formerly M. immunogen, is a newly described RGM first recognized in contaminated metalworking fluids (110). It is closely related to M. chelonae and M. abscessus but readily distinguished by genetic methods (212). The organism was named for its potential relationship to cases of hypersensitivity pneumonitis in factory workers (metal grinders) who used mycobacterium-contaminated metal-grinding fluids for lubrication and cooling of their machines. By 16S rDNA sequencing, the ATCC type strain ATCC 700505T differs by only 8 bp from M. abscessus and by 10 bp from M. chelonae. It is morphologically similar to M. abscessus but has a different drug susceptibility pattern and a different PCR restriction analysis pattern of the hsp65 Telenti fragment (212).
Type of disease.
(i) Community-acquired disease.
For several years prior to the current molecular microbiology era, the M. chelonae-abscessus group was referred to collectively as M. chelonei or M. chelonae without further differentiation of species (see “Taxonomy and clinical significance above”). The M. chelonae-abscessus group has been associated with a variety of different diseases.
The most common clinical disease is probably chronic lung disease, usually in elderly women with bronchiectasis or young adults with cystic fibrosis (CF). The M. chelonae-abscessus group is responsible for approximately 95% of disseminated cutaneous infections caused by the RGM. Unlike patients with a localized infection, patients with disseminated cutaneous disease have multiple painful draining small abscesses that involve the arms and legs. Localized cellulitis, osteomyelitis, and small-joint arthritis are also commonly associated with the M. chelonae-abscessus group.
(ii) Health care-associated disease.
Sporadic (single) cases of otitis media, following tympanostomy tube placement, catheter infections, and postsurgical wound infections following a variety of surgical procedures (especially plastic surgery) also have involved this group of RGM (53, 98, 130, 135, 205). The M. chelonae-abscessus group has been involved in several health care-associated disease outbreaks including post-cardiac surgery sternal wound infections and vein graft site infections (87). Other outbreaks of M. chelonae-abscessus group infection have involved plastic surgery (52, 146), hemodialysis, and miscellaneous outbreaks including wound infections following laparoscopy, liposuction (107), and post-tympanostomy tube placement (98). Additionally, postinjection abscess outbreaks following the use of multidose vials (15, 62, 129) or contaminated biologicals (30, 33, 57, 183) also have been reported. Vaccine-related outbreaks involving M. chelonae-abscessus as contaminants also are recorded (59, 119). (Isolates from most outbreaks since 1980 were restudied at a later date and shown to be M. chelonae or M. abscessus. These outbreaks are detailed under the specific species.)
In addition to these true outbreaks of infection, several health care-associated pseudo-outbreaks have been described in conjunction with contaminated or malfunctioning bronchoscopes (66, 116, 121, 211; K. Petersen, N. Bus, V. Walter, and C. Chenoweth, Abstr. Infect. Control Hosp. Epidemiol., abstr. S-32, 1994), automated endoscope-cleaning machines (54, 55), and contaminated laboratory reagents (90).
Geography.
The M. chelonae-abscessus group is a collection of ubiquitous organisms found in soil and water worldwide. Outbreaks of M. chelonae-abscessus group disease have occurred primarily in the United States in southern coastal states and have been reported in North Carolina (54, 98), Louisiana, Georgia, Florida, and Texas (198). Generally, almost all of the states in the southern United States have reported disease with M. abscessus (70, 224). Outside the United States, isolates have been recovered from Hungary (205), Japan, Germany (6), Canada (53), France, Italy, Sweden, Australia, Belgium, Switzerland, Colombia, South America (183), and the United Kingdom (70; unpublished data).
Individual taxa.
(i) M. chelonae.
M. chelonae is one of the most antibiotic- resistant species of the pathogenic RGM. Like M. abscessus, M. chelonae is involved in several different types of community-acquired infections.
(a) Community-acquired disease. Pulmonary disease. Unlike M. abscessus and M. fortuitum, M. chelonae is only rarely a cause of chronic lung disease. In the series of 154 patients with chronic lung disease due to RGM reported by Griffith et al. (63), only 1 of 146 isolates identified to species was an M. chelonae.
Disseminated disease. M. chelonae causes three basic types of cutaneous disease (see Table 1). The most common type is disseminated cutaneous disease, which occurs when the host is chronically immunosuppressed (4). Wallace et al. (195, 206) reported that 53% of 100 clinical skin and/or soft tissue and/or bone isolates of M. chelonae were from patients with disseminated cutaneous infections (193). These infections were seen in patients receiving long-term corticosteroids and/or chemotherapy, primarily because of underlying organ transplantation, rheumatoid arthritis, or other autoimmune disorders (206). Chronic lung disease, solid-tumor malignancies and other disorders were less frequently associated with this disease (185, 222). McWhinney et al. (104) described three cases of M. chelonae in febrile neutropenic patients receiving chemotherapy. Infections with M. chelonae have occurred predominantly in patients with drug-induced immunocompromised status. In contrast, disease states which lead to immune suppression, such as AIDS, have not been significant risk factors for the development of disseminated M. chelonae infection.
Localized infections. The second type of infection seen with M. chelonae involves community-acquired localized infections following trauma (62, 205, 206, 210). These infections range from localized cellulitis or abscess to osteomyelitis. In the series by Wallace et al., 35% of the infections caused by M. chelonae were of this group (i.e., localized wound infections) (Table 1) (193).
(b) Health care-associated disease. Sporadic localized wound infections following medical or surgical procedures including needle injections can occur with M. chelonae but are rare compared to infections with M. fortuitum and M. abscessus. Health care-associated outbreaks due to M. chelonae are also rare and have been observed only following injection with contaminated syringes or needles, the implantation of contaminated porcine heart valves (55, 79, 189), and, recently, the use of liposuction (107). In this last outbreak, the organism was recovered from tap water connected to the suction cannulas.
The third, and least common, type of infection caused by M. chelonae, yet the most common type of health care-associated disease, is that of catheter-related infections (Table 1). In 1992, Wallace et al. reported that 8 of 100 clinical isolates of M. chelonae were associated with intravenous catheters, an additional 3 involved chronic peritoneal dialysis catheters, and 1 involved a hemodialysis shunt (193). They found that both the use of corticosteroids and renal failure were risk factors for these catheter-related infections (193).
(ii) M. abscessus.
M. abscessus and M. chelonae are probably the most antibiotic resistant species of the pathogenic RGM. Like M. chelonae, M. abscessus is involved in a variety of different types of community-acquired infections.
(a) Community-acquired disease. Pulmonary disease. Pulmonary disease accounts for most clinical isolates of this species (Table 1). According to Griffith et al. (63), the majority (82%) of the 146 disease-associated pulmonary RGM isolates identified to species over a 15-year period by a Texas reference laboratory were M. abscessus. In patients with M. abscessus pulmonary disease, underlying diseases included bronchiectasis, CF (40), gastroesophageal disorders, and prior granulomatous disease such as sarcoidosis or tuberculosis. The analysis by Griffith et al. (63) of M. abscessus pulmonary disease emphasized striking similarities to pulmonary M. avium complex lung disease of the type known as nodular bronchiectasis. The latter presents as an indolent course, occurs predominantly in elderly nonsmoking female patients, and exhibits a possible geographic disposition (i.e., southern coastal states have 69 to 75% of the cases). These similarities suggest a common pathogenicity or host susceptibility (63). Like patients infected with M. avium complex, most patients with pulmonary disease due to M. abscessus have underlying bronchiectasis of a type known as nodular bronchiectasis (196). Approximately 20% of patients with M. abscessus infection will also develop infection or disease due to M. avium complex (63), again emphasizing similar if not identical risk factors. There is controversy about whether M. abscessus can be a “colonizer” in the lungs. These authors believe that true colonization does not exist and that patients with minimal symptoms just have minimal disease. Repeated isolation of M. abscessus from the respiratory tract is usually associated with significant lung disease.
Pulmonary disease in patients with CF. M. abscessus patients with underlying CF deserve some special comments. The recovery of M. abscessus from the respiratory tracts of patients with CF is being noted with increasing frequency. Patients with CF are predisposed to airway and parenchymal infections for several reasons, including the nature of CF disease and the usual associated bronchiectasis (48). The primary risk factor that makes patients with CF more susceptible to mycobacterial disease is thought to be bronchiectasis. Evaluation of the significance of the mycobacterial infection can be complicated because isolation of other organisms such as Pseudomonas aeruginosa often makes isolation and interpretation of the clinical significance of the RGM difficult. After M. avium complex, M. abscessus is the second most common species of nontuberculous mycobacteria recovered from respiratory specimens in patients with CF (149).
Lung transplantation may be considered a therapeutic option in some CF patients. However, the posttransplantation immunosuppressive therapy increases the risk of both the development and the dissemination of nontuberculous mycobacterial infections. Patients with CF and M. abscessus lung disease carry the risk of developing disseminated infections, including cervical adenitis, following transplantation (48, 168).
Extrapulmonary disease. After M. fortuitum, M. abscessus is the second most common RGM species in clinical specimens; it also produces a wide variety of extrapulmonary diseases. Wallace et al. (205) studied a series of 59 nonrespiratory isolates belonging to the M. chelonae-abscessus group and found that M. abscessus cases outnumbered M. chelonae cases more than 2:1 (30 and 12 cases, respectively). Among the 30 cases of nonpulmonary disease caused by M. abscessus, 43% were postsurgical or postinjection wound infections, 23% were localized community-acquired wound infections, 20% were disseminated cutaneous infections, and 13% were miscellaneous types of infections including keratitis and prosthetic valve endocarditis.
Of the 23% of nonpulmonary disease cases resulting in localized infection, most characteristically developed following a break in the skin surface and subsequent direct contact with contaminated water or soil. Localized trauma with a resulting pyogenic abscess is sometimes followed by a sporotrichoid appearance of ascending lymphadenitis predominantly in immunocompromised patients (81). Other examples of localized M. abscessus wound infections include a soft tissue infection of the cheek following an insect bite (26) and a case of vertebral osteomyelitis (102).
(b) Health care-associated disease. M. abscessus and M. fortuitum are the most common mycobacterial species causing nosocomial disease, especially sporadic and clustered outbreaks of surgical wound infections. As noted above (205), surgical wound infections represented 43% of clinical cases of nonpulmonary infections due to this species. Disease outbreaks have been described after augmentation mammaplasty, facial plastic surgery, cardiac surgery, injections of alternative medicines, steroid injections, and miscellaneous types of surgery (55, 223).
In the largest outbreak of RGM-mediated postinjection abscesses, which occurred in an alternative medicine clinic in Colombia, South America (183), 205 (59%) of 350 of patients developed localized cutaneous abscesses or cellulitis due to M. abscessus. Another large M. abscessus outbreak in the United States resulted from the injection of an unlicensed product sold as adrenal cortex extract (ACE) (57). Of 140 persons known to have received the ACE injections, 87 subjects (62%) from 16 states were identified as infected. M. abscessus was cultured from seven vials of ACE, six of which were unopened. Isolates from both patients and opened and unopened vials of ACE were typed by MEE and pulsed-field gel electrophoresis and shown to be identical (57). Pseudo-outbreaks related to contaminated bronchoscopes have also been attributed to M. abscessus (55).
Although disseminated M. abscessus disease is relatively unusual, it is serious. Most cases have occurred in chronically immunosuppressed patients receiving corticosteroids, and the disease has no apparent portal of entry. The disease presents as multiple draining cutaneous nodules, usually involving the lower extremities. Patients with disseminated infection have rarely included detectable bacteremia and endocarditis (41), and these cases can occur as a complication of localized infections. This is especially true in patients on hemodialysis. Bolan et al. (12) reported 25 infections due to M. abscessus in a hemodialysis center in Louisiana (see the section on M. mucogenicum [above] for details). Nine of these patients had widely disseminated disease. Subsequent molecular studies using random amplified polymorphic DNA-PCR showed that the M. abscessus strains from the water supply and the clinical isolates were identical (224).
This hemodialysis outbreak not only served to show the potential virulence of RGM disease in this setting but also pointed out the relative resistance of these organisms to commonly used disinfectants, a fact which increases the risk of health care-associated infections. In the Louisiana outbreak, investigators discovered that formaldehyde concentrations lower than 2% were used in disinfecting the reusable hemodialyzers. Failure to maintain a 2% concentration probably played a large role in this outbreak because this concentration had been previously established (31) as the minimum concentration to which M. abscessus was susceptible in vitro (12). Five years later, Lowry et al. (97) reported M. abscessus infection in five patients receiving dialysis with reusable dialysis tubing at another outpatient hemodialysis clinic. Again, the disinfectant used (2.5% Renalin) appeared to play a role, since at this concentration it did not completely kill the M. abscessus recovered from the patients and from the dialyzers that were manually reprocessed (97).
(iii) M. immunogenum.
In 2000, Moore et al. (110), described an outbreak of hypersensitivity pneumonitis among workers in an industrial plant that was undergoing extensive remodeling and renovation. The workers utilized cutting, drilling, and grinding machines and worked with a semisynthetic metalworking fluid that was sprayed on the machines to keep them cool. Part of the outbreak investigation involved performance of cultures of the metalworking fluid for mycobacteria. Twenty- five isolates were recovered from different samples throughout the plant that were similar to M. chelonae-abscessus complex but with a unique hsp65 PRA pattern. This finding launched a search for other M. chelonae-abscessus-like RGM isolates with the same RFLP pattern. Isolates with this PRA pattern were identified from unrelated nosocomial pseudo-outbreaks involving contaminated endoscopes and from patients with serious infections. Although these strains exhibited overlapping biochemical and HPLC features with M. chelonae and M. abscessus, they differed from clinical and reference strains of both these species (54, 212) and most isolates had a unique susceptibility pattern of resistance to both cefoxitin and tobramycin (212). Molecular examination that included DNA homology studies showed that these isolates belonged to a separate species, which has been proposed as M. immunogenum (212). M. immunogenum organisms are able to grow and survive in degraded metalworking fluid (110), although it has not yet been established whether these organisms can metabolize any of the constituents of the fluid or additive materials for nutrition. The presence of other microorganisms (especially aerobic gram-negative bacilli) in degraded metal-grinding fluids and the use of biocides probably facilitates fluid degradation and subsequent growth of this species (110).
(a) Clinical disease. In the only detailed study of clinical disease, 11 isolates of M. immunogenum were identified from patients (212). Three came from cultures of blood from patients with catheter- or pacemaker-related sepsis; two came from cutaneous cultures of samples from a liver transplant recipient and an infant with severe combined immunodeficiency syndrome with disseminated cutaneous infections; two came from catheter exit sites; and one each came from fluid a septic joint in a hand, bronchoalveolar lavage fluid from a patient with chronic pneumonia, a cornea from a patient with suspected keratitis, and urine from a patient with an unknown diagnosis (212).
Additionally, two pseudo-outbreaks have been reported from Kentucky and Missouri, involving contaminated automated bronchoscopic washing machines which ultimately led to contaminated bronchoscopes (54, 55, 100, 207), retrospect, these infections were found to be due to M. immunogenum (212).
M. smegmatis Group
The M. smegmatis group currently is composed of M. smegmatis sensu stricto and the recently described M. wolinskyi (17) and M. goodii (17). For purposes of this study, isolates identified specifically as one of the three recent taxonomic groups are referred to by their current species name(s). Strains not so recognized are referred to as the M. smegmatis group. Characteristic features of the M. smegmatis group include a negative 3-day arylsulfatase, growth at 45°C, a positive nitrate reductase, a positive iron uptake, often a very smooth colony type, utilization of mannitol, inositol, and sorbitol as carbon sources, and a unique characteristic PRA pattern of the Telenti fragment of the hsp65 gene with BstEII (17, 199). A late (7- to 10- day) yellow to orange pigmentation (most but not all isolates of M. smegmatis sensu stricto and M. goodii; the M. wolinskyi isolates are nonpigmented) on Middlebrook 7H10 agar is often seen. Isolates of some of these taxa have been recovered from the environment, and all have been recovered from patients, most of whom had clinical disease (17, 215).
One important distinguishing feature of isolates of the M. smegmatis group, in contrast to the M. fortuitum group and the M. chelonae-abscessus group, is their general lack of susceptibility to the new macrolides, including clarithromycin (17). Since clarithromycin has been considered the cornerstone of antimicrobial therapy for RGM disease, it becomes vital to identify RGM isolates to exclude groups like the M. smegmatis group and the M. fortuitum third biovariant complex sorbitol-positive group, which are intrinsically resistant to this class of drugs.
Historical perspective.
The M. smegmatis group, first isolated by Lustgarten in 1885, was named for the genital secretions (smegma) from which it was recovered in a patient with a penile ulcer (99). The first well-described case of human disease caused by the M. smegmatis group involved the lungs and pleura of a patient with underlying exogenous lipoid pneumonia and was reported less than 15 years ago (184).
Type of disease.
(i) Community-acquired disease.
The first series of clinical patients was reported by Wallace et al. in 1988 (199) when they characterized 22 clinical isolates. The authors noted that the isolates were heterogeneous, and fell into three groups with different antibiotic susceptibility patterns. Later, in 1999, these three groups were studied in greater detail, including DNA homologies, and were found to be three distinct species: M. smegmatis sensu stricto, M. wolinskyi, and M. goodii (17). The last two names honored Emanuel Wolinsky and Robert Good, two early leaders in the field of nontuberculous mycobacteriology (17).
The three species are separated with approximately 90% accuracy on the basis of tobramycin susceptibility. M. smegmatis sensu stricto is tobramycin susceptible (MIC, ≤1μg/ml; agar disk diffusion zone, >30 mm). M. goodii has intermediate susceptibility to tobramycin (MIC, 2 to 8 μg/ml; agar disk diffusion zone, 11 to 30 mm), and M. wolinskyi is resistant to tobramycin (MIC, >8 μg/ml; agar disk diffusion zone, ≤10 mm) (17).
The HPLC patterns produced by M. smegmatis sensu stricto, M. wolinskyi, and M. goodii can be differentiated from those produced by members of the M. fortuitum group (16; K. C. Jost, Jr., S. H. Chiu, R. B. Dunlap, L. B. Elliott, B. A. Brown, V. A. Steingrube, R. W. Wilson, and R. J. Wallace Jr., Abstr. 99th Gen. Meet. Amer. Soc. Microbiol. 1999, abstr. U-36, 1999), and the three species have different patterns. The overlap between patterns of all mycobacterial species makes identification of the individual M. smegmatis species difficult, however, when evaluating individual clinical isolates.
The most accurate separation of the three species within the M. smegmatis group is achieved by molecular techniques including PRA of the Telenti fragment of the hsp65 gene and 16S rRNA gene sequence analysis (17).
Until a case of lung disease proven by lung biopsy was reported in 1986 (184), the M. smegmatis group was considered to be an environmental saprophyte of no clinical significance. Community-acquired disease due to M. smegmatis group is now known to involve cellulitis, localized abscesses, and/or osteomyelitis of a wound site following a traumatic event. Newton et al. (113), reported that the M. smegmatis group was the causative agent of two cases of infection following motor vehicle accidents, with cellulitis and extensive soft tissue and periosteal necrosis evident at the time of surgical debridement. A few cases of lipoid pneumonia (pneumonia resulting from inhalation or aspiration of lipid-containing medicinals or food particles) with secondary mycobacterial infection have also been reported to be caused by the M. smegmatis group (17, 199). No case of disseminated cutaneous disease due to the M. smegmatis group has been reported to date.
(ii) Health care-associated disease.
Health care-associated infections involving the M. smegmatis group have included sporadic cases of catheter sepsis, infected pacemaker site, sternal wound infection with possible osteomyelitis following cardiac surgery, and infections following plastic surgery (breast reduction surgery and a face-lift) (17, 199). No health care-associated disease outbreak or pseudo-outbreak due to the M. smegmatis group has yet been reported.
Geography.
Isolates of the M. smegmatis group have a wide geographic distribution. Isolates have been recovered in the United States from Texas, Alabama, California, Florida, Illinois, Indiana, Massachusetts, Minnesota, Mississippi, Missouri, North Carolina, Ohio, Oklahoma, South Carolina, Utah, and Wyoming (17, 56, 199). Outside the United States, isolates have been reported from Australia, Russia, Canada, and Switzerland (17, 127, 199).
Individual taxa.
(i) M. smegmatis sensu stricto.
In 1988, Wallace et al. (199), reported a series of 21 patients with infections due to the M. smegmatis group. In the latter taxonomic study of these isolates published in 1999 (17), 52% of the 21 original clinical isolates matched the type strain and additional ATCC reference strains of M. smegmatis and hence were renamed M. smegmatis sensu stricto. With expansion of the number of clinical isolates which met the criteria for the M. smegmatis group to 71, 49% were M. smegmatis sensu stricto (17). These isolates had a unique mycolic acid pattern and were susceptible to tobramycin agar disk diffusion (zones, >30 mm with a 10 μg commercial disk). Additionally, 16S rRNA gene sequence analysis and PRA of the 439-bp hsp65 gene sequence were unique to this species (17). Isolates of M. smegmatis sensu stricto have been reported from several states including Florida, Wyoming, South Carolina, Texas, Mississippi, and Illinois and, outside the United States, in Australia (199).
M. smegmatis sensu stricto has been incriminated in community-acquired cases of lymphadenitis, cellulitis, osteomyelitis, wound infections and, rarely, respiratory disease, usually associated with exogenous lipoid pneumonia (17, 199). It has been recovered from health care-associated infections, including sternal wound sites following cardiac surgery, bacteremia from intravenous catheter placement, and breast abscess following augmentation mammaplasty (17, 199).
(ii) M. goodii.
As mentioned previously, the Wallace study in 1988 (208) launched a second study, published in 1999 (17), which identified 8 of 21 isolates reported in the 1988 publication and 20 new isolates in the second study which proved to be M. goodii. These 28 isolates represented 39% of the 71 total isolates of the M. smegmatis group studied. They had a mycolic acid pattern that differed from the other two species in the group, were intermediately susceptible to tobramycin by agar disk diffusion (zones, 11 to 30 mm), had a unique 16S rRNA gene sequence, and a unique PRA pattern (159). M. goodii is the second most frequently isolated species within the M. smegmatis group.
Isolates of M. goodii have been recovered from California, Texas, Florida, Alabama, Minnesota, Utah, Oklahoma, Missouri, Indiana, Ohio, North Carolina, and Massachusetts (17, 199). Outside of the United States, isolates have been reported from Russia (17, 56), Australia (17, 199) and Canada (Sylvia Chomyc, Provincial Laboratory of Public Health, Alberta, Canada, personal communication).
Isolates of M. goodii have been recovered from cases of cellulitis, bursitis, and osteomyelitis after open (compound) fracture or penetrating trauma (17, 56, 199). A few cases of respiratory disease due to M. goodii have been reported. Most have been associated with underlying exogenous lipoid pneumonia with pulmonary infiltrates, similar to M. smegmatis sensu stricto (17, 199).
M. goodii has been involved in several types of sporadic health care-associated disease, including bacteremia with catheter sepsis (199), cardiac bypass infection with osteomyelitis, infected pacemaker site, and infection following breast reduction surgery (17).
(iii) M. wolinskyi.
Of 21 isolates from the 1988 study by Wallace et al. (199), 2 matched 6 other isolates in the second study (17) in their 16S rRNA gene sequence patterns, PRA patterns, and mycolic acid patterns. These eight strains collectively were named M. wolinskyi (17, 199) and represented 11% of the total 71 clinical isolates identified as belonging to the M. smegmatis group. Isolates of M. wolinskyi have been recovered from Texas, California, and Switzerland (17, 127, 199).
Clinical histories were available for seven of the eight reported isolates of M. wolinskyi (17, 199). Four (57%) of these seven isolates were associated with community-acquired infections that included cellulitis and osteomyelitis following a traumatic event and cellulitis and localized abscess following a motor vehicle accident (17, 199).
The remaining three isolates (43%) of M. wolinskyi were associated with sporadic health care-associated infections. These included sternal wound infection and osteomyelitis following cardiac surgery, surgical wound infection following facial plastic surgery (127), and an infected arteriovenous shunt in a patient on hemodialysis.
Other nonpigmented RGM of uncertain clinical (human) significance.
M. chitae was described by Tsukamura in 1966 (177, 178). Four strains of this species were recovered from soil samples collected near manure heaps. The organisms are not known to be associated with disease.
M. agri was described by Tsukamura (179). The only isolate was recovered from an alkali-treated soil sample and was not known to be associated with disease. Differential characteristics of both these species may be found in Bergey's Manual of Systematic Bacteriology, vol. 2 (208).
M. porcinum also was described by Tsukamura et al. (179a) in 1983, when the authors characterized 10 strains recovered from pigs with submandibular lymphadenitis.
TREATMENT OF INFECTION
Antimicrobial Treatment
General.
Antimicrobial therapy for RGM, unlike chemotherapy used for most slowly growing mycobacterial diseases, may vary depending on the nature of the disease. For example, single-drug therapy for localized or minor disease due to RGM is often sufficient, with minimal risk of development of mutational drug resistance. In contrast, disseminated cutaneous disease and pulmonary disease usually require multiple antimicrobials, including both intravenous and oral medications. The newer oral antimicrobials linezolid and gatifloxacin offer great promise as alternatives to injectable medicines, but clinical experience with them is very limited (21; B. A. Brown-Elliott, R. J. Wallace Jr., and C. J. Crist, Program Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-723, 2001). Table 2 summarizes antimicrobial treatment of the frequently encountered pathogenic RGM.
TABLE 2.
Antimicrobial activity for the three most frequently isolated species of nonpigmented RGM
| Species | Antimicrobial agenta
|
|
|---|---|---|
| >90% susceptible or intermediate (% if <100%) | <90% susceptible or intermediate (approx. % susceptibility/intermediate) | |
| M. fortuitum group | Amikacin, cefoxitin,c ciprofloxacin, gatifloxacin, imipenem, levofloxacin, linezolid (96%),f sulfamethoxazole or trimethoprim-sulfamethoxazole | Clarithromycin (80%),b doxycycline (46%),d vancomycin (38%)e |
| M. abscessus | Amikacin (98%),d cefoxitin (95%),h clarithromycinb | Ciprofloxacin (<1%),g doxycycline (4%),d imipenem (57%),h linezolid (48%) |
| M. chelonae | Amikacin (97%), clarithromycin,b gatifloxacin (96%),i linezolid (94%),f tobramycin | Ciprofloxacin (19%),g doxycycline (26%),d imipenem (40%)h |
Data given in exact percentages are referenced in the publications in the footnotes; otherwise, approximate percent positive is given.
Reference 20.
Excludes M. fortuitum third biovar sorbitol positive (proposed M. houstonense), of which >90% are resistant (194).
Reference 171.
C. J. Crist, R. J. Wallace Jr., B. A. Brown-Elliott, and L. B. Mann, Abstr. 101st Gen. Meet. Am. Soc. Microbiol 2001, abstr. U-35, 2001.
Reference 195.
Reference 187.
Reference 192.
B. A. Brown-Elliott, R. J. Wallace Jr., and C. J. Crist, Program Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-723, 2001.
M. fortuitum group.
Since the M. fortuitum group is much less drug resistant than M. abscessus and M. chelonae, treatment has been much easier and generally more effective. The usual therapeutic drugs recommended for infections with the former group include amikacin, cefoxitin, imipenem, sulfamethoxazole, and fluoroquinolones (171, 194, 202, 203). Doxycycline has proven to be an effective oral agent (203), but only about 50% of isolates of M. fortuitum are susceptible to ≤4 μg/ml (171).
In vitro susceptibility to clarithromycin within the M. fortuitum group is variable (20). Approximately 80% of isolates of M. fortuitum are susceptible to ≤4 μg/ml. Isolates of (proposed species) M. bonickei (sorbitol-negative third biovariant group) and M. peregrinum are all susceptible to ≤4 μg/ml, while isolates of the M. fortuitum third biovariant sorbitol- positive group (e.g., M. houstonense [proposed] and M. mageritense), are uniformly resistant to clarithromycin. Wallace et al. (196, 197, 203) have recommended a sulfonamide or doxycycline as acceptable agents for oral monotherapy of localized wound infections caused by the M. fortuitum group (based on in vitro susceptibilities). A major problem associated with quinolone monotherapy, however, was the development of mutational resistance with treatment failure or relapse (187, 192). Therefore, if a quinolone is used for therapy, an additional antimicrobial agent is usually necessary. Acquired mutational resistance of the M. fortuitum group to clarithromycin, doxycycline, and trimethoprim-sulfamethoxazole has not been reported, and we have not observed it (unpublished observations).
For serious disease with M. fortuitum, the aminoglycoside amikacin, combined with a β-lactam (cefoxitin or imipenem) (192) or a quinolone (ciprofloxacin or l-ofloxacin), has been used for initial therapy (187, 192).
Linezolid offers excellent potential as an oral or intravenous (i.v.) therapeutic agent, since 96% of isolates of all members of the M. fortuitum group (M. fortuitum, M. peregrinum, and the proposed new species M. houstonense and M. bonickei) are susceptible or intermediate in vitro (21, 195). However, there is no reported clinical experience with this drug for the M. fortuitum group.
M. chelonae-abscessus group.
The treatment of localized infections due to M. chelonae or M. abscessus is currently managed by using the newer macrolide clarithromycin as the cornerstone of therapy (20). Azithromycin appears to work as well, but there is much less clinical experience with it.
Acquired mutational resistance to the macrolides has not been observed when treating localized infections (186, 191). However, more serious disease should be treated, for at least the first 2 weeks, with clarithromycin in combination with one of the injectable agents (102, 196).
For serious, extensive extrapulmonary disease or disseminated infections involving M. chelonae, the injectable agents tobramycin plus imipenem have been used for the first 2 to 6 weeks in combination with clarithromycin to avoid or minimize the development of drug resistance to the macrolide (182). For M. chelonae, cefoxitin is not used, since isolates of M. chelonae are resistant to this agent (MIC, >128 μg/ml), and tobramycin is preferred to amikacin because of its greater in vitro activity (171).
Potential alternative oral agents are available which make injectable therapy less mandatory than in years past. The tetracycline analogues minocycline and doxycycline are effective against about 20% of M. chelonae (193, 222). Newer potential oral agents being considered for use in combination with clarithromycin are the 8-methoxyfluoroquinolones (e.g., gatifloxacin) and/or linezolid. However, there is little experience with these newer agents (see “Newer drugs” below).
For serious, extensive extrapulmonary disease or for disseminated disease due to M. abscessus, amikacin plus cefoxitin or imipenem is used for the first 2 to 6 weeks in combination with clarithromycin. Approximately 90% of M. abscessus isolates are susceptible or intermediate to amikacin (32 μg/ml) and cefoxitin (64 μg/ml) and about 50% are susceptible or intermediate to imipenem (8 μg/ml) and linezolid (16 μg/ml) (193,195). Whether linezolid could be used for this species has not yet been studied.
For patients with pulmonary disease due to M. abscessus, treatment options are few. Generally, the only oral antimicrobials to which M. abscessus is susceptible are the new macrolides clarithromycin and azithromycin. M. abscessus also is usually susceptible in vitro to amikacin, cefoxitin, and imipenem (194), i.v. medicines whose administration is limited by cost, toxicity, and the need for frequent administration (the β-lactams). (As with extrapulmonary disease, the potential usefulness of linezolid has not been studied.) The optimal treatment for M. abscessus lung disease is probably combination therapy using a macrolide and parenteral antibiotics. Most patients improve when given parenteral therapy for 2 to 4 weeks, but few can tolerate these medicines for longer periods. Unfortunately, to date, permanent sputum conversion along with permanent symptomatic improvement when using the combination of oral clarithromycin and parenteral agents for M. abscessus is rare. Only surgical resection (with localized disease) of the lung has produced long-term conversion of sputum cultures to negative and complete resolution of symptoms in patients with M. abscessus infectious (63). Better drugs are clearly needed for this species.
M. smegmatis group.
Treatment of disease has generally involved the same drugs as for treatment of the M. fortuitum group, with doxycycline and trimethoprim-sulfamethoxazole being the most common oral agents. Injectable agents have usually included amikacin and/or imipenem. All isolates of the M. smegmatis group are uniformly susceptible to sulfonamides, doxycycline, imipenem, and amikacin (17, 199). They exhibit intermediate susceptibility to the older fluoroquinolones (ciprofloxacin and ofloxacin) and variable susceptibility to cefoxitin and clarithromycin. The M. smegmatis group is the only nonpigmented RGM pathogenic for humans that is susceptible to ethambutol. With this exception, none of the other first-line antituberculosis drugs is efficacious against any of the RGM (17, 199).
Newer drugs.
Several new alternative antimicrobials have emerged for the management of RGM disease, with the most active in vitro being linezolid, the 8-methoxyfluoroquinolones moxifloxacin and gatifloxacin (195; B. A. Brown-Elliott, R. J. Wallace Jr., and C. J. Crist, Program Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-723, 2001), and tigecycline (formerly GAR-936) (R. J. Wallace, Jr., B. A. Brown- Elliott, C. J. Crist, L. Mann, and R. W. Wilson, submitted for publication). The oxazolidinone linezolid (Zyvox; Pharmacia) offers great potential in the treatment of RGM disease, primarily against isolates of the M. fortuitum group, the M. smegmatis group, M. mucogenicum, and M. chelonae (195). Wallace et al. (195) reported that 96% of 74 isolates of M. fortuitum group were susceptible to ≤16 μg of linezolid per ml, with a modal MIC of 4 μg/ml. Most (94%) of 50 isolates of M. chelonae had linezolid MICs of 4 to 16μg/ml, with a mode of 8 μg/ml. In contrast, more isolates of M. abscessus were resistant, with only 24 of 98 (48%) isolates being susceptible or intermediate (MIC, ≤16μg/mL) (195). A single patient with disseminated cutaneous disease due to M. chelonae resistant to clarithromycin was successfully treated with a 3-month course of linezolid monotherapy (21).
The recently FDA-approved 8-methoxyfluoroquinolones gatifloxacin and moxifloxacin have greater activity against the M. fortuitum group and M. chelonae than do the older fluoroquinolones such as ciprofloxacin. In a study by Brown-Elliott et al. (41st ICAAC), 100% of 26 isolates of M. fortuitum group and 1 isolate each of M. smegmatis, M. wolinskyi, M. goodii, and M. immunogenum were susceptible, with gatifloxacin and ciprofloxacin MICs for 90% of isolates an (MIC90) of ≤0.12 L and 1 μg/ml, respectively. In the same study, 96% of 27 isolates of M. chelonae were susceptible or intermediate to gatifloxacin (MIC, ≤4 μg/ml), in contrast to only 8% susceptible or intermediate to ciprofloxacin (MIC, ≤2 μg/ml). No isolates of M. abscessus were susceptible or intermediate to ciprofloxacin, and only 10% (of 20 isolates) were susceptible or intermediate to gatifloxacin.
The new glycylcycline (a tetracycline analog) tigecycline (formerly Gar-936) is in phase III clinical trials. In vitro studies with RGM have shown all pathogenic species of the nonpigmented RGM to be highly susceptible to this agent, with MICs of ≤0.25 μg/ml (R. J. Wallace, Jr., et al., submitted).
Duration of therapy.
Generally, the length of treatment with any of the current antimicrobials for most RGM skin, soft tissue, or bone disease has been 4 months for mild disease and 6 months for serious disease. Treatment with injectable agents is usually limited to the first 2 to 6 weeks of therapy, in an attempt to minimize cost and drug toxicity. The exception for total length of therapy is that pulmonary disease is usually treated for at least 12 months. However, microbiological cure has not been possible in most cases of pulmonary disease caused by M. abscessus, even after lengthy antimicrobial treatment (196).
Surgical Treatment
Minor wound infections with RGM may resolve spontaneously or after surgical debridement. In an early study by Wallace et al. (203), 13 (17%) of 76 and 3 (6%) of 47 extrapulmonary cases of infection with the M. fortuitum group and the M. chelonae-abscessus group were successfully treated by surgery alone. The authors also reported that surgery alone for more serious wound infections, without antimicrobial therapy, was followed by healing but that relapse often occurred within 4 to 6 weeks of the surgery. In contrast, surgical excision and/or debridement of the wound site combined with appropriate antimicrobial therapy resulted in healing without relapse (131, 194, 203, 205). Likewise, when a foreign body such as breast implant, percutaneous catheter, etc., is involved, removal of the foreign body appears to be essential to recovery (38, 41, 97, 185, 196).
In a 1985 report (203), the success rate for the treatment of M. fortuitum wound infections was about 90% even though the macrolides, the newer quinolones, and imipenem were not yet available. In 1985, that was not the picture for wound infections due to M. chelonae or M. abscessus even with surgical intervention. Until the arrival of the newer macrolides (in approximately 1993), serious disease caused by M. abscessus or M. chelonae was difficult to treat, even with a combination of antimicrobials and surgery, and infection with M. chelonae-abscessus could persist for years (185). No highly active oral agents were then available for treatment. The available drugs (amikacin, tobramycin, and cefoxitin) were too toxic and/or too expensive to continue long enough to cure most serious infections, and their effects were worse than those of minor disease. The introduction of clarithromycin almost certainly improved the response rate for M. chelonae and M. abscessus and has decreased the need for routine surgery in patients infected by these species.
In a series of two patients infected with the M. smegmatis group, Newton et al. (113) expressed the opinion that extensive surgical debridement of soft tissue and bone followed by skin grafting has been necessary for cure of these organisms. We would agree with surgical debridement in patients with evidence of tissue necrosis or extensive disease, but we have rarely had to resort to skin grafting for the M. smegmatis group or other RGM.
Lung infections caused by M. abscessus have remained difficult to treat and incurable with currently available drugs. Patients with localized unilateral disease, who were surgical candidates, were treated with amikacin and cefoxitin before surgical resection was performed. Unfortunately, most patients with M. abscessus lung disease have bilateral disease and are not surgical candidates.
CLINICAL FEATURES OF INFECTIONS
Posttraumatic Wound Infections
The M. fortuitum group and the M. chelonae-abscessus group are most commonly associated with skin and soft tissue infections, usually following some type of penetrating trauma. Accidental puncture wounds, especially due to stepping on nails contaminated with soil or water, are often seen in this group of infections (9, 118, 169, 188). Of the 34 RGM infections discussed by Wallace et al. (205), 12 (35%) directly involved the foot and 9 of these 12 (75%) were associated with stepping on a nail.
In a study exclusively of M. chelonae, localized cellulitis, subcutaneous abscess, or osteomyelitis usually followed similar types of traumatic injury (193). Use of corticosteroids predisposed this group to this type of infection. (Among the pathogenic RGM species, this species is the one most commonly associated with chronic corticosteroid use.)
Other species of RGM which have been associated with posttraumatic wound infection include M. wolinskyi and M. goodii (17, 199). Brown et al. (17) reported that four (50%) of eight isolates of M. wolinskyi were associated with cellulitis following a traumatic event. Two of the four patients also progressed to osteomyelitis. Brown et al. (17) reported that 7 (25%) of 28 patients had infections with M. goodii that were associated with posttraumatic cellulitis. Of the seven patients, four were thought to have complicating osteomyelitis.
For mild localized posttraumatic wound infections, monotherapy with an oral agent given for 4 to 6 months has been very successful. For extensive disease, surgical debridement combined with initial combination drug therapy based on in vitro susceptibilities followed by oral therapy to complete 6 months of treatment has also proven effective.
Bone and Joint Infection
Bone and/or joint disease is not an infrequent complication of infection with the RGM. As with bacterial disease, osteomyelitis may follow open bone fractures, puncture wounds, and hematogenous spread from another source. The most common setting is an open fracture of the femur, often followed by orthopedic correction that includes open reduction and internal fixation. The most common pathogen in this setting is M. fortuitum and the M. fortuitum third biovariant complex (proposed species M. houstonense and M. bonickei) (194; Schinsky et al., submitted).
Osteomyelitis due to M. chelonae or M. abscessus may be seen in patients receiving long-term steroid therapy. Maxson et al. (102) reported a case of osteomyelitis due to M. abscessus in a patient with systemic lupus erythematosus who had been treated with prednisone for 10 years. The patient presented with lumbar pain and an area of caseous abscess formation, and compression fractures were discovered after several incorrect diagnoses were made. Operative cultures revealed M. abscessus. Previously, Wallace et al. had reported at least two cases of multifocal osteomyelitis with disseminated M. chelonae cutaneous disease in patients receiving corticosteroids (193). In a later series involving 76 patients, Wallace reported (203) another 14 cases of osteomyelitis.
A case of vertebral osteomyelitis in a patient with a history of intravenous drug abuse was described by Sarria et al. (151) computed tomography guided aspiration yielded a pure culture of M. abscessus. Although RGM rarely cause vertebral osteomyelitis. Sarria and colleagues identified 15 cases and found clinical information on six of the cases (151). Four of the six cases of RGM vertebral osteomyelitis were due to M. fortuitum (133, 151). All of the patients had some type of underlying condition such as systemic lupus erythamatosus, chronic granulomatous disease, achalasia, back injury, mental retardation, and/or IV drug abuse.
Osteomyelitis secondary to a puncture wound is probably the second most common cause of osteomyelitis after open fractures and has been reported with M. peregrinum and M. fortuitum (35, 106, 109). Both et al. (14) reported two cases of M. fortuitum infections in patients with septic arthritis associated with joint prostheses. Both patients responded favorably to surgical drainage and removal of the prosthesis. Later, Herold et al. (71) reviewed five other cases of M. fortuitum involved in prosthetic knee and joint infections.
Brown et al. (17) recently reported that 13 (36%) of 36 patients with infection caused by two new species (M. goodii and M. wolinskyi, belonging to the M. smegmatis group) were diagnosed with osteomyelitis. These new species were more often associated with osteomyelitis than were infections involving M. smegmatis sensu stricto.
Generally, cures of osteomyelitis due to RGM have been accomplished by surgical wound debridement combined with drug therapy (based on in vitro susceptibilities of the isolate) given for a minimum of 6 months (203). Surgical debridement is of greater benefit to patients with disease due to the M. chelonae-abscessus group since drug therapy is so much more difficult.
Postsurgical Wound Infections
The RGM have been recognized for over 20 years as causative agents of sporadic nosocomial or health care-associated infections including disease that involves renal dialysis; punch biopsy surgery; augmentation mammaplasty; other forms of plastic surgery including face-lifts and liposuction; sternal wound infections following cardiac surgery; and postinjection abscesses (12, 22, 38, 57, 97, 107, 183, 198). In one series of 44 postsurgical RGM infections, 31 cases involved M. fortuitum and 13 cases involved the M. chelonae-abscessus group (203).
RGM infections following augmentation mammaplasty are well known. In a 3-year time span, Clegg et al. (38) collected 17 cases of infections with M. fortuitum group and M. chelonae-abscessus group following implantation of breast prostheses. All of these infections remained localized, and most were unilateral. In 1989, Wallace et al. (201) reported an additional 37 cases of RGM wound infections that occurred after augmentation mammaplasty. Most of these infections (70%) were caused by M. fortuitum. Successful therapy of these infections involved removal of the infected implant, 6 months of drug therapy, then reimplantation of the breast if desired. Shorter causes of therapy (e.g., 4 months) might be effective but have not been studied.
In addition, RGM are well recognized as a cause of cardiac surgical infections. These may involve the sternal wound site, the saphenous vein site, or even an inserted prosthetic valve. Of 36 isolates in one series of patients with sporadic infection, 26 were M. fortuitum and the remaining 10 were placed in one of several groups: the M. chelonae-abscessus group (4 cases), M. smegmatis group, later identified as M. wolinskyi (4 cases), and 2 cases of pigmented RGM (17, 199). Thus, two (25%) of eight isolates of the newly proposed M. wolinskyi were seen with postsurgical infections (17, 127). At least 5 (18%) of 28 other cases of the newly proposed M. goodii were associated with postsurgical infections including infected pacemakers, breast reduction surgery, and cardiac surgery (17). Additionally, one case of M. abscessus associated with an infected pacemaker was reported by Cutay et al. (41).
Treatment of sternal wound infections (osteomyelitis) has invariably included surgical debridement, an initial period of i.v. combination antibiotics that usually includes amikacin and a β-lactam (cefoxitin or imipenem), and subsequent oral therapy (when possible) to complete at least 6 months of therapy (196, 203). Patients with M. abscessus sternal wound infections often undergo removal of the sternum (sternectomy) because drug therapy for this species is so much more difficult than for the M. fortuitum group or the M. smegmatis group. Treatment of prosthetic valve endocarditis requires removal of the infected valve.
RGM infections in hemodialysis patients have also been recognized. Bolan et al. (12) reported on 27 of 140 patients infected with RGM in a center in Louisiana. Three had soft tissue infections, 9 had disseminated disease, 1 had an access-graft infection, and 14 had bacteremia alone. Of 26 identified isolates, 25 (96%), were M. abscessus, and 1 (4%) was identified as M. mucogenicum. Environmental cultures of the water system showed heavy contamination with RGM. Later, Lowry et al. (97), described five patients with M. abscessus who were dialyzed at an outpatient clinic in California. Four of the five patients had arteriovenous graft infections, and two of them died during antimicrobial therapy. These authors stressed the need for complete graft removal to ensure total recovery. These studies antedated the availability of many of the newer antibiotics, so optimal drug therapy for these types of infection is not known.
Postinjection Abscesses
RGM also have been associated with a large number of outbreaks of postinjection abscesses. Two of the largest case studies recently were published. The first large outbreak (183) in Baranquilla, Colombia, involved the isolation of M. abscessus subsequent to local injections of lidocaine in a physician's office. Over a 5-month period, 350 of about 2,000 patients (18%) developed localized abscesses or cellulitis. Therapy with a combination of surgical excision and 3 to 6 months of clarithromycin administration was successful in 95% of the treated patients. Fewer than one-third of the patients who received either surgical therapy alone or clarithromycin alone were cured. An identical outcome was also seen in an earlier single case of M. chelonae infection following excision and closure surgery for a basal cell carcinoma (147,183). Galil et al. (57) reported the largest outbreak of M. abscessus infection in the United States, which followed injections of ACE. From 1995 to 1996, 87 persons were identified with postinjection abscesses attributable to M. abscessus from contaminated ACE.
Similar to the Colombia outbreak, surgical excision or drainage combined with clarithromycin therapy for 3 to 6 months appeared to be the optimal therapy for M. chelonae or M. abscessus postinjection abscesses.
Catheter-Related Infections
Catheter-related infections are a relatively common form of RGM disease and the most common form of health care-associated disease. The most common mycobacterial pathogen is M. fortuitum. However, M. chelonae, M. abscessus, M. immunogenum, the M. smegmatis group, M. peregrinum, M. mucogenicum, and some pigmented species have also been associated with catheter-related infections (17, 196, 199, 200, 203, 205, 212). Most clinically significant cases of M. mucogenicum disease have involved catheter-related infections (200).
The organisms may produce exit site infections, tunnel infections (where the catheter is tunneled under the skin), and/or bacteremia. Rarely, hematogenous dissemination to sites such as the lungs or liver (granulomatous hepatitis) also occurs (16, 61). In a large study by Wallace et al. (193) of M. chelonae infections, 12 of 100 cases (12%) were catheter related. Immunosuppressive disorders such as transplantation, leukemia, and corticosteroid therapy are most commonly associated with RGM catheter-related infections (94, 134). Most commonly, long-term central indwelling catheters (51, 76) such as Hickman or Broviac catheters are incriminated (1, 37, 94, 134).
Examples of these infections are provided by Burns et al. (26), who described two cases of RGM catheter infection. The first was a 5-year-old boy with end-stage renal disease who had received peritoneal dialysis for 4 years. M. chelonae was cultured from a peritoneal fluid aspirate. The peritoneal dialysis catheter was removed, and treatment with amikacin, cefoxitin, and clarithromycin was instituted. After 3 weeks, cefoxitin and clarithromycin administration was stopped and the child was treated successfully for 3 months with intravenous amikacin. The second case involved a 7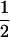 -year-old girl with a congenital defect in her gastrointestinal system that had required total parenteral nutrition since birth. She had utilized multiple subclavian Hickman catheters over the years. She ultimately developed a tunnel site infection associated with one of the catheters. The Hickman catheter was removed with implantation of a Broviac catheter but with no resolution of symptoms. Cultures made from the catheter site grew out M. chelonae. The infected tissue in the chest wall was excised down to the muscle. A new central line was placed using a new tunnel and exit site. The patient recovered after the debridement and administration of multiple antibiotics over 6 months, including ciprofloxacin, amikacin, and clarithromycin for 2 months followed by an additional 4 months of clarithromycin therapy.
-year-old girl with a congenital defect in her gastrointestinal system that had required total parenteral nutrition since birth. She had utilized multiple subclavian Hickman catheters over the years. She ultimately developed a tunnel site infection associated with one of the catheters. The Hickman catheter was removed with implantation of a Broviac catheter but with no resolution of symptoms. Cultures made from the catheter site grew out M. chelonae. The infected tissue in the chest wall was excised down to the muscle. A new central line was placed using a new tunnel and exit site. The patient recovered after the debridement and administration of multiple antibiotics over 6 months, including ciprofloxacin, amikacin, and clarithromycin for 2 months followed by an additional 4 months of clarithromycin therapy.
Successful therapy of these catheter-related infections involves removal of the catheter and antimicrobial therapy, usually for 2 to 4 months. Although disease due to M. fortuitum may resolve if the catheter is removed, reinsertion of another catheter in a similar location without drug therapy usually results in disease recurrence (as in the above case).
Disseminated Cutaneous Disease
Disseminated cutaneous disease associated with RGM is unusual but serious. More than 90% of patients with disseminated cutaneous disease have identified risk factors such as chronic renal failure, renal transplantation, and, especially, a history of chronic corticosteroid therapy (185, 206). Only rarely is the patient with disseminated cutaneous disease due to RGM also human immunodeficiency virus positive (193). Most documented cases of RGM disseminated cutaneous disease have been associated with the M. abscessus-chelonae group. In a series of 100 nonpulmonary isolates of M. chelonae, more than 50% were associated with disseminated cutaneous disease (193). Similar data for M. fortuitum and M. abscessus are much more limited. Of nine cases reported by Wallace et al. (205), six (67%) were caused by M. abscessus, two (22%) were caused by M. fortuitum, and one (11%) was caused by M. chelonae. Of these nine patients with disseminated RGM disease, four had multiple, recurrent episodes of chronic skin and soft tissue abscesses, usually involving the upper extremities (173, 205), but did not appear systemically ill. The other five patients were acutely ill, and organisms were recovered from multiple sources including blood. Only two of the nine patients were not immunosuppressed, and one died (205). Later, it was noted that the development of clarithromycin mutational resistance with clinical relapses of M. chelonae-abscessus was rare except in the setting of disseminated disease (191). Tebas et al. (173) reported a case of clarithromycin resistance leading to multiple relapses in a patient with disseminated disease with M. chelonae following heart transplantation.
An unusual group of 16 cases of M. chelonae-abscessus disseminated disease that included chronic cervical lymphadenitis were reported from a university hospital in northeastern Thailand. Most patients also had serious infections with pathogens other than the RGM (36). The duration of symptoms in the 16 patients persisted from 1 month to 5 years, with a mean of 15.6 months, before diagnosis of the RGM was established. There were 12 patients who had other multiple organ systems involved including sinuses (6 patients), lungs (4 patients), liver (4 patients), spleen (3 patients), skin (3 patients), bone and joint (2 patients), and tonsils (2 patients). Eleven patients had reactive skin disease based on pathology (36).
In the 1992 series (193) of 53 patients with disseminated skin disease due to M. chelonae, most patients had multiple nodular, subcutaneous, draining lesions. Evidence of spread to other parts of the body was rare. Only three patients had positive blood cultures; no patients had any associated systemic disease. Unlike post-traumatic wound infections, in which a portal of entry for the infection is found, no source was evident in the patients. In addition, osteomyelitis was rare in this group, with only two patients having multifocal osteomyelitis of the extremities and draining lesions secondary to primary bone involvement.
Disseminated disease with M. fortuitum, unlike the M. chelonae-abscessus group, is rare but does occur. An unusual case of disseminated M. fortuitum in a 76-year-old male with chronic lung disease, in which the RGM infection began in the lungs and spread to the bone and skin, was described by Burns et al. (24). Additionally, Horsburg and Selik (75) reported five cases of disseminated M. fortuitum infection in AIDS patients. No further details were documented. Until 1979, only 11 cases of disseminated M. fortuitum disease were documented, and exact taxonomy of the RGM was not very good at that time, making it likely that some of these isolates may have actually been M. chelonae-abscessus (213).
Treatment of disseminated cutaneous disease involves drainage of abscesses and, for the M. chelonae-abscessus group, the use of clarithromycin for at least 6 months. Wallace et al. (206), studied 14 patients with cutaneous infections due to M. chelonae, most of which were disseminated. Of 11 patients who completed therapy, all had complete resolution with no relapses within 6 months of clarithromycin monotherapy. Because of the risk of the development of clarithromycin resistance with monotherapy of disseminated disease (estimated to be about 10 to 20% in this setting), initial therapy for the first 3 to 6 weeks should include other drugs based on in vitro susceptibilities whenever possible.
Pulmonary Disease
Approximately 80% of the chronic pulmonary disease caused by the RGM is due to M. abscessus (63, 185). Although some heterogeneity occurs, the typical patient with M. abscessus lung disease is a nonsmoking female in her 60s, with disease symptoms that have existed for at least 2 years. Many patients have a diagnosis of bronchiectasis with chronic lung disease (185). The symptoms most frequently observed are cough and chronic fatigue, although sputum production, hemoptysis (coughing up blood), and weight loss also occur. As mentioned above, the cases are seen clustered in the southeast coastal United States (63). CF patients comprise about 10 to 15% of patients with M. abscessus lung disease (6). The CF patient is at risk for developing infections due to M. abscessus and M. avium complex because of existing obstructive airway disease and underlying bronchiectasis (48). They are most often identified because of episodic fever that is poorly controlled with anti-pseudomonal therapy.
Treatment of M. abscessus lung disease remains a disappointment, since the use of amikacin, cefoxitin, and clarithromycin often produces clinical improvement but is insufficient to cure the disease. There is a desperate need for newer and better oral drugs. Linezolid offers potential to some patients, but the MICs are relatively high and there is little clinical experience. The new glycycycline, tigecycline, offers exciting potential, but is not yet FDA approved and is an i.v. drug.
The remaining 20% of cases of lung infection not due to M. abscessus are due to the less commonly encountered M. smegmatis group and the M. fortuitum group. Of 28 cases due to the newly reported species M. goodii, 6 (21%) were pulmonary disease (17). M. fortuitum is also associated with pulmonary disease, with approximately 50% of cases associated with esophageal achalasia (a disease of severe dilation and loss of function of the esophagus), lipoid pneumonia, and diseases with chronic vomiting (182) and aspiration (107, 205). Rare cases of lung abscess and infections in CF patients as a result of M. fortuitum have also been seen, although accurate species identification for this species has been readily available only in recent years (47, 83, 120, 180). Although M. fortuitum may be isolated frequently from respiratory specimens, it is usually a single positive culture and, compared to M. abscessus, only rarely is a significant pathogen (3, 139).
Treatment of M. fortuitum lung disease with such antimicrobials as the quinolones, sulfonamides, doxycycline, amikacin, and cefoxitin has usually been successful (64, 78, 120, 180, 220).
Central Nervous System Disease
Central nervous system disease involving RGM is rare but serious. Most cases have been associated with M. fortuitum (32). The first report of central nervous system infection with M. fortuitum was in 1970. An 8-year-old boy was thought to have “aseptic meningitis” after routine cerebrospinal fluid (CSF) cultures were negative. However, after repeated spinal taps with negative cultures, surgery was performed and a vertebral abscess was discovered and drained. Cultures subsequently grew M. fortuitum. The patient was treated and recovered (68). Dalovisio et al. (43) described a case of M. fortuitum meningitis in a child with a history of chronic otitis media who subsequently developed an obstructive hydrocephalus and had a ventriculoperitoneal shunt placed. His condition improved when amikacin injections were administered every 48 h directly into the ventricles through the shunt. Cultures of spinal fluid at 8 months were negative for M. fortuitum. The patient's illness resolved except for residual decreased hearing. Another case involved infection of a ventriculoatrial shunt inserted for spontaneous cerebral hematoma with obstructive hydrocephalus in a 60-year-old woman. After placement of the shunt, the patient had persistent fever and her CSF grew M. fortuitum. The patient's condition continued to deteriorate, in spite of therapy with amikacin and ofloxacin, until the shunt was removed (35). A study from India reviewed 50 cases of brain abscesses in which one specimen grew M. fortuitum (91). Details of this case were not given.
Another isolation of M. fortuitum from spinal fluid occurred in a teenage boy who had been in a motor vehicle accident. During the accident, he sustained a wound in the sacral region, which harbored a foreign body. The patient ultimately responded to trimethoprim-sulfamethoxazole and surgical drainage (150). A case of M. fortuitum meningitis was reported in a patient with AIDS. M. fortuitum was recovered from skin biopsy specimens, bone marrow, and CSF. Unfortunately, therapy with amikacin and doxycycline was not successful, and the patient died (157).
The only other RGM which have been reported to cause meningitis are M. mucogenicum and M. goodii. In a series of 20 cases of M. mucogenicum, 1 patient with AIDS had multiple CSF samples that were positive on smear and culture for M. mucogenicum. No other details were given (200). Similarly, M. goodii was recovered from the CSF of one patient but was of unknown clinical significance (17).
Finally, Flor et al. (50) recently reviewed the literature for cases of nontuberculous mycobacteria associated with meningitis and found that 12% of 52 isolates were identified as M. fortuitum.
Treatment of CNS infections due to RGM has been difficult and often requires prolonged therapy (6 to 12 months) with multiple drugs.
Miscellaneous Diseases
Various other diseases have been associated with the RGM. A few of these unusual situations are discussed here.
Otitis media.
Lowry et al. (98) reported an outbreak of 17 cases of otitis media caused by M. abscessus in two ear-nose-throat clinics. It is not clear if the index patient from the first clinic “spread” the strain into the other clinic and contaminated the equipment in the second clinic or if the strain was already present. However, all but 1 of the 14 isolates obtained from the 17 patients were aminoglycoside resistant, as were isolates obtained from the suction tubing and suction-sink water in both clinics. The resistance was secondary to a mutation in the 16S rRNA gene, which implies that the original outbreak isolate was from a patient with chronic otitis rather than from the environment. In addition to surgery, all patients who had tympanostomy tubes had them removed and received erythromycin (premacrolide era). Of the 14 patients, 13 had clinical resolution of drainage (98). Franklin et al. (53) later detailed clinical information on 21 sporadic cases of chronic otitis media with local drainage caused by RGM, which followed ear tube placement. Some of these patients also had mastoiditis. Of these, 20 of 21 cases (95%) were caused by M. abscessus. Therapy included surgical debridement, removal of the tympanostomy tubes, and antibiotic therapy. Initial therapy included amikacin (if susceptible) and either cefoxitin or imipenem for 3 to 6 weeks, followed by long-term (6 months) oral therapy with erythromycin (pre-newer macrolide era) or clarithromycin (53). Approximately 50% of the isolates from these isolates were also aminoglycoside resistant secondary to the use of chronic aminoglycoside ear drops.
Only a few cases of M. fortuitum chronic otitis media or mastoiditis have been reported (2, 43, 131). The earliest report of otitis media due to M. fortuitum was described by Austin and Lockey (2). A 63-year-old man with mastoiditis also had a subperiosteal abscess at the surgical site from which M. fortuitum was cultured.
Dalovisio et al. (43) reported finding M. fortuitum in a 10-year-old boy with chronic recurrent otitis media and chronic mastoiditis, who also had a subdural empyema and meningitis. The patient was treated with amikacin for 4 months and underwent drainage procedures and placement of ventriculoperitoneal shunt before the infection resolved.
Plemmons et al. (131) reported a case of persistent otitis media in a 14-year-old girl who underwent right-side mastoidectomy. Prior to surgery, she had purulent otorrhea unresponsive to topical and systemic antibiotics and steroids. During surgery, a specimen was obtained from which M. fortuitum was grown. Cultures of fluid draining from the ear after surgery also grew M. fortuitum. She was given clarithromycin, and later, since the site was still draining, oral trimethoprim-sulfamethoxazole was added. The otitis media resolved and her mastoidectomy site completely healed after a 12-month course of antibiotics.
Corneal infections (keratitis).
Ocular infections due to RGM have become more prevalent over the last 20 years. A comment must be made that before 1978, all of the cases reported were identified as “M. fortuitum.” This probably reflects the inadequacies in taxonomy that existed at that time. Often in pre-1978 literature, “M. fortuitum” was used to designate M. fortuitum complex, and most publications did not give sufficient data to classify the organisms to species (as M. fortuitum, M. chelonae, or M. abscessus). We have attempted here to ascertain species (i.e., M. abscessus versus M. chelonae) when possible. This often was evident if susceptibility patterns of the organisms were reported.
Before 1978, when Gangadharam et al. (58), reported a case of ulcerative keratitis caused by M. chelonae, the few prior reports had involved only M. fortuitum (154). Gangadharam et al. detailed the ophthalmological, bacteriological, and histopathological examinations of a puncture wound in a patient who was misdiagnosed as having herpes simplex keratitis. The patient received unsuccessful treatment for 2 months with topical, subconjunctival, and systemic antibiotics including gentamicin, kanamycin, and erythromycin. The ulcer regressed only after keratoplasty. After the Gangadharam et al. report, several other cases of keratitis caused by “M. chelonae” were noted (43, 82, 140, 197). In most cases, it was still unclear whether the authors actually differentiated between M. chelonae and M. abscessus. Several reports of keratitis due to M. abscessus were described (65, 105, 132, 142, 158), and the organism was identified to species. Other authors only identified their isolates as M. chelonae (or the former term M. chelonei), but from the text (susceptibility patterns), we were able to differentiate the organisms as M. abscessus (23, 103, 108). Additionally, Bullington et al. (23) did an extensive review of the literature involving keratitis with nontuberculous mycobacteria from 1965 to 1992. He found that 21 of 38 (55%) isolates were M. fortuitum, 16 of 38 (42%) were M. chelonae-abscessus group, and 1 (2%) was identified only as group IV mycobacteria (92).
The most widely used antimicrobial treatment in the patients with keratitis in this review involved the use of topical aminoglycoside and/or systemic amikacin, gentamicin, kanamycin, or neomycin. Almost half of the patients required some type of surgical intervention including keratoplasty, keratoectomy, corneal graft, and several surgical debridement procedures (23).
Since the review by Bullington et al. (23), a few additional cases of keratitis due to RGM have been recorded. Recently, an interesting case concerned a soft-contact-lens wearer with a corneal infiltrate. The patient had received oral steroids, systemic antibiotics including ampicillin and co-trimoxazole, and also topical chloromycetin, tobramycin, and miconazole. Subsequent corneal scrapings revealed M. chelonae. The isolate was initially misidentified as Nocardia (161) and treated with ciprofloxacin, erythromycin, and fusidic acid. About 6 months later, the patient experienced a recurrence of the infiltrate, which initially improved with amikacin eye drops and systemic erythromycin but subsequently recurred and necessitated keratoplasty. Cultures of the excised cornea were still positive for M. chelonae. The patient was treated again with amikacin, erythromycin, and co-trimoxazole for 2 months. Eighteen months later, the graft had cleared and no evidence of infection remained (82). This patient's case, as others, was presumably complicated by the use of steroids (136).
Since the early 1990s, other reports of eye disease due to RGM have been recorded, including postkeratoplasty and following laser (LASIK) surgery for correction of myopia (136, 170). Medical therapy often is ineffective, due to delayed or incorrect diagnosis, difficulty in drug penetration (the cornea has no blood vessels and antibiotic entrance is only by surface diffusion), resistance to conventional antibiotics, and, rarely, emergence of drug resistant strains. Often, lamellar keratoectomy or keratoplasty (removal of the cornea) is the suggested treatment for patients with nonresponsive disease.
These studies illustrate that, although the RGM are unusual causes of keratitis, infection can be severe and the outcome is often unfavorable.
Cervical lymphadenitis.
The most common mycobacterial cause of lymphadenitis is M. tuberculosis in adults and M. avium complex in children. However, other mycobacteria including M. fortuitum, have been reported to cause lymphadenitis. Most RGM disease has involved the cervical lymph nodes, and has followed a dental procedure. Although M. fortuitum is a rare cause of lymphadenitis in adults or children, at least 19 cases have been reported in the literature.
Cervical adenitis caused by M. fortuitum has been reported in patients with AIDS (148). Butt (28) described two cases of lymphadenitis in patients with AIDS who were successfully treated with initial surgical drainage and antibiotics. Details were also extracted from seven other patients with of lymphadenitis caused by M. fortuitum, including one patient with generalized lymphadenopathy, two with bilateral involvement, and one each with unilateral submandibular nodes, submental lymph nodes, supraclavicular lymph nodes and cervical lymph nodes (28). Three of the nine patients listed (33%) had a history of dental procedure or extraction up to 6 months pre-dating the diagnosis of M. fortuitum. Four of the nine patients (44%) were treated with surgical incision and drainage followed by administration of combination antibiotics. Only two of the nine patients (22%) were treated with excision alone. Six of the nine patients (67%) improved or resolved their symptoms, and three (33%) died, with two of the three deaths being unrelated to the infection. At autopsy, however, disseminated disease with M. fortuitum was discovered in the third patient (28).
Generally, it is accepted that abscess formation in lymphadenitis due to M. fortuitum is best treated by incision and drainage followed by combination antibiotics that includes amikacin (initially) and concludes with one or more oral antibiotics based on in vitro susceptibilities. Therapy should continue for at least 6 months (28, 196).
LABORATORY ASPECTS OF THE RAPIDLY GROWING MYCOBACTERIA
General
Traditional clinical laboratory identification of the RGM involved relatively few tests and was based mainly on growth rate, selected biochemical tests, pigmentation, and colony morphology. The RGM are defined as mycobacteria that grow within 7 days (most within 3 to 4 days) (208, 209). Standard biochemical tests include iron uptake, nitrate reductase activity, tolerance to 5% NaCl, and arylsulfatase reaction. All members of the M. fortuitum group and the M. chelonae-abscessus group exhibit strong arylsulfatase activity at 3 days (208). Members of the M. smegmatis group are similar in growth rate but do not exhibit arylsulfatase activity at 3 days. The latter group is the only one of the three that produces pigmentation (17). Approximately 95% of isolates of M. smegmatis sensu stricto and 78% of isolates of M. goodii develop yellow-orange pigmentation after prolonged (7 to 10 days) incubation on Middlebrook 7H10 agar (17). (Because the pigmentation is so late and often occurs only on select media, it is often missed. It should always be sought in RGM with a negative arylsulfatase that are not obviously pigmented.)
These relatively uncomplicated methods have proved inadequate for recognition of some older species (e.g., M. peregrinum) and many of the newer species (e.g., M. mucogenicum, M. immunogenum, M. goodii, and members of the M. fortuitum third biovariant complex such as the proposed species M. houstonense). Carbohydrate utilization tests and molecular studies have enabled a more accurate laboratory identification of the RGM species and groups. Unfortunately, in most clinical laboratories and many reference laboratories, identification of RGM to the species level has been relegated to low priority. Isolates often are identified to the group level only (e.g., M. chelonae-abscessus group, M. fortuitum-smegmatis group). However, it is no longer acceptable among good clinical and reference laboratories to fail to identify disease- producing RGM isolates to the species level, especially the separation of M. chelonae from M. abscessus.
Types of clinical disease and antimicrobial susceptibilities often differ for individual species of RGM. The most common taxonomic error is the failure to separate the two distinct species, M. chelonae and M. abscessus. Although once thought to be subspecies within the species M. chelonae, they are, in fact, two distinct species. M. chelonae (formerly M. chelonae subspecies chelonae) is most often associated with disseminated skin and soft tissue infections with multiple painful draining lesions in immunosuppressed persons and is rarely a cause of chronic lung disease. Although M. abscessus is a cause of skin and soft tissue infections in patients (some of whom are immunosuppressed), it is also responsible for more than 80% of the chronic lung disease caused by RGM (63, 64, 205). The aminoglycoside preferred for treatment of M. chelonae is tobramycin, while amikacin is the preferred aminoglycoside for M. abscessus. Resistance to cefoxitin is one of the key differences between these two species, since M. chelonae is highly resistant (MIC, ≥256 μg/ml) while M. abscessus is intermediately susceptible, with modal cafoxitin MICs of 32 μg/ml. M. chelonae appears to be much more susceptible to the newer antimicrobials linezolid and gatifloxacin than is M. abscessus (see the discussions of the two species above). Thus, the two species differ in clinical disease presentation as well as in susceptibility to drugs and in optimal therapeutic regimens, hence the importance of separating the two species in the laboratory.
Implementation of additional laboratory methods including carbohydrate utilization and molecular diagnostics are now required to enable accurate species identification of the nonpigmented and late-pigmenting RGM (Tables 3 and 4). Although highly accurate at identifying slowly growing nontuberculous mycobacteria to species, HPLC has proven to be ineffective in identifying these organisms to the species level.
TABLE 3.
Currently recognized taxa or species of nonpigmented (or late-pigmenting) RGM
| Taxon or species |
|---|
| Common human pathogens |
| M. chelonae |
| M. abscessus |
| M. fortuitum |
| Infrequent but proven human pathogens |
| M. fortuitum third biovariant complex |
| Sorbitol positive |
| M. mageritense |
| M. houstonense (proposed species) |
| Sorbitol negative |
| M. septicum |
| M. bonickei (proposed species) |
| M. mucogenicum |
| M. immunogenum |
| M. smegmatis group |
| M. smegmatis (sensu stricto)a |
| M. goodiia |
| M. wolinskyi |
| M. peregrinum |
| Type 1 (pipemidic acid susceptible) |
| Type 2 (pipemidic acid resistant) |
| Unproven human pathogens |
| M. agri |
| M. chitae |
| M. senegalense |
| M. porcinum |
Some strains produce a late pigment.
TABLE 4.
Laboratory phenotypic features of the 12 most clinically important species of nonpigmented or late-pigmenting RGMa
| Species or complex | Prior designations | Pigment | 3-Day aryl- sulfatase | Nitrate reduction | Iron uptake | Utilization of:
|
5% NaCl | Unique PRA (hsp65) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mannitol | Inositol | Citrate | Sorbitol | ||||||||
| M. chelonae-abscessus group | |||||||||||
| M. abscessus | M. chelonae subsp. abscessus | − | + | − | − | − | − | − | − | + | + |
| M. chelonae | M. borstelense, M. chelonei, M. chelonae subsp. chelonae | − | + | − | − | − | − | + | − | − | + |
| M. immunogenum | M. immunogen | − | + | − | − | − | − | − | − | − | + |
| M. fortuitum group | |||||||||||
| M. fortuitum | M. ranae, M. fortuitum biovar fortuitum | − | + | + | + | − | − | − | − | + | + |
| M. peregrinum (type 1) | M. fortuitum biovar peregrinum (pipemidic acid susceptible) | − | + | + | + | + | − | − | − | + | −c |
| M. peregrinum (type 2) | M. fortuitum biovar peregrinum (pipemidic acid resistant) | − | + | + | + | + | − | − | − | + | −c |
| M. fortuitum third biovariant complex | |||||||||||
| M. houstonense (proposed) | M. fortuitum third biovar sorbitol positive | − | + | + | + | + | + | − | + | + | −c |
| M. bonickei (proposed) | M. fortuitum third biovar sorbitol negative | − | + | + | + | + | + | − | − | + | −c |
| M. mucogenicum | MCLO | − | + | ± | −b | + | − | + | − | − | + |
| M. smegmatis group | |||||||||||
| M. smegmatis sensu stricto | M. smegmatis | ± | − | + | + | + | + | ± | + | + | + |
| M. wolinskyi | M. smegmatis | − | − | + | + | + | + | ± | + | + | + |
| M. goodii | M. smegmatis | ± | − | + | + | + | + | ± | + | + | + |
Modified from reference 188. Symbols: ±, variable or late; +, ≥90%; −, ≤10%; ±, 11 to 89%.
Tan appearance.
M. peregrinum (type 1) has the same PRA pattern as the proposed M. bonickei (M. fortuitum third biovariant, sorbitol negative), whereas M. peregrinum (type 2) has the same PRA pattern as the proposed M. houstonense (M. fortuitum third biovariant, sorbitol positive). Biochemical testing is necessary for differentiation of these species and taxa.
Biochemical and Phenotypic Identification
After establishing a clinical isolate as an RGM, the best combination of traditional tests for recognition of the most commonly encountered species include the 3-day arylsulfatase test, iron uptake, nitrate reductase, and utilization of the carbohydrates mannitol, inositol, and citrate (Table 4). A number of additional nonmolecular tests have also been utilized. A disk diffusion test using polymyxin B can also distinguish between the M. fortuitum group and the M. chelonae-abscessus group. Isolates of the M. fortuitum group exhibit a partial or complete zone of growth inhibition of 10 mm or greater around the polymyxin disk, whereas isolates of the M. chelonae-abscessus group show no partial or complete zone of inhibition (204). A previous IWGMT study (86) showed that growth in 5% NaCl could reliably differentiate strains of M. abscessus (100% positive) from M. chelonae (17% positive). The citrate test was also found to be another useful biochemical test in that approximately 80% of M. abscessus isolates were citrate negative and 100% of M. chelonae isolates were citrate positive (86). (In our hands, the citrate utilization test has proven highly reliable.) Additionally, of the M. fortuitum group, only the unnamed third biovariant complex is positive for inositol. Utilizing molecular methods as the standard of identification, positive citrate tests with M. abscessus are rare (219).
Many of the published studies of biochemical testing of RGM have utilized in-house prepared media or tests. There have been few studies of currently available commercial test systems to see if they are equivalent to these other methods. Knowing this, commercial systems could be used but require careful in-house validation. For example, Conville and Witebsky noted problems with such systems in identifying isolates of M. mucogenicum (38).
High-Performance Liquid Chromatography
HPLC of mycobacterial cell wall mycolic acids also is used routinely in many reference laboratories as a means of identifying isolates of RGM (27, 175). Recently, a comparison of the HPLC patterns obtained from the pathogenic members of the RGM was performed by Jost et al. (Abstr. 99th Gen. Meet. Am. Soc. Microbiol.). A standardized modified fluorescence detection (FL-HPLC) method was used for the analysis. FL-HPLC and UV detection methods (UV-HPLC) were analogous, and the study concluded that only under standardized conditions of culture medium, incubation time, and temperatures could “most” isolates and “most” species of the RGM be differentiated to species level by either method. The use of standardized methods is stressed by Chiu et al. (S. H. Chiu, K. C. Jost, Jr., D. F. Dunbar, and L. B. Elliott, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. U-76, 1998), because differences in growth conditions (e.g., medium, temperature, and harvest time) can cause a diversity of patterns of the mycolic acid peaks and present difficulties in species identification (27; Chiu, et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998).
Thus, under nonstandardized conditions, even group identification of the RGM by HPLC is clearly problematic, in contrast to the general ease by which this method is able to differentiate the slowly growing mycobacteria into species. By routine HPLC and FL-HPLC, most isolates of M. chelonae and M. abscessus cannot be differentiated from one another. The new species M. immunogenum is also not separable by routine HPLC from the other two species in the M. chelonae-abscessus group (212). Similarly, members of the M. fortuitum group including M. fortuitum, M. peregrinum, and the unnamed third biovariant complex are not separable from each other or from the M. smegmatis group. They are generally grouped as the M. fortuitum-smegmatis group. Although the three species of the M. smegmatis group can be separated from each other and from M. fortuitum (17), the pattern overlap of all the M. fortuitum group members is too great to identify an unknown isolate to species. Thus, current studies show that HPLC is an acceptable method for separation of isolates of the M. fortuitum-smegmatis group from those of the M. chelonae-abscessus group but cannot identify isolates accurately to species. HPLC also works well when comparing a new taxon with already established one, since minor differences between the taxa may be readily apparent. It can be helpful for identification of members of the RGM only when used in conjunction with other methods and when used under specifically standardized testing conditions. Thus, identification of the RGM based solely on HPLC is not adequate.
Molecular Identification
Nucleic acid probes.
No commercial DNA or RNA probes are currently available in the United States for any of the RGM. A kit (INNO-LIPA Mycobacteria; Innogenetics, Ghent, Belgium) based on reverse hybridization, in which the mycobacterial 16S-23S internal transcribed spacer region is amplified by PCR and amplicons are subsequently hybridized with probes for several species of RGM, is currently available in Europe but not in the United States (141). However, molecular methods for the identification of mycobacteria have been evolving rapidly and are now used in some specialized reference laboratories. Hybridization techniques with species-specific nucleotide probes, PRA, or direct sequencing of PCR-amplified products based on the polymorphism of the 16S rRNA gene have been useful in the identification of slowly growing mycobacterial species. However, because of the low level of variability within the 16S rRNA gene between some RGM species (e.g., M. chelonae and M. abscessus differ by only 4 bp in the entire 16S gene and have an identical hypervariable region A), a more variable gene sequence such as the hsp65 gene has proven helpful to distinguish between closely related species such as M. chelonae and M. abscessus (84, 89).
PCR-restriction enzyme analysis.
Several investigators have evaluated the hsp65 gene, present in all mycobacteria, for its value in the identification of RGM (45). Ringuet et al. (138) found a less than 2% difference between the three most common RGM pathogenic species (M. abscessus, M. chelonae, and M. fortuitum) when the base sequences of the three type strains were studied. The base pair diversity was still much greater compared to the 16S rRNA gene. For example, the M. chelonae and M. abscessus sequences differ by almost 30 nucleotides, whereas their 16S rRNA genes differ by only 4 nucleotides. Thus, the hsp gene sequence is much more advantageous for the accurate identification of these two species than is 16S rRNA gene sequencing. The hsp65 sequences are highly conserved within a species and thus can be used for taxonomic studies.
Telenti et al. (174) demonstrated that a 439-bp portion of the hsp65 gene could be used for PRA and showed the patterns for most slowly growing mycobacteria and selected RGM. Steingrube et al. (165) provided the most detailed PRA study to date of the RGM. They reported the PRA patterns from the 439-bp Telenti segment of the hsp65 gene for 129 clinical and reference strains of RGM belonging to 10 taxonomic groups. The authors found that among 24 endonucleases evaluated, PRA patterns produced by HaeIII and BstEII gave the best separation. More than half of the RGM were differentiated using HaeIII digestion alone. Single unique patterns were observed using both HaeIII and BstEII for M. fortuitum, M. smegmatis, M. mucogenicum, the sorbitol-negative third biovariant of M. fortuitum (100%), M. abscessus (96%), and M. chelonae (94%). Using another restriction endonuclease, AciI, RFLP patterns among clinical isolates of the M. smegmatis group supported the presence of the recently named two new species within the M. smegmatis group (M. goodii and M. wolinskyi) (17). Currently, the Telenti fragment of the hsp65 gene is the most widely used sequence for PCR-based identification of the RGM and is highly accurate for the M. fortuitum group, the M. chelonae-abscessus group, and the M. smegmatis group. It has not been studied for the identification of pigmented RGM.
Vaneechoutte et al. (181) devised another system of enzymatic amplification and restriction analysis using the entire 16S rRNA gene sequence. They studied 18 different species of Mycobacterium including strains of M. fortuitum and M. chelonae. They used different restriction enzymes (CfoI, MboI, and RsaI) from Telenti et al. (174) and Steingrube et al. (165) (both of whom used BstEII and HaeIII) and called their method ARDRA (amplified rDNA restriction analysis).
Other target sequences have been studied for the identification of mycobacteria by using PRA or sequencing. These include the 32-kDa protein gene (159), the internal transcribed spacer of the 16S-23S rRNA gene (141), the superoxide dismutase gene (226), and the DNA J gene (172). However, to date, only selected slowly growing species have been extensively studied using these gene sequences and only one or two isolates of RGM have been tested. Preliminary data suggest that for the RGM, most of these gene sequences are much more variable and perhaps less useful for species identification than is the hsp65 gene.
PRA seems particularly useful for identifying clinical isolates which gave equivocal results between compared species when other identification techniques were used. Although computerized analysis of PRA patterns has been recommended, visual inspection of the profiles is satisfactory when appropriate or comparative control strains are used (45).
Recently, a related method for the identification of mycobacteria (including nontuberculous mycobacteria and M. tuberculosis), using amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers, was described (122). A total of 87 isolates of nontuberculous mycobacteria from 10 species, including M. fortuitum, M. chelonae, and M. abscessus, were evaluated and identified using this system. The authors (122) commented that the spacer sequences of the slowly growing mycobacteria are approximately 75 nucleotides shorter than those of RGM. Detailed studies of multiple strains of each RGM were not done and, to date, have been done only with the Telenti et al. 439-bp fragment of the hsp65 gene sequence (165).
Plasmid profiles.
One of the earliest molecular tools used to help differentiate RGM was plasmid profiling. DNA probing with a plasmid-associated probe has also been performed and found to be a potentially useful guide for comparison of strains of RGM (29, 198, 221). Genetic relatedness of plasmids also may be studied further by RFLP of the plasmid or hybridization with defined or repetitive sequences. However, because these methods focus on extrachromosomal DNA, they do not provide conclusive evidence that strains are related. In fact, isolates having similar plasmid profiles may belong to different biovariant groups and vice versa (221). Thus, the use of plasmid profiles for species identification is currently of limited value, since some isolates of RGM do not contain detectable plasmids, plasmid profiles may change with time, and completely different plasmids may be of the same size (143, 198, 221). In general, other molecular methods that focus on chromosomal DNA have replaced this early type of analysis.
Ribotyping.
Ribotyping is another potential molecular tool for strain comparison. However, it is probably more useful in delineating taxonomic rather than epidemiological relatedness of strains. Yew et al. (221) stated two caveats to remember when applying this method. First, mutational changes can alter restriction patterns, and second, strains with small numbers of bands require restriction enzyme analysis with at least two different endonucleases. This method has been applied to only one major outbreak of RGM, and so knowledge of its usefulness is limited.
Hybridization.
Another genetic technique which has been applied to the identification of some species of RGM is DNA amplification and oligonucleotide-specific hybridization (44). The system is based on selective amplification by PCR using mycobacterial DNA primers based on genes coding for 16S rRNA. During PCR, a label (digoxigenin-11-dUTP) is incorporated with the amplicon. After amplification, the amplicon is hybridized with species-specific oligonucleotides. After specific hybridization, enzyme immunoassay is used to show the specifically bound completer and thus identify the mycobacterial isolate. De Beenhouwer et al. (44) reported that four of five M. chelonae-abscessus group strains were positive with their prepared probe using this technique and the fifth strain was positive with genus-specific probes but negative with all species-specific probes. None of the M. fortuitum strains studied hybridized with the species-specific probe. Hybridization with species-specific probes requires the development and availability of probes for every species. Thus, although this method has been useful for studying some species of mycobacteria, a detailed analysis of the RGM has been problematic. Patel et al. (123) also studied a method which specifically hybridized a 5′-fluorescein-labeled strand of DNA to a species probe and was colorimetrically detected with an antifluorescein-enzyme conjugate. The method was able to correctly identify 10 species of mycobacteria, including some species of RGM.
Direct detection in paraffin-embedded tissue.
Detection of nontuberculous mycobacteria by direct detection of nontuberculous mycobacteria in paraffin-embedded tissue by using amplified nucleic acid probes is an important advance in the diagnosis of mycobacterial disease. The technique is especially useful in clinical areas where the diagnosis is uncertain. Additionally, this technique has been advantageous when the mycobacteria fail to grow in culture because of small numbers or the fastidious nature of the organism, as well as in cases wherein mycobacterial disease was not initially considered. Moreover, an amplification assay can potentially give a result much faster (within 2 to 3 days) than can culture (up to 6 weeks) (60, 126).
PCR for detection of M. tuberculosis, from such clinical specimens as sputum, fluid aspirates, and tissue homogenates, has also been helpful in establishing a more rapid diagnosis of tuberculosis (114). Recently, human tissue samples stored as formalin-fixed, paraffin-embedded blocks have been used together with PCR methods to detect and identify the mycobacteria present (67).
Briefly, as described by Marchetti et al. (101), DNA is extracted from formalin-fixed paraffin-embedded tissues. This method was originally developed to detect M. tuberculosis but, with some modification, can also detect nontuberculous mycobacteria. Paraffin is removed by adding xylene, vortex mixing, incubating at room temperature, and centrifuging. To facilitate pelleting and hydration of samples, ethanol is added and the supernatant fluid (xylene layer) is removed from the sedimented pellet. The pellet is air dried and resuspended in a special digestion buffer. Proteinase K is inactivated, and DNA is extracted from the emulsified tissue by adding phenol, vortex mixing, and centrifuging the mixture. A nested PCR of four assays, which uses three different concentrations of DNA, is performed. When a sample yields a positive result when amplified with primers homologous to sequences shared by a variety of mycobacterial species other than M. tuberculosis, it can be identified presumptively as containing a nontuberculous mycobacterial species.
Shafran and Chui (155) described a similar method of DNA extraction from paraffin-fixed skin biopsy tissue using a modified protocol described by Telenti et al. (174) for detection of M. tuberculosis. No mention was made of detection of nontuberculous mycobacteria by this technique.
Earlier reports by Ghossein et al. (60), described a method in which amplified fragments from paraffin-embedded tissue, as well as cultures of M. tuberculosis, M. avium complex, and “saprophytic mycobacteria,” were identified by PCR of a 383-bp segment of the gene encoding the 65-kDa mycobacterial surface antigen and subsequent digestion with NarI. Unfortunately, the “saprophytic mycobacteria” were not further identified by the authors (60).
Later, Perosio and Frank (128) described a proteinase K digestion using a freeze-fracture extraction method developed by Ghossein and associates which enhances detection of mycobacterial DNA in clinical samples. Subsequently, a nested PCR with primers for the mycobacterial 65-kDa antigen gene was performed. Their primers were within a genus-specific region conserved among several nontuberculous mycobacteria including M. avium complex, M. gordonae, M. kansasii, and M. fortuitum. By comparing their primers with those used in previous studies and using available sequence data for different species, they predicted that their primers would amplify DNA from most pathogenic Mycobacterium species (128). Using PCR, mycobacterial DNA was detected in 7 of 7 wedge specimens and 9 of 18 transbronchial biopsy specimens. Restriction enzyme digestion of the amplified PCR product differentiated the species. Bascuñana and Belak (7) developed another nested PCR technique to amplify a 424-bp segment of the gene encoding the 65-kDa surface antigen of mycobacteria by using a restriction enzyme analysis procedure. The authors found that the location of the binding sites of the PCR primers in highly conserved parts of the 65-kDa antigen gene was important in the detection of all mycobacterial species without sacrificing the sensitivity of the test (7). The authors stated, however, that identification of similar restriction patterns such as with the RGM is difficult without special equipment and computer programs. Furthermore, nonspecific bands, which may be present with clinical samples, may interfere with the restriction enzyme analysis.
Fluorescence in situ hybridization assay.
Another useful technique for direct detection of mycobacteria in situ was reported by Stender and colleagues (H. Stender, O. F. Rasmussen, K. Lund, K. H. Petersen, P. Hongmanee, H. Miörner, and S. E. Godtfredsen, Abstr. 29th World Conf. Int. Union TB Lung Dis., abstr. 170-PPDisc, 1998). This procedure, known as fluorescence in situ hybridization assay, uses peptide nucleic acids to penetrate mycobacterial cell walls and hybridize specifically to target rRNA. The authors state that the procedure includes a probe for a “range of other mycobacteria” including M. avium complex, M. gordonae, M. kansasii, and M. tuberculosis, but no specific mention of RGM is made.
In summary, PRA using the hsp65 gene sequence and direct sequencing of the 16S rRNA gene to include the hypervariable regions, especially region A, are the best genetic methods at present for RGM species identification. PRA is the more practical and cost-efficient of the two methods.
Susceptibility Testing for Taxonomic Purposes
The RGM may also be differentiated taxonomically using some standard antimicrobial susceptibility results. Both broth microdilution MICs and agar disk diffusion may be useful. Overall, the most useful agents have been polymyxin B, relative susceptibility to amikacin and kanamycin, and susceptibility to cefoxitin (164, 204).
The M. fortuitum group is easily separated from the M. chelonae-abscessus group by polymyxin B disk susceptibility. As noted above, members of the M. fortuitum group are inhibited by polymyxin B whereas the M. chelonae-abscessus group is resistant to polymyxin, with no complete or partial zone of inhibition (204). The uniform susceptibility of the M. fortuitum group to the sulfonamides, the fluoroquinolones, amikacin (with low MICs), and other drugs is distinctly different from the situation for the very resistant M. chelonae-abscessus group and also helps in separating the two groups.
Furthermore, in the separation of M. chelonae from M. abscessus, susceptibilities to both cefoxitin and tobramycin are useful. As discussed above, isolates of M. chelonae have cefoxitin MICs of >256 μg/ml and tobramycin MICs generally of ≤4μg/ml. In contrast, isolates of M. abscessus have cefoxitin MICs in the range from 16 to 64μg/ml, with 32 μg/ml being the modal MIC. Also, tobramycin MICs tend to be higher (usually ≥16 μg/ml) than those of amikacin. Unlike the M. fortuitum group, all wild strains of M. chelonae and M. abscessus are clarithromycin susceptible.
The newly proposed species M. immunogenum is similar to M. chelonae in that isolates are resistant to cefoxitin (MICs, >256 μg/ml). However, in contrast, the MICs of tobramycin are also high, usually ≥8 μg/ml, which is more like those for M. abscessus. When using the agar disk diffusion susceptibility test, it was noted that the diameters of the zones of inhibition of both amikacin and kanamycin were equivalent for M. immunogenum, while M. abscessus and M. chelonae are more susceptible to kanamycin than to amikacin (212).
Another species of RGM which has equivalent zone sizes for amikacin and kanamycin is M. mucogenicum. This species is highly drug susceptible, as are other members of the M. fortuitum group, but the MICs of amikacin are much lower than those of kanamycin for the other members (M. fortuitum, etc.). Isolates of M. mucogenicum are distinguished from the other nonpigmented RGM by the presence of a zone of inhibition of cephalothin for 90% of clinical isolates (200).
Finally, tobramycin susceptibility is a useful test for the two newly proposed species within the M. smegmatis group compared to each other and M. smegmatis sensu stricto. By disk diffusion, isolates of M. smegmatis sensu stricto have zones of inhibition of >30 mm, M. goodii isolates have zones of inhibition of 11 to 30 mm, and isolates of the other new species, M. wolinskyi, have no zones of inhibition of tobramycin. Likewise, by broth microdilution, these three members of the M. smegmatis group have tobramysin MICs of ≤1, 2 to 8, and >8 μg/ml, respectively. Isolates of the M. smegmatis group, unlike most of the other species of RGM, are usually resistant to clarithromycin (17).
Susceptibility Testing for Clinical Purposes
For almost 20 years, susceptibility testing of RGM has been used as another tool for taxonomic separation of the RGM. Most species of RGM have a unique drug susceptibility pattern, and these patterns not only are important for therapeutic reasons but also can be used for taxonomic purposes to help identify the organism.
Four different methods have been used for suspectibility testing isolates of RGM. These methods are agar disk diffusion, broth microdilution, E test, and agar disk elution. Each method has proved useful, but until recently, none of the methods has been well standardized and each method has both advantages and disadvantages that must be considered. In December 2000 (218), members of the NCCLS (National Committee for Clinical Laboratory Standards) Mycobacterial Subcommittee on Antimicrobial Susceptibility recommended the use of MIC determinations by using broth microdilution as the “gold standard” for susceptibility testing of the RGM. The eight antimicrobials initially recommended for MIC testing were amikacin, cefoxitin, ciprofloxacin, clarithromycin, doxycycline, imipenem, sulfamethoxazole, and tobramycin. These selections were made by the Mycobacterial Subcommittee after a review and statistical analysis of two interlaboratory studies that involved multiple susceptibility test methods and representative test organisms (217).
Since some laboratories may perform susceptibility testing using one of the other methods, we briefly address each method.
Agar disk diffusion.
Agar disk diffusion (204) applies the Kirby-Bauer technique to the testing of RGM. Briefly, a suspension of the test organisms is prepared to match a McFarland 0.5 standard and inoculated onto plates of Mueller-Hinton agar supplemented with 5% oleic acid-albumin-dextrose. Commercial antibiotic disks are then applied, and plates are read after 72 h of incubation at 30°C (204).
This method has not been recommended for several years due to inherent technical problems. The major problems with this method are the absence of ready availability of some disks (e.g., sulfamethoxazole and doxycycline) and interpretation of “partial zones” of inhibition, which are observed when the concentration of the drug is near the MIC breakpoint between susceptibility and resistance to the drug. For example, the cefoxitin disk contains 30 μg of drug. However, the intermediate breakpoint is 32 to 64 μg/ml, so that the amount of drug in the disk is near the breakpoint and thus an area of colony growth often occurs within the primary zone of inhibition. Another major disadvantage is that susceptibility by disk diffusion for many of the newer drugs (e.g., fluoroquinolones, clarithromycin, and imipenem) has not been validated against an MIC method. The major advantages of the method are its ease and simplicity of set-up, plus the opportunity to look at colony morphology and to exclude the presence of a mixed culture. However, the above-mentioned problems outweigh the advantages, and thus the agar disk diffusion method is no longer recommended as anything other than a “screening tool” or taxonomic test. Therapeutic decisions should not be made on the basis of this method alone (19).
Agar disk elution.
Agar disk elution has been used mostly by laboratory personnel who test limited numbers of isolates and do so only infrequently. This method uses commercial susceptibility disks from which the drug is eluted into the oleic acid-albumin-dextrose and then mixed with melted agar to produce specific drug concentrations. Failure to elute the drug prior to addition of the melted agar results in uneven drug distribution and often growth of susceptible organisms at the edges of the agar, where the drug concentration is the lowest. This method is optimally performed using six round-well tissue culture plates which hold about 5 ml each (18).
Susceptibility testing is then performed by applying the method of proportions with a inoculum that averages 100 to 400 CFU/ml (167). The advantages of this method are that it utilizes materials and commercial drug disks readily available in most susceptibility laboratories and that the plates may be prepared on demand. It is not advisable to store plates for more than 3 days because of antimicrobial degradation (18). The method also correlates well with MIC tests performed in broth or agar for older drugs such as amikacin, doxycycline, cefoxitin, and sulfonamides. Some disadvantages of this method include the following: (i) preparation of the plates is tedious; (ii) the inoculum suspension must be carefully adjusted so that overinoculation of the wells does not occur (this is especially important and easily detected with drugs like the sulfonamides); (iii) there have been no MIC correlation or validation studies with agar disk elution and such newer antimicrobials as the fluoroquinolones, imipenem, linezolid, and clarithromycin; (iv) some desired concentrations for some drugs are not attainable due to the amount of drug in the commercial disks; (v) “trailing end points” can be a major problem with erythromycin and presumably the newer macrolides, because many strains that are susceptible in broth may produce fine, dysgonic colonies on agar or the drug may be slowly bactericidal (or bacteristatic), and since the end point for agar disk elution is either “growth” or “no growth”, this means that isolates could be misinterpreted as resistant by the agar disk elution method when they appear susceptible by broth microdilution (202).
E test.
The E test (AB Biodisk), or gradient MIC test, actually combines the agar diffusion technique with an exponential gradient of antimicrobial dilutions to produce an MIC result. The main advantages of this system include ease of set-up and use of standard agar media (8, 73, 85). However, recent interlaboratory studies by members of the Mycobacterial Subcommittee for Antimicrobial Susceptibility Testing of the NCCLS indicated that interpretation and reproducibility of the E-test MICs with RGM were often difficult. Isolates that were susceptible in broth were often interpreted as resistant by the E test in the four separate laboratories involved in the study (216). Diffuse elliptical edges and trailing growth or end points often made the determination of a precise MIC difficult for several drugs including ciprofloxacin, clarithromycin, imipenem, and cefoxitin. Because no standard RGM strain was found that exhibited reproducibility for all drugs tested, no breakpoints could be established and no control strains could be suggested for use in this method. Therefore, the NCCLS decided that further studies using the E test with the RGM were necessary before a recommendation for its usage could be endorsed (216).
Broth microdilution MIC.
The broth microdilution method is the only method currently recommended by the NCCLS for antimicrobial susceptibility testing of RGM (218).
The drug dilutions are prepared in cation-supplemented Mueller-Hinton broth using serial twofold dilutions of each drug (19). Several colonies of an isolate of RGM are suspended in Mueller-Hinton or Trypticase soy broth to reach a turbidity equivalent to the 0.5 McFarland standard. The organisms are diluted and inoculated into the drug wells of a 96-well microtiter plate. The panels are covered and incubated at 30°C for 3 days in room air. End point MICs of all drugs except sulfonamides, which are read at 80% inhibition, are read as the first well in which there is no growth. The plates may be prepared in-house by the user with 96-well microtiter plates and automated dispensing equipment such as the Mini-Quick Spense System (Dynatech Inc., Chantilly, Va.). Commercially prepared custom-made panels for mycobacteria are also available from Trek Diagnostic Systems, Inc. (Columbus, Ohio). Commercial MIC panels for routine bacterial susceptibility testing, can be used but do not provide the optimal combinations and concentrations of antimicrobials that should be tested. For example, the concentrations of cefoxitin are too low (≤16 μg/ml) and panels that contain clarithromycin or doxycycline are not readily available. The eight drugs recommended by the NCCLS for the panel include those previously listed (218).
A number of recommendations about test results were also made (218). As noted in Table 3, tobramycin should be reported only for isolates of M. chelonae since it has been recommended for therapy and validated by interlaboratory study only for this species. Any isolate of M. abscessus with an amikacin MIC of ≥64 μg/ml should be retested and/or sent to a reference laboratory if the repeat result is the same since validated resistance is unusual (although mutational resistance involving the 16S rRNA gene does occur) (191). Imipenem MICs should not be reported for isolates of M.chelonae and M. abscessus because the results are not reproducible. Also, from that same study, it was decided that isolates of the M. fortuitum group with imiperen MICs of >8 μg/ml should also be repeat tested, with careful attention being paid to use of a maximum incubation time of 3 days, since all isolates should have imiperen MICs of ≤8 μg/ml and the drug is notoriously unstable over time. Isolates of M. chelonae and M. abscessus do not exhibit good reproducibility in tests with imipenem and thus are not recommended for testing against this drug. This lack of reproducibility among the four laboratories was one of the major findings in the 1999 NCCLS study reported by Woods et al. (217).
Another problem which was noted during the study was the trailing end point for M. fortuitum isolates when tested against clarithromycin. Therefore, this study recommended that isolates of M. fortuitum with clear end points should have their clarithromycin MICs reported; isolates that exhibit trailing end points to macrolides should currently be considered resistant until a better or different method is available. If confirmation is necessary, the isolate should be sent to a qualified reference laboratory.
Finally, as stated above, MICs of sulfamethoxazole are read using 80% inhibition of growth as the susceptibility end point, not the 100% inhibition used for other antimicrobials. The 80% growth inhibition is usually the well in which a marked, definite decrease in the growth button is observed. Because few (if any) isolates of the M. fortuitum group are resistant to sulfonamides, testing of the resistance of this group of organisms to sulfonamides may not be necessary. Overinoculation of the drug panels is often most obvious with the sulfonamide wells. An inexperienced laboratorian may interpret the sulfonamide MIC as resistant when, in reality, the inoculum was too heavy; in this case, the MIC test should be repeated with a lower inoculum.
In the same NCCLS study (217), the remaining antimicrobials—cefoxitin, ciprofloxacin, and doxycycline—exhibited few discrepancies in reproducibility and accuracy among laboratories. Additionally, Woods et al. (217) proposed several breakpoint changes from the current NCCLS criteria for aerobic bacteria. The newly recommended RGM resistance breakpoint for cefoxitin is ≥128 μg/ml, compared to its breakpoint of 32 μg/ml for other aerobic bacteria (218).
For doxycycline the intermediate breakpoint is 8 μg/ml for other aerobic bacteria; however, for the RGM, the recommended susceptible MIC is ≤1 μg/ml, with an intermediate range of 2 to 8 μg/ml. The resistance breakpoint is unchanged at ≥16μg/ml. The newly recommended NCCLS breakpoints for the RGM (218) are shown in Table 5.
TABLE 5.
Suggested broth microdilution breakpoints for susceptibility testing of RGMa
| Drug | MIC (μg/ml) for category:
|
||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Recommended test drugsa | |||
| Amikacin | ≤16 | 32 | ≥64 |
| Cefoxitin | ≤16 | 32-64 | ≥128 |
| Ciprofloxacin | ≤1 | 2 | ≥4 |
| Clarithromycin | ≤2 | 4 | ≥8 |
| Doxycycline | ≤1 | 2-8 | ≥16 |
| Imipenemb | ≤4 | 8 | ≥16 |
| Sulfamethoxazolec | ≤32 | ≥64 | |
| Tobramycind | ≤4 | 8 | ≥16 |
| Secondary test drugse | |||
| Cefmetazole | ≤16 | 32 | ≥64 |
| Gatifloxacin | ≤2 | 4 | ≥8 |
| Levofloxacin | ≤2 | 4 | ≥8 |
| Linezolid | ≤8 | 16 | ≥32 |
| Moxifloxacin | ≤1 | 2 | ≥4 |
| Vancomycin | ≤4 | 8-16 | ≥32 |
Some additional drugs not yet approved by the NCCLS should also be considered for testing. These include the new 8-methoxyfluoroquinolones (gatifloxacin and moxifloxacin), cefmetazole (not currently available in the United States), levofloxacin, vancomycin, and linezolid (195; C. J. Crist, R. J. Wallace, Jr., B. A. Brown- Elliott, and L. B. Mann, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. 2001, abstr. U-35, 2001). For these agents, the breakpoints are those for aerobic organisms in NCCLS M100- S11 (49, 112), except for linezolid, for which tentative breakpoints for RGM were recently proposed (195). These values are listed in Table 5.
CONCLUSIONS
The RGM have continued to emerge as important human pathogens that can cause a variety of diseases from localized cutaneous infections to disseminated disease. The RGM have been responsible for a number of health care-associated outbreaks and pseudo-outbreaks.
The recent advances in antimicrobial therapy, including the new macrolides, fluoroquinolones, and oxazolidinones (linezolid), have improved the therapeutic options for the clinician and the prognosis of disease due to these organisms for the patient. This is especially true for M. chelonae and the M. fortuitum group. There is still, however, an urgent and compelling need for the development of better, more effective, and safe oral antimicrobials for the treatment of disease caused by the RGM, especially M. abscessus. M. abscessus lung disease, for example, is still generally incurable with currently available drug therapy. Susceptibility testing of the RGM is essential to choose optimal effective drug therapy and to monitor for the development of mutational drug resistance, which may occur with prolonged therapy. NCCLS tentative standards for such testing by broth microdilutions were published for the first time in December 2000.
Taxonomically, the pathogenic RGM have undergone dramatic changes in the past few years. Multiple new species such as M. goodii, M. immunogenum, and M. houstonense (proposed) have been introduced, and some former subspecies or subgroups have attained species status. These taxonomic advances have been attributed primarily to the use of HPLC and the new molecular techniques such as PRA, ribotyping, hybridization, and 16S RNA gene sequence analysis. These molecular methods, especially PRA, are rapidly replacing conventional biochemical testing in the large reference laboratory. Identification of the RGM by these molecular diagnostic methods may not only improve the correct recognition of current RGM species as well as identity previously uncharacterized species, but will also decrease the traditional laboratory delays in species identification and hence will lead to a more rapid and accurate diagnosis of disease. This should result in earlier and more effective therapy for many of these infections. Hopefully, time will prove both of these speculations to be true.
Acknowledgments
We gratefully acknowledge the many coinvestigators who have supported numerous studies on these organisms, especially Vella Silcox, Kenneth Jost, Véronique Vincent, Zeta Blacklock, Michio Tsukamura, and June Brown. We also acknowledge George Kubica for his thoughtful review of this paper and Joanne Woodring for preparation of the manuscript.
REFERENCES
- 1.Al Shaalan, M., B. J. Law, S. J. Israels, P. Pianosi, A. G. Lacson, and R. Higgins. 1997. Mycobacterium fortuitum interstitial pneumonia with vasculitis in a child with Wilms' tumor. Pediatr. Infect. Dis. J. 16:996-1000. [DOI] [PubMed] [Google Scholar]
- 2.Austin, W. K., and M. W. Lockey. 1976. Mycobacterium fortuitum mastoiditis. Arch Otolaryngol. 102:558-560. [DOI] [PubMed] [Google Scholar]
- 3.Awe, R. J., P. R. Gangadharam, and D. E. Jenkins. 1973. Clinical significance of Mycobacterium fortuitum infections in pulmonary disease. Am. Rev. Respir. Dis. 108:1230-1234. [DOI] [PubMed] [Google Scholar]
- 4.Azadian, B. S., A. Beck, J. R. Curtis, L. E. Cherrington, P. E. Gower, M. Phillips, J. B. Eastwood, and J. Nicholls. 1981. Disseminated infection with Mycobacterium chelonei in a haemodialysis patient. Tubercle 62:281-284. [DOI] [PubMed] [Google Scholar]
- 5.Band, J. D., J. I. Ward, D. W. Fraser, N. J. Peterson, V. A. Silcox, R. C. Good, P. R. Ostrey, and J. Kennedy. 1982. Peritonitis due to a Mycobacterium chelonei-like organism associated with intermittent chronic peritoneal dialysis. J. Infect. Dis. 145:9-17. [DOI] [PubMed] [Google Scholar]
- 6.Bange, F.-C., B. A. Brown, C. Smaczny, R. J. Wallace Jr., and E. C. Böttger. 2001. Lack of transmission of Mycobacterium abscessus among patients with cystic fibrosis attending a single clinic. Clin. Infect. Dis. 32:1648-1650. [DOI] [PubMed] [Google Scholar]
- 7.Bascuñana, C. R., and K. Belák. 1996. Detection and identification of mycobacteria in formalin-fixed, paraffin-embedded tissues by nested PCR and restriction enzyme analysis. J. Clin. Microbiol. 34:2351-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biehle, J. R., S. J. Cavalieri, M. A. Saubolle, and L. J. Getsinger. 1995. Evaluation of Etest for susceptibility testing of rapidly growing mycobacteria. J. Clin. Microbiol. 33:1760-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blacklock, Z. M., and D. J. Dawson. 1979. Atypical mycobacteria causing non-pulmonary disease in Queensland. Pathology 11:283-287. [DOI] [PubMed] [Google Scholar]
- 10.Bojalil, L. F., and J. Cerbón. 1961. Taxonomic analysis of nonpigmented, rapidly growing mycobacteria. J. Bacteriol. 81:338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bojalil, L. F., J. Cerbón, and A. Trujillo. 1962. Adansonian classification of mycobacteria. J. Gen. Microbiol. 28:333-346. [DOI] [PubMed] [Google Scholar]
- 12.Bolan, G., A. L. Reingold, L. A. Carson, V. A. Silcox, C. L. Woodley, P. S. Hayes, A. W. Hightower, L. McFarland, J. W. Brown III, N. J. Petersen, M. S. Favero, R. C. Good, and C. V. Broome. 1985. Infections with Mycobacterium chelonei in patients receiving dialysis and using processed hemodialyzers. J. Infect. Dis. 152:1013-1019. [DOI] [PubMed] [Google Scholar]
- 13.Bönicke, R. 1966. The occurrence of atypical mycobacteria in the environment of man and animal. Bull. Int. Union Tuberc. Lung Dis. 37:361-368. [Google Scholar]
- 14.Booth, J. E., J. A. Jacobson, T. A. Kurrus, and T. W. Edwards. 1979. Infection of prosthetic arthroplasty by M. fortuitum. Two case reports. J. Bone Joint Surg. 61A:300. [PubMed] [Google Scholar]
- 15.Borghaus, J. G. A., and J. L. Stanford. 1973. Mycobacterium chelonei in abscesses after injection of diphtheria-pertussis-tetanus-polio vaccine. Am. Rev. Respir. Dis. 107:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Brannan, D. P., R. E. DuBois, M. J. Ramirez, M. J. R. Ravry, and E. O. Harrison. 1984. Cefoxitin therapy for Mycobacterium fortuitum bacteremia with associated granulomatous hepatitis. South. Med. J. 77:381-382. [DOI] [PubMed] [Google Scholar]
- 17.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, S. H. Chiu, G. O. Onyi, E. C. Böttger, and R. J. Wallace, Jr. 1999. Description of Mycobacterium wolinskyi and Mycobacterium goodii, two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. [DOI] [PubMed] [Google Scholar]
- 18.Brown, B. A., J. M. Swenson, and R. J. Wallace Jr. 1992. Agar disk elution test for rapidly growing mycobacteria, p. 5.10.1-5.10.11. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, section 5: antimicrobial susceptibility testing, vol. 1. American Society for Microbiology, Washington, D.C.
- 19.Brown, B. A., J. M. Swenson, and R. J. Wallace, Jr. 1992. Broth microdilution MIC test for rapidly growing mycobacteria, p. 5.11.1-5.11.9. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, section 5: antimicrobial susceptibility testing, vol. 1. American Society for Microbiology Washington, D.C.
- 20.Brown, B. A., R. J. Wallace, Jr., G. Onyi, V. DeRosas, and R. J. Wallace III. 1992. Activities of four macrolides including clarithromycin against Mycobacterium fortuitum, Mycobacterium chelonae, and Mycobacterium chelonae-like organisms. Antimicrob. Agents Chemother. 36:180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown-Elliott, B. A., R. J. Wallace Jr., R. Blinkhorn, C. J. Crist, and L. M. Mann. 2001. Successful treatment of disseminated Mycobacterium chelonae infection with linezolid. Clin. Infect. Dis. 33:1433-1434. [DOI] [PubMed] [Google Scholar]
- 22.Buckley, R., M. W. Cobb, S. Ghurani, N. F. Brock, and R. R. Harford. 1997.Mycobacterium fortuitum infection occurring after a punch biopsy procedure. Pediatr. Dermatol. 14:290-292. [DOI] [PubMed] [Google Scholar]
- 23.Bullington, R. H., J. D. Lanier, and R. L. Font. 1992. Nontuberculous mycobacterial keratitis. Arch. Ophthalmol. 110:519-524. [DOI] [PubMed] [Google Scholar]
- 24.Burns, D. N., P. K. Rohatgi, R. Rosenthal, M. Seiler, and F. M. Gordin. 1990. Disseminated Mycobacterium fortuitum successfully treated with combination therapy including ciprofloxacin. Am. Rev. Respir. Dis. 142:468-470. [DOI] [PubMed] [Google Scholar]
- 25.Burns, D. N., R. J. Wallace, Jr., M. E. Schultz, Y. Zhang, S. Q. Zubairi, Y. Pang, C. L. Gibert, B. A. Brown, E. S. Noel, and F. M. Gordin 1991. Nosocomial outbreak of respiratory tract colonization with Mycobacterium fortuitum: demonstration of the usefulness of pulsed-field gel electrophoresis in an epidemiologic investigation. Am. Rev. Respir. Dis. 144:1153-1159. [DOI] [PubMed] [Google Scholar]
- 26.Burns, J. L., U. Malhotra, J. Lingappa, and S. Smith. 1997. Unusual presentations of nontuberculous mycobacterial infections in children. Pediatr. Infect. Dis. J. 16:802-806. [DOI] [PubMed] [Google Scholar]
- 27.Butler, W. R., and J. O. Kilburn. 1990. High-performance liquid chromatography patterns of mycolic acids as criteria for identification of Mycobacterium chelonae, Mycobacterium fortuitum, and Mycobacterium smegmatis. J. Clin. Microbiol. 28:2094-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butt, A. A. 1998. Cervical adenitis due to Mycobacterium fortuitum in patients with acquired immunodeficiency syndrome. Am. J. Med. Sci. 315:50-55. [DOI] [PubMed] [Google Scholar]
- 29.Campagnaro, R. L., H. Teichtahl, and B. Dwyer. 1994. A pseudoepidemic of Mycobacterium chelonae: contamination of a bronchoscope and autocleaner. Aust. N. Z. J. Med. 24:693-695. [DOI] [PubMed] [Google Scholar]
- 30.Carmago, D., C. Saad, F. Ruiz, M. E. Ramirez, M. Lineros, G. Rodriguez, E. Navarro, B. Pulido, and L. C. I. Orozco. 1996. Iatrogenic outbreak of M. chelonae skin abscesses. Epidemiol. Infect. 117:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carson, L. A., L. B. Cusick, L. A. Bland, and M. S. Favero. 1988. Efficacy of chemical dosing methods for isolating nontuberculous mycobacteria from water supplies of dialysis centers. Appl. Environ. Microbiol. 54:1756-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cegielski, J. P., and R. J. Wallace Jr. 1997. Infections due to nontuberculous mycobacteria, p. 445-461. In W. M. Scheld, R. J. Whitley, and D. T. Durack (ed.), Infections of the central nervous system, 2nd ed., Lippincott-Raven Publishers, Philadelphia, Pa.
- 33.Centers for Disease Control and Prevention. 1996. Infection with Mycobacterium abscessus associated with intramuscular injection of adrenal cortex extract—Colorado and Wyoming, 1995-1996. Morb. Mortal. Wkly. Rev. 45:713-715. [PubMed] [Google Scholar]
- 34.Chamoiseau, G. 1979. Etiology of farcy in African bovines: nomenclature of the causal organisms Mycobacterium farcinogenes Chamoiseau and Mycobacterium senegalense (Chamoiseau) comb. nov. Int. J. Syst. Bacteriol. 29:407-410. [Google Scholar]
- 35.Chang, M. J., and L. L. Barton. 1974. Mycobacterium fortuitum osteomyelitis of the calcaneus secondary to a puncture wound. J. Pediatr. 85:517-519. [DOI] [PubMed] [Google Scholar]
- 36.Chetchotisakd, P., P. Mootsikapun, S. Anunnatsiri, K. Jirarattanapochai, C. Choonhakarn, A. Chaiprasert, P. N. Ubol, L. J. Wheat, and T. E. Davis. 2000. Disseminated infection due to rapidly growing mycobacteria in immunocompetent hosts presenting with chronic lymphadenopathy: a previously unrecognized clinical entity. Clin. Infect. Dis. 32:29-34. [DOI] [PubMed] [Google Scholar]
- 37.Choueiry, M. A., P. L. Scurto, P. M. Flynn, B. N. Rao, and W. T. Hughes. 1998. Disseminated infection due to Mycobacterium fortuitum in a patient with desmoid tumor. Clin. Infect. Dis. 26:237-238. [DOI] [PubMed] [Google Scholar]
- 38.Clegg, H. W., M. T. Foster, W. E. Sanders, Jr., and W. B. Baine. 1983. Infection due to organisms of the Mycobacterium fortuitum complex after augmentation mammaplasty: clinical and epidemiologic features. J. Infect. Dis. 147:427-433. [DOI] [PubMed] [Google Scholar]
- 38a.Conville, P. S., and F. G. Witebsky. 2001. Lack of usefulness of carbon utilization tests for identification of Mycobacterium mucogenicum. J. Clin. Microbiol. 39:2725-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cullen, A. R., C. L. Cannon, E. J. Mark, and A. A. Colin. 2000. Mycobacterium abscessus infection in cystic fibrosis. Am J. Respir. Crit. Care Med. 161:641-645. [DOI] [PubMed] [Google Scholar]
- 41.Cutay, A. M., H. W. Horowitz, R. W. Pooley, K. Van Horn, and G. P. Wormser. 1998. Infection of epicardial pacemaker wires due to Mycobacterium abscessus. Clin. Infect. Dis. 26:520-521. [DOI] [PubMed] [Google Scholar]
- 42.da Costa Cruz, J. C. 1938. Mycobacterium fortuitum: um novo bacilo acido- resistente patogenico para o homen (new acid fast bacillus pathogenic for man). Acta Med. (Rio de Jåneiro) 1:298-301. [Google Scholar]
- 43.Dalovisio, J. R., G. A. Pankey, R. J. Wallace, Jr., and D. B. Jones. 1981. Clinical usefulness of amikacin and doxycycline in the treatment of infection due to Mycobacterium fortuitum and Mycobacterium chelonei. Rev. Infect. Dis. 3:1068-1074. [DOI] [PubMed] [Google Scholar]
- 44.De Beenhouwer, H., Z. Liang, P. de Rijk, C. van Eekeren, and F. Portaels. 1995. Detection and identification of mycobacteria by DNA amplification and oligonucleotide-specific capture plate hybridization. J. Clin. Microbiol. 33:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devallois, A., K. S. Goh, and N. Rastogi. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 35:2969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domenech, P., M. S. Jimenez, M. C. Menendez, T. J. Bull, S. Samper, A. Manrique, and M. J. Garcia. 1984. Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47:535-540. [DOI] [PubMed] [Google Scholar]
- 47.Efthimiou, J., M. J. Smith, M. E. Hodson, and J. C. Batten. 1984. Fatal pulmonary infection with Mycobacterium fortuitum in cystic fibrosis. Br. J. Dis. Chest 78:299-302. [PubMed] [Google Scholar]
- 48.Fauroux, B., B. Delaisi, A. Clément, C. Saizou, D. Moissenet, C. Truffot- Pernot, G. Tournier, and H. Vu Thien. 1997. Mycobacterial lung disease in cystic fibrosis: a prospective study. Pediatr. Infect. Dis. J. 16:354-358. [DOI] [PubMed] [Google Scholar]
- 49.Ferraro, M. J., W. A. Craig, G. Eliopoulos, J. Fung-Tomc, S. L. Hansen, D. W. Hecht, J. Hindler, L. B. Reller, D. F. Sahm, J. M. Swenson, F. C. Tenover, R. T. Testa, and M. A. Wikler. 1998. Performance standards for antimicrobial susceptibility testing; eighth informational supplement. Standard M100-S8. NCCLS, Wayne, Pa.
- 50.Flor, A., J. A. Capdevila, N. Martin, J. Gavaldà, and A. Pahissa. 1996. Nontuberculous mycobacterial meningitis: report of two cases and review. Clin. Infect. Dis. 23:1266-1273. [DOI] [PubMed] [Google Scholar]
- 51.Flynn, P. M., B. Van Hooser, and F. Gigliotti. 1988. Atypical mycobacterial infections of Hickman catheter exit sites. Pediatr. Infect. Dis. J. 7:510-513. [DOI] [PubMed] [Google Scholar]
- 52.Foz, A., C. Roy, J. Jurado, E. Arteaga, J. M. Ruiz, and A. Moragas. 1978. Mycobacterium chelonei iatrogenic infections. J. Clin. Microbiol. 7:319-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franklin, D. J., J. R. Starke, M. T. Brady, B. A. Brown, and R. J. Wallace, Jr. Chronic otitis media after tympanostomy tube placement caused by Mycobacterium abscessus: a new clinical entity? Am. J. Otol. 15:313-320. [PubMed]
- 54.Fraser, V. J., M. Jones, P. R. Murray, G. Medoff, Y. Zhang, and R. J. Wallace, Jr. 1992. Contamination of flexible fiberoptic bronchoscopes with Mycobacterium chelonae linked to an automated bronchoscope disinfection machine. Am. Rev. Respir. Dis. 145:853-855. [DOI] [PubMed] [Google Scholar]
- 55.Fraser, V., and R. J. Wallace, Jr. 1996. Nontuberculous mycobacteria, p. 1224. In C. G. Mayhall (ed.), Hospital epidemiology and infection control. The Williams & Wilkins Co., Baltimore, Md.
- 56.Friedman, N. D., and D. J. Sexton. 2001. Bursitis due to Mycobacterium goodii, a recently described, rapidly growing mycobacterium. J. Clin. Microbiol. 39:404-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galil, K., L. A. Miller, M. A. Yakrus, R. J. Wallace Jr., D. G. Mosley, B. England, G. Huitt G, M. M. McNeill, and B. A. Perkins. 1999. Abscesses due to Mycobacterium abscessus linked to injection of unapproved alternative medication. Emerg. Infect. Dis. 5:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gangadharam, P. R. J., J. D. Lanier, D. E. Jones. 1978. Keratitis due to Mycobacterium chelonei. Tubercle 59:55-60. [DOI] [PubMed] [Google Scholar]
- 59.Georgia Department of Human Resources. 1990. Abscesses in an allergy practice due to M. chelonae. Georgia Epidemiol. Rep. 6:2. [Google Scholar]
- 60.Ghossein, R. A., D. G. Ross, R. N. Salmon, and A. R. Rabson. 1992. Rapid detection and species identification of mycobacteria in paraffin-embedded tissues by polymerase chain reaction. Diagn. Mol. Pathol. 1:185-191. [PubMed] [Google Scholar]
- 61.Goldblatt, M. R., and J. A. Ribes. 2002. Mycobacterium mucogenicum isolated from a patient with granulomatous hepatitis. Arch. Pathol. Lab. Med. 126:73-75. [DOI] [PubMed] [Google Scholar]
- 62.Gremillion, D. H., S. B. Mursch, and C. J. Lerner. 1983. Injection site abscesses caused by Mycobacterium chelonei. Infect. Control 4:25-28. [DOI] [PubMed] [Google Scholar]
- 63.Griffith, D. E., W. M. Girard, and R. J. Wallace, Jr. 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria: analysis of 154 patients. Am. Rev. Respir. Dis. 147:1271-1278. [DOI] [PubMed] [Google Scholar]
- 64.Griffith, D. E., and R. J. Wallace, Jr. 1988. Pulmonary disease due to rapidly growing mycobacteria. Semin. Respir. Med. 9:505-513. [PubMed] [Google Scholar]
- 65.Grigg, J., L. W. Hirst, M. Whitby, K. Stallard, and P. Gnanaharan. 1992. Atypical Mycobacterium keratitis. Aust. N. Z. J. Opthalmol. 20:257-261. [DOI] [PubMed] [Google Scholar]
- 66.Gubler, J. G. H., M. Salfinger, and A. von Graevenitz. 1992. Pseudoepidemic of nontuberculous mycobacteria due to a contaminated bronchoscope cleaning machine: report of an outbreak and review of the literature. Chest 101:1245-1249. [DOI] [PubMed] [Google Scholar]
- 67.Gyimesi, Z. S., I. H. Stalis, J. M. Miller, and C. O. Thoen. 1999. Detection of Mycobacterium avium subspecies avium in formalin-fixed, paraffin-embedded tissues of captive exotic birds using polymerase chain reaction. J. Zoo Wild. Med. 30:348-353. [PubMed] [Google Scholar]
- 68.Hand, W. L., and J. P. Sanford. 1970. Mycobacterium fortuitum—a human pathogen. Ann. Intern. Med. 73:971-977. [DOI] [PubMed] [Google Scholar]
- 69.Hayes, P. S., D. L. McGiboney, J. D. Band, and J. C. Feeley. 1982. Resistance of Mycobacterium chelonei-like organisms to formaldehyde. Appl. Environ. Microbiol. 43:722-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hector, J. S. R., Y. Pang, G. H. Mazurek, Y. Zhang, B. A. Brown, and R. J. Wallace, Jr. 1992. Large restriction fragment patterns of genomic Mycobacterium fortuitum DNA as strain-specific markers and their use in epidemiologic investigation of four nosocomial outbreaks. J. Clin. Microbiol. 30:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herold, R. C., P. A. Lotke, and R. R. MacGregor. 1987. Prosthetic joint infections secondary to rapidly growing Mycobacterium fortuitum. Clin. Orthop. Relat. Res. 216:183-187. [PubMed] [Google Scholar]
- 72.Hoffman, P. C., D. W. Fraser, F. Robiesek, P. R. O'Bar, and C. U. Mauney. 1981. Two outbreaks of sternal wound infections due to organisms of the Mycobacterium fortuitum complex. J. Infect. Dis. 143:533-542. [DOI] [PubMed] [Google Scholar]
- 73.Hoffner, S. E., L. Klintz, B. Olsson-Lijequist, and A. Bolmström. 1994. Evaluation of Etest for rapidly growing susceptibility testing of Mycobacterium chelonae and M. fortuitum. J. Clin. Microbiol. 32:1846-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hogg, G. G., M. F. Schinsky, M. M. McNeil, B. A. Lasker, V. A. Silcox, and J. M. Brown. 1994. Central line sepsis in a child due to a previously unidentified mycobacterium. J. Clin. Microbiol. 37:1193-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horsburgh, C. R., and R. M. Selik. 1989. The epidemiology of disseminated nontuberculous mycobacterial infection in the acquired immunodeficiency syndrome (AIDS). Am. Rev. Respir. Dis. 139:4-7. [DOI] [PubMed] [Google Scholar]
- 76.Hoy, J., K. Rolston, and R. L. Hopfer. 1987. Pseudoepidemic of Mycobacterium fortuitum in bone marrow cultures. Am. J. Infect. Control 15:268-271. [DOI] [PubMed] [Google Scholar]
- 77.Hoy, J. F., K. V. I. Rolston, R. L. Hopfer, and G. P. Bodey. 1987. Mycobacterium fortuitum bacteremia in patients with cancer and long-term venous catheters. Am. J. Med. 83:213-217. [DOI] [PubMed] [Google Scholar]
- 78.Ichiyama, S., and M. Tsukamura. 1987. Ofloxacin and the treatment of pulmonary disease due to Mycobacterium fortuitum. Chest 92:1110-1112. [DOI] [PubMed] [Google Scholar]
- 79.Jackson, P. G., H. Keen, C. J. Noble, and N. A. Simmons. 1981. Injection abscesses due to Mycobacterium chelonei occurring in a diabetic patient. Tubercle 62:277-279. [DOI] [PubMed] [Google Scholar]
- 80.Jarvis, W. R., and the Epidemiology Branch, Hospital Infections Program. 1991. Nosocomial outbreaks: the Centers for Disease Control's Hospital Infections Program Experience, 1980-1990. Am. J. Med. 91(Suppl. 3B):101S. [DOI] [PubMed] [Google Scholar]
- 81.Kelley, L. C., K. C. Deering, and E. T. Kaye. 1995. Cutaneous Mycobacterium chelonei presenting in an immunocompetent host: case report and review of the literature. Cutis 56:293-295. [PubMed] [Google Scholar]
- 82.Khooshabeh, R., J. M. Grange, M. D. Yates, A. C. E. McCartney, and T. A. Casey. 1994. A case report of Mycobacterium chelonae keratitis and a review of mycobacterial infections of the eye and orbit. Tubercle Lung Dis. 75:377-382. [DOI] [PubMed] [Google Scholar]
- 83.Kilby, J. M., P. H. Gilligan, J. R. Yankaskas, W. E. Highsmith, Jr., L. J. Edwards, and M. R. Knowles. 1992. Nontuberculous mycobacteria in adult patients with cystic fibrosis. Chest 102:70-75. [DOI] [PubMed] [Google Scholar]
- 84.Kirschner, P., M. Kiekenbeck, D. Meissner, J. Wolters, and E. C. Böttger. 1992. Genetic heterogeneity within Mycobacterium fortuitum complex species: genotypic criteria for identification. J. Clin. Microbiol. 30:2772-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koontz, F. P., M. E. Erwin, M. S. Barrett, and R. N. Jones. 1994. Etest for routine clinical antimicrobial susceptibility testing of rapidly-growing mycobacteria isolates. Diagn. Microbiol. Infect. Dis. 19:183-186. [DOI] [PubMed] [Google Scholar]
- 86.Kubica, G. P., I. Baess, R. E. Gordon, P. A. Jenkins, J. B. G. Kwapinski, C. McDurmont, S. R. Pattyn, H. Saito, V. Silcox, J. L. Stanford, K. Takeya, and M. Tsukamura. 1972. A co-operative numerical analysis of rapidly growing mycobacteria. J. Gen. Microbiol. 73:55-70. [DOI] [PubMed] [Google Scholar]
- 87.Kuritsky, J. N., M. G. Bullen, C. V. Broome, V. A. Silcox, R. C. Good, and R. J. Wallace Jr. 1983. Sternal wound infections and endocarditis due to organisms of the Mycobacterium fortuitum complex. Ann. Intern. Med. 98:938-939. [DOI] [PubMed] [Google Scholar]
- 88.Küster, E. 1905. Ueber kaltblutertuberkulose. Muench. Med. Wochenschr. 57:57-59. [Google Scholar]
- 89.Kusunoki, S., and T. Ezaki. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42:240-245. [DOI] [PubMed] [Google Scholar]
- 90.Lai, K. K., B. A. Brown, J. A. Westerling, S. A. Fontecchio, Y. Zhang, and R. J. Wallace, Jr. 1998. Long-term laboratory contamination by Mycobacterium abscessus resulting in two pseudo-outbreaks: recognition with use of random amplified polymorphic DNA (RAPD) polymerase chain reaction. Clin. Infect. Dis. 27:169-175. [DOI] [PubMed] [Google Scholar]
- 91.Lakshmi, V., R. R. Rao, and I. Dinakar. 1993. Bacteriology of brain abscess—observations on 50 cases. J. Med. Microbiol. 38:187-190. [DOI] [PubMed] [Google Scholar]
- 92.Lauring, L. M., F. L. Wergeland, and G. E. Sack. 1969. Anonymous Mycobacterium keratitis. Am. J. Ophthalmol. 67:130-133. [DOI] [PubMed] [Google Scholar]
- 93.Laussucq, S., A. L. Baltch, R. P. Smith, R. W. Smithwick, B. J. Davis, E. K. Desjardin, V. A. Silcox, A. B. Spellacy, R. T. Zeimis, H. M. Gruft, R. C. Good, and M. L. Cohen. 1988. Nosocomial Mycobacterium fortuitum colonization from a contaminated ice machine. Am. Rev. Respir. Dis. 138:891-894. [DOI] [PubMed] [Google Scholar]
- 94.Levendoglu-Tugal, O., J. Munoz, A. Brudnicki, M. F. Ozkaynak, C. Sandoval, and S. Jayabose. 1998. Infections due to nontuberculous mycobacteria in children with leukemia. Clin. Infect. Dis. 27:1227-1230. [DOI] [PubMed] [Google Scholar]
- 95.Lévy-Frébault, V., M. Daffé, K. S. Goh, M. A. Lanéelle, C. Asselineau, and H. L. David. 1983. Identification of Mycobacterium fortuitum and Mycobacterium chelonei. J. Clin. Microbiol. 17:744-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lévy-Frébault, V., F. Grimont, P. A. D. Grimont, and H. L. David. 1986. Deoxyribonucleic acid relatedness study of the Mycobacterium fortuitum- Mycobacterium chelonae complex. Int. J. Syst. Bacteriol. 36:458-460. [Google Scholar]
- 97.Lowry, P. W., C. M. Beck-Sague, L. A. Bland, S. M. Aguero, M. J. Arduino, A. N. Minuth, R. A. Murray, J. M. Swenson, and W. R. Jarvis. 1990. Mycobacterium chelonae infection among patients receiving high-flux dialysis in a hemodialysis clinic in California. J. Infect. Dis. 161:85-90. [DOI] [PubMed] [Google Scholar]
- 98.Lowry, P. W., W. R. Jarvis, A. D. Oberle, L. A. Bland, R. Silberman, J. A. Bocchini Jr., H. D. Dean, J. M. Swenson, and R. J. Wallace, Jr. 1988. Mycobacterium chelonae causing otitis media in an ear-nose-and-throat practice. N. Engl. J. Med. 319:978-982. [DOI] [PubMed] [Google Scholar]
- 99.Lustgarten, S. 1885. The bacillus of syphilis. Lancet i:609-610. [Google Scholar]
- 100.Maloney, S., S. Welbel, B. Daves, K. Adams, S. Becker, L. Bland, M. Arduino, R. J. Wallace, Jr., Y. Zhang, G. Buck, P. Risch, and W. Jarvis. 1994. Mycobacterium abscessus pseudoinfection traced to an automated endoscope washer: utility of epidemiologic and laboratory investigation. J. Infect. Dis. 169:1166-1169. [DOI] [PubMed] [Google Scholar]
- 101.Marchetti, G., A. Gori, L. Catozzi, L. Vago, M. Nebuloni, M. C. Rossi, A. D. Esposti, A. Bandera, and F. Franzetti. 1998. Evaluation of PCR in detection of Mycobacterium tuberculosis from formalin-fixed, paraffin-embedded tissues: comparison of four amplification assays. J. Clin. Microbiol. 36:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maxson, S., G. E. Schutze, and R. F. Jacobs. 1994. Mycobacterium abscessus osteomyelitis: treatment with clarithromycin. Infect. Dis. Clin. Pract. 3:203-204. [Google Scholar]
- 103.McClellan, K. A., P. J. Bernard, L. P. Robinson, K. V. Meades, G. W. Aylward, and F. A. Billson. 1989. Atypical mycobacterial keratitis. Aust. N. Z. J. Ophthalmol. 17:103-105. [DOI] [PubMed] [Google Scholar]
- 104.McWhinney, P. H. M., M. Yates, H. G. Prentice, M. Thrussell, S. H. Gillespie, and C. C. Kibbler. 1992. Infection caused by Mycobacterium chelonae: a diagnostic and therapeutic problem in the neutropenic patient. Clin. Infect. Dis. 14:1208-1212. [DOI] [PubMed] [Google Scholar]
- 105.Meisler, D. M., M. H. Friedlaender, and M. Okumoto. 1982. Mycobacterium chelonei keratitis. Am. J. Ophthalmol. 94:398-401. [DOI] [PubMed] [Google Scholar]
- 106.Meredith, F. T., and D. J. Sexton. 1996. Mycobacterium abscessus osteomyelitis following a plantar puncture wound. Clin. Infect. Dis. 23:651-653. [DOI] [PubMed] [Google Scholar]
- 107.Metcalf, J. F., J. F. John, Jr., G. B. Wilson, H. H. Fudenberg, and R. A. Harley. 1981. Mycobacterium fortuitum pulmonary infection associated with an antigen-selective defect in cellular immunity. Am. J. Med. 71:485-492. [DOI] [PubMed] [Google Scholar]
- 107a.Meyers, H., B. A. Brown-Elliott, D. Moore, J. Curry, C. Truong, Y. Zhang, and R. J. Wallace, Jr. 2002. An outbreak of Mycobacterium chelonae following liposuction. Clin. Infect. Dis. 34:1500-1507. [DOI] [PubMed] [Google Scholar]
- 108.Mirate, D. J., D. S. Hull, J. H. Steel, Jr., and M. J. Carter. 1983. Mycobacterium chelonei keratitis: a case report. Br. J. Ophthalmol. 67:324-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miron, D., E. A. Lev, M. Zuker, D. Lumelsky, M. Murph, M. M. Floyd, and J. M. Brown. 2000. Mycobacterium fortuitum osteomyelitis of the cuboid after nail puncture wound. Pediatr. Infect. Dis. J. 19:483-485. [DOI] [PubMed] [Google Scholar]
- 110.Moore, J. S., M. Christensen, R. W. Wilson, R. J. Wallace, Jr., Y. Zhang, D. R. Nash, and B. Shelton. 2000. Mycobacterial contamination of metalworking fluids: involvement of a possible new taxon of rapidly growing mycobacteria. Am. Ind. Hyg. Assoc. J. 61:205-213. [DOI] [PubMed] [Google Scholar]
- 111.Moore, M., and J. B. Frerichs. 1953. An unusual acid fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region: report of a case study with a study of the organism, Mycobacterium abscessus. J. Investig. Dermatol. 20:133-169. [DOI] [PubMed] [Google Scholar]
- 112.NCCLS. 2001. Performance standards for antimicrobial susceptibility testing. Eleventh informational supplement, M100-S11. NCCLS, Wayne, Pa.
- 113.Newton, J. A., Jr., P. J. Weiss, W. A. Bowler, and E. C. Oldfield III. 1993. Soft-tissue infection due to Mycobacterium smegmatis: report of two cases. Clin. Infect. Dis. 16:531-533. [DOI] [PubMed] [Google Scholar]
- 114.Ninet, B., O. Rutschmann, K. Burkhardt, C. Metral, B. Borisch, B. Hirschel, and the Swiss HIV Cohort Study. 1999. Detection of mycobacterial nucleic acids by polymerase chain reaction in fixed tissue specimens of patients with human immunodeficiency virus infection. Diagn. Mol. Pathol. 8:145-151. [DOI] [PubMed] [Google Scholar]
- 115.Nolan, C. M., P. A. Hashisaki, and D. F. Dundas. 1991. An outbreak of soft- tissued infections due to Mycobacterium fortuitum associated with electromyography. J. Infect. Dis. 163:1150-1153. [DOI] [PubMed] [Google Scholar]
- 116.Nye, K., D. K. Chadha, P. Hodgkin, C. Bradley, J. Hancox, and R. Wise. 1990. Mycobacterium chelonei isolation from broncho-alveolar lavage fluid and its practical implications. J. Hosp. Infect. 16:257-261. [DOI] [PubMed] [Google Scholar]
- 117.O'Brien, R. J., L. J. Geiter, and D. E. Snider. 1987. The epidemiology of nontuberculous mycobacterial diseases in the United States: results from a national survey. Am. Rev. Respir. Dis. 135:1007-1014. [DOI] [PubMed] [Google Scholar]
- 118.Offer, R. C., A. G. Karlson, and J. A. Spittell, Jr. 1971. Infection caused by Mycobacterium fortuitum. Mayo Clin. Proc. 46:747-749. [PubMed] [Google Scholar]
- 119.Owen, M., A. Smith, and J. Coultras. 1963. Granulomatous lesions occurring at site of injections of vaccines and antibiotics. South. Med. J. 56:949-952. [DOI] [PubMed] [Google Scholar]
- 120.Pacht, E. R. 1990. Mycobacterium fortuitum lung abscess: resolution with prolonged trimethoprim/sulfamethoxazole therapy. Am. Rev. Respir. Dis. 141:1599-1601. [DOI] [PubMed] [Google Scholar]
- 121.Pappas, S. A., D. M. Schaaff, M. B. DiCostanzo, F. W. King, Jr., and J. T. Sharp. 1983. Contamination of flexible fiberoptic bronchoscopes. Am. Rev. Respir. Dis. 127:381-392. [DOI] [PubMed] [Google Scholar]
- 122.Park, H., H. Jang, C. Kim, B. Chung, C. L. Chang, S. K. Park, and S. Song. 2000. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers. J. Clin. Microbiol. 38:4080-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Patel, S., M. Yates, and N. A. Saunders. 1997. PCR-enzyme-linked immunosorbent assay and partial rRNA gene sequencing: a rational approach to identifying mycobacteria. J. Clin. Microbiol. 35:2375-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pattyn, S. R., M. Magnusson, J. L. Stanford, and J. M. Grange. 1974. A study of Mycobacterium fortuitum (ranae). J. Med. Microbiol. 7:67-76. [DOI] [PubMed] [Google Scholar]
- 125.Pattyn, S. R., J. Vandepitte, F. Portaels, and A. De-Muynck. 1971. Cases of M. borstelense and M. abscessus infection observed in Belgium. J. Med. Microbiol. 4:145-149. [DOI] [PubMed] [Google Scholar]
- 126.Peneau, A., D. Moinard, I. Berard, O. Pascal, and J. P. Moisan. 1992. Detection of mycobacteria using the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 11:270-271. [DOI] [PubMed] [Google Scholar]
- 127.Pennekamp, A., G. E. Pfyffer, J. Wüest, C. A. George, and C. Ruef. 1997. Mycobacterium smegmatis infection in a healthy woman following a facelift: case report and review of the literature. Ann. Plast. Surg. 39:80-83. [DOI] [PubMed] [Google Scholar]
- 128.Perosio, P. M., and T. S. Frank. 1993. Detection and species identification of mycobacteria in paraffin sections of lung biopsy specimens by the polymerase chain reaction. Am. J. Clin. Pathol. 100:643-647. [DOI] [PubMed] [Google Scholar]
- 129.Pettini, B., P. Hellstrand, and M. Erickson. 1980. Infection with M. chelonae following injections. Scand. J. Infect. Dis. 12:237-238. [DOI] [PubMed] [Google Scholar]
- 130.Plaus, W. J., and G. Hermann. 1991. The surgical management of superficial infections caused by atypical mycobacteria. Surgery 110:99-103. [PubMed] [Google Scholar]
- 131.Plemmons, R. M., C. K. McAllister, D. A. Liening, and M. C. Garces. 1996. Otitis media and mastoiditis due to Mycobacterium fortuitum: case report, review of four cases, and a cautionary note. Clin. Infect. Dis. 22:1105-1106. [DOI] [PubMed] [Google Scholar]
- 132.Pope, J., Jr., P. Sternberg, Jr., N. J. McLane, D. W. Potts, and R. D. Stulting. 1989. Mycobacterium chelonae scleral abscess after removal of a scleral buckle. Am. J. Ophthalmol. 107:557-558. [DOI] [PubMed] [Google Scholar]
- 133.Pruitt, T. C., L. O. Hughes, R. D. Blasir, R. E. McCarthy, C. M. Glasier, and G. J. Roloson. 1993. Atypical mycobacterial vertebral osteomyelitis in a steroid-dependent adolescent. Spine 18:2553-2555. [DOI] [PubMed] [Google Scholar]
- 134.Raad, I. I., S. Vartivarian, A. Khan, and G. P. Bodey. 1991. Catheter-related infections caused by the Mycobacterium fortuitum complex: 15 cases and review. Rev. Infect. Dis. 13:1120-1125. [DOI] [PubMed] [Google Scholar]
- 135.Rappaport, W., G. Dunington, L. Norton, D. Ladin, E. Peterson, and J. Ballard. 1990. The surgical management of atypical mycobacterial soft-tissue infections. Surgery 108:36-39. [PubMed] [Google Scholar]
- 136.Reviglio, V., M. L. Rodriguez, G. S. Picotti, M. Paradello, J. D. Luna, and C. P. Juárez. 1998. Mycobacterium chelonae keratitis following laser in situ keratomileusis. J. Refrac. Surg. 14:357-360. [DOI] [PubMed] [Google Scholar]
- 137.Ridell, M., and M. Goodfellow. 1983. Numerical classification of Mycobacterium farcinogenes, Mycobacterium senegalense and related taxa. J. Gen. Microbiol. 129:599-611. [DOI] [PubMed] [Google Scholar]
- 138.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rolston, K. V. I., P. G. Jones, V. Fainstein, and G. P. Bodey. 1985. Pulmonary disease caused by rapidly growing mycobacteria in patients with cancer. Chest 87:503-506. [DOI] [PubMed] [Google Scholar]
- 140.Rootman, D. S., M. S. Insler, and D. E. Wolfley. 1989. Canaliculitis caused by Mycobacterium chelonae after lacrimal intubation with silicone tubes. Can. J. Ophthalmol. 24:221-222. [PubMed] [Google Scholar]
- 141.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Roussel, T. J., W. H. Stern, D. F. Goodman, and J. P. Whitcher. 1989. Postoperative mycobacterial endophthalmitis. Am. J. Ophthalmol. 107:403-406. [DOI] [PubMed] [Google Scholar]
- 143.Rubens, C. E., W. E. Farrar, Jr., Z. A. McGee, and W. Schaffner. 1981. Evolution of a plasmid mediating resistance to multiple antimicrobial agents during a prolonged epidemic of nosocomial infections. J. Infect. Dis. 143:170-181. [DOI] [PubMed] [Google Scholar]
- 144.Runyon, H. 1972. Conservation of the specific epithet fortuitum in the name of the organism known as Mycobacterium fortuitum da Costa Cruz. Int. J. Syst. Bacteriol. 22:50-51. [Google Scholar]
- 145.Sack, J. B. 1990. Disseminated infection due to Mycobacterium fortuitum in a patient with AIDS. Rev. Infect. Dis. 12:961-963. [DOI] [PubMed] [Google Scholar]
- 146.Safranek, T. J., W. R. Jarvis, L. A. Carson, L. B. Cusick, L. A. Bland, J. M. Swenson, and V. A. Silcox. 1987. Mycobacterium chelonae wound infections after plastic surgery employing contaminated gentian violet skin-marking solution. N. Engl. J. Med. 317:197-201. [DOI] [PubMed] [Google Scholar]
- 147.Saluja, A., N. T. Peters, L. Lowe, and T. M. Johnson. 1997. A surgical wound infection due to Mycobacterium chelonae successfully treated with clarithromycin. Dermatol. Surg. 23:539-543. [DOI] [PubMed] [Google Scholar]
- 148.Samuel, J. J., G. Alangaden, and P. H. Chandrasekar. 2000. Cervical adenitis due to Mycobacterium fortuitum in a patient with AIDS. Infect. Dis. Clin. Pract. 9:221-222. [Google Scholar]
- 149.Sanguinetti, M., F. Ardito, E. Fiscarelli, M. La Sorda, P. D'Argenio, G. Ricciotti, and G. Fadda. 2001. Fatal pulmonary infection due to multidrug- resistant Mycobacterium abscessus in a patient with cystic fibrosis. J. Clin. Microbiol. 39:816-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Santamaría-Jaúregui, J., J. Sanz-Hospital, J. Berenguer, D. Muñoz, E. Gómez-Mampaso, and E. Bouza. 1984. Meningitis caused by Mycobacterium fortuitum. Am. Rev. Respir. Dis. 130:136-137. [DOI] [PubMed] [Google Scholar]
- 151.Sarria, J. C., N. B. Chutkan, J. E. Figueroa, and A. Hull. 1998. Atypical mycobacterial vertebral osteomyelitis: case report and review. Clin. Infect. Dis. 26:503-505. [DOI] [PubMed] [Google Scholar]
- 152.Schinsky, M. F., M. M. McNeil, A. M. Whitney, A. G. Steigerwalt, B. A. Lasker, M. M. Floyd, G. C. Hogg, D. J. Brenner, and J. M. Brown. 2000. Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteraemia. Int. J. Syst. E vol. Microbiol. 50:575-581. [DOI] [PubMed] [Google Scholar]
- 153.Schulze-Röbbecke, R., B. Janning, and R. Fischeder. 1992. Occurrence of mycobacteria in biofilm samples. Tubercle Lung Dis. 73:141-144. [DOI] [PubMed] [Google Scholar]
- 154.Sexton, R. R. 1970. Mycobacterium fortuitum infection of the cornea, p. 25-28. In F. M. Polack (ed.), Corneal and external disease of the eye. Charles C Thomas, Springfield, Ill.
- 155.Shafran, S. D., and L. Chui. 2000. Erythema induratum as a form of active cutaneous tuberculosis: case and review. Infect. Dis. Clin. Pract. 9:33-36. [Google Scholar]
- 156.Silcox, V. A., R. C. Good, and M. M. Floyd. 1981. Identification of clinically significant Mycobacterium fortuitum complex isolates. J. Clin. Microbiol. 14:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Smith, M. B., M. C. Boyars, and G. L. Woods. 1996. Fatal Mycobacterium fortuitum meningitis in a patient with AIDS. Clin. Infect. Dis. 23:1327-1328. [DOI] [PubMed] [Google Scholar]
- 158.Smith, R. E., J. J. Salz, R. Moors, D. Silverstein, and W. Lewis. 1980. Mycobacterium chelonei and orbital granuloma after tear duct probing. Am. J. Ophthalmol. 89:139-141. [DOI] [PubMed] [Google Scholar]
- 159.Soini, H., and M. K. Viljanen. 1997. Diversity of the 32-kilodalton protein gene may form a basis for species determination of potentially pathogenic mycobacterial species. J. Clin. Microbiol. 35:769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Springer, B., E. C. Böttger, P. Kirschner, and R. J. Wallace, Jr. 1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 161.Staneck, J. L., P. T. Frame, W. A. Altemeier, and E. H. Miller. 1981. Infection of bone by Mycobacterium fortuitum masquerading as Nocardia asteroides. Am. J. Clin. Pathol. 76:216-222. [DOI] [PubMed] [Google Scholar]
- 162.Stanford, J. L., and W. J. Gunthopre. 1969. Serological and bacteriological investigation of Mycobacterium ranae (fortuitum). J. Bacteriol. 98:375-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stanford, J. L., S. R. Pattyn, F. Portaels, and W. J. Gunthorpe. 1972. Studies of Mycobacterium chelonei. J. Med. Microbiol. 5:177-182. [DOI] [PubMed] [Google Scholar]
- 164.Steele, L. C., and R. J. Wallace, Jr. 1987. Ability of ciprofloxacin but not pipemidic acid to differentiate all three biovariants of Mycobacterium fortuitum from Mycobacterium chelonae. J. Clin. Microbiol. 25:456-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Steingrube, V. A., R. J. Wallace, Jr., L. C. Steele, and D. R. Nash. 1991. Mercuric reductase activity and evidence of broad spectrum mercury resistance among clinical isolates of rapidly growing mycobacteria. Antimicrob. Agents Chemother. 35:819-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stone, M. S., R. J. Wallace, Jr., J. M. Swenson, C. Thornsberry, and L. A. Christensen. 1983. Agar disk elution method for susceptibility testing of Mycobacterium marinum and Mycobacterium fortuitum complex to sulfonamides and antibiotics. Antimicrob. Agents Chemother. 24:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stratford, S., R. Gonzalez-Rothi, M. Lauzardo, and A. Stecenko. 1992. A six-year cross-sectional study of mycobacterial infection in cystic fibrosis. Am. Rev. Respir. Dis. 145:A116. [Google Scholar]
- 169.Subbarao, E. K., M. M. Tarpay, and M. I. Marks. 1987. Soft-tissue infections caused by Mycobacterium fortuitum complex following penetrating injury. Am. J. Dis. Child. 141:1018-1020. [DOI] [PubMed] [Google Scholar]
- 170.Sudesh, S., E. J. Cohen, L. W. Schwartz, and J. S. Myers. 2000. Mycobacterium chelonae infection in a corneal graft. Arch. Ophthalmol. 118:294-295. [DOI] [PubMed] [Google Scholar]
- 171.Swenson, J. M., R. J. Wallace Jr., V. A. Silcox, and C. Thornsberry. 1985. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob. Agents Chemother. 28:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Takewaki, S. I., K. Okuzumi, I. Manabe, M. Tanimura, K. Miyamura, K. I. Nakahara, Y. Yazaki, A. Ohkubo, and R. Nagai. 1994. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int. J. Syst. Bacteriol. 44:159-166. [DOI] [PubMed] [Google Scholar]
- 173.Tebas, P., F. Sultan, R. J. Wallace, Jr., and V. Fraser. 1995. Rapid development of resistance to clarithromycin following monotherapy for disseminated Mycobacterium chelonae infection in a heart transplant patient. Clin. Infect. Dis. 20:443-444. [DOI] [PubMed] [Google Scholar]
- 174.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thibert, L., and S. LaPierre. 1993. Routine application of high-performance liquid chromatography for identification of mycobacteria. J. Clin. Microbiol. 31:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tsang, A. Y., V. L. Barr, J. K. McClatchy, M. Goldberg, I. Drupa, and P. J. Brennan. 1984. Antigenic relationships of the Mycobacterium fortuitum- Mycobacterium chelonae complex. Int. J. Syst. Bacteriol. 34:35-44. [Google Scholar]
- 177.Tsukamura, M. 1966. Mycobacterium chitae, a new species. A preliminary report. Med. Biol. (Tokyo) 73:203-205. (In Japanese.) [PubMed] [Google Scholar]
- 178.Tsukamura, M. 1967. Mycobacterium chitae: a new species. Jpn. J. Microbiol. 11:43-47. [DOI] [PubMed] [Google Scholar]
- 179.Tsukamura, M. 1972. Mycobacterium agri Tsukamura sp. nov. A new relatively thermophilic Mycobacterium. Med. Biol. (Tokyo) 85:153-156. (In Japanese.)
- 179a.Tsukamura, M., H. Nemoto, and H. Yugi. 1983. Mycobacterium porcinum sp. nov., a porcine pathogen. Int. J. Syst. Bacteriol. 33:162-165. [Google Scholar]
- 180.Vadakekalam, J., and M. J. Ward. 1991. Mycobacterium fortuitum lung abscess treated with ciprofloxacin. Thorax 46:737-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Vaneechoutte, M., H. De Beenhouwer, G. Claeys, G. Verschraegen, A. De Rouck, N. Paepe, A. Elaichouni, and F. Portaels. 1993. Identification of Mycobacterium species using amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 31:2061-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Varghese, G., R. Shepherd, P. Watt, and J. H. Bruce. 1988. Fatal infection with Mycobacterium fortuitum associated with esophageal achalasia. Thorax 43:151-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Villaneuva, A., R. V. Calderon, B. A. Vargas, F. Ruiz, S. Aguero, Y. Zhang, B. A. Brown, and R. J. Wallace, Jr. 1997. Report on an outbreak of post-injection abscesses due to Mycobacterium abscessus, including management with surgery and clarithromycin therapy and comparison of strains by random amplified polymorphic DNA polymerase chain reaction. Clin. Infect. Dis. 24:1147-1153. [DOI] [PubMed] [Google Scholar]
- 184.Vonmoos, S., P. H. Leuenberger, V. Beer, and R. de Haller. 1986. Infection pleuro-pulmonaire à Mycobacterium smegmatis. Schweiz. Med. Wochenschr. 116:1852-1856. [PubMed] [Google Scholar]
- 185.Wallace, R. J., Jr. 1989. The clinical presentation, diagnosis, and therapy of cutaneous and pulmonary infections due to the rapidly growing mycobacteria, M. fortuitum and M. chelonae. Clin. Chest Med. 10:419-429. [PubMed] [Google Scholar]
- 186.Wallace, R. J., Jr. 1996. Treatment of infections caused by rapidly growing mycobacteria in the era of the newer macrolides. Res. Microbiol. 147:30-35. [DOI] [PubMed] [Google Scholar]
- 187.Wallace, R. J., Jr., G. Bedsole, G. Sumter, C. V. Sanders, L. C. Steele, B. A. Brown, J. Smith, and D. R. Graham. 1990. Activities of ciprofloxacin and ofloxacin against rapidly growing mycobacteria with demonstration of acquired resistance following single-drug therapy. Antimicrob. Agents Chemother. 34:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wallace, R. J., Jr., and B. A. Brown. 1999. Mycobacterium fortuitum, chelonae, abscessus, p. 372-379. In D. Schlossberg (ed.), Tuberculosis and nontuberculous mycobacterial infections. The W. B. Saunders Co., Philadelphia, Pa.
- 189.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 2:453-490. [DOI] [PubMed] [Google Scholar]
- 190.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr. 2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wallace, R. J., Jr., A. Meier, B. A. Brown, Y. Zhang, P. Sander, G. O. Onyi, and E. C. Böttger. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40:1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wallace, R. J., Jr., B. A. Brown, and G. Onyi. 1991. Susceptibilities of Mycobacterium fortuitum biovar fortuitum and the two subgroups of Mycobacterium chelonae to imipenem, cefmetazole, cefoxitin, and amoxicillin- clavulanic acid. Antimicrob. Agents Chemother. 35:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wallace, R. J., Jr., B. A. Brown, and G. Onyi. 1992. Skin, soft tissue, and bone infections due to Mycobacterium chelonae subspecies chelonae—importance of prior corticosteroid therapy, frequency of disseminated infections, and resistance to oral antimicrobials other than clarithromycin. J. Infect. Dis. 166:405-412. [DOI] [PubMed] [Google Scholar]
- 194.Wallace, R. J., Jr., B. A. Brown, V. A. Silcox, M. Tsukamura, D. R. Nash, L. C. Steele, V. A. Steingrube, J. Smith, G. Sumter, Y. Zhang, and Z. Blacklock. 1991. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J. Infect. Dis. 163:598-603. [DOI] [PubMed] [Google Scholar]
- 195.Wallace, R. J., Jr., B. A. Brown-Elliott, S. C. Ward, C. J. Crist, L. B. Mann, and R. W. Wilson. 2001. Activities of linezolid against rapidly growing mycobacteria. Antimicrob. Agents Chemother. 45:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wallace, R. J., Jr., J. L. Cook, J. Glassroth, D. E. Griffith, K. N. Olivier, and F. Gordin. 1997. American Thoracic Society Statement: diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. Respir. Crit. Care Med. 156:S1-S25. [DOI] [PubMed] [Google Scholar]
- 197.Wallace, R. J., Jr., D. B. Jones, and K. Wiss. 1981. Sulfonamide activity against Mycobacterium fortuitum and Mycobacterium chelonei. Rev. Infect. Dis. 3:898-904. [DOI] [PubMed] [Google Scholar]
- 198.Wallace, R. J., Jr., J. M. Musser, S. I. Hull, V. A. Silcox, L. C. Steele, G. D. Forrester, A. Labidi, and R. K. Selander. 1989. Diversity and sources of rapidly growing mycobacteria associated with infections following cardiac bypass surgery. J. Infect. Dis. 159:708-716. [DOI] [PubMed] [Google Scholar]
- 199.Wallace, R. J., Jr., D. R. Nash, M. Tsukamura, Z. M. Blackock, and V. A. Silcox. 1988. Human disease due to Mycobacterium smegmatis. J. Infect. Dis. 158:52-59. [DOI] [PubMed] [Google Scholar]
- 200.Wallace, R. J., Jr., V. A. Silcox, M. Tsukamura, B. A. Brown, J. O. Kilburn, W. R. Butler, and G. Onyi. 1993. Clinical significance, biochemical features, and susceptibility patterns of sporadic isolates of the Mycobacterium chelonae-like organism. J. Clin. Microbiol. 31:3231-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wallace, R. J., Jr., L. C. Steele, A. Labidi, and V. A. Silcox. 1989. Heterogeneity among isolates of rapidly growing mycobacteria responsible for infections following augmentation mammaplasty despite case clustering in Texas and other southern coastal states. J. Infect. Dis. 160:281-288. [DOI] [PubMed] [Google Scholar]
- 202.Wallace, R. J., Jr., J. M. Swenson, and V. A. Silcox. 1985. The rapidly growing mycobacteria: characterization and susceptibility testing. Antimicrob. Newsl. 2:85-92. [Google Scholar]
- 203.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, and M. G. Bullen. 1985. Treatment of non-pulmonary infections due to Mycobacterium fortuitum and Mycobacterium chelonei on the basis of in vitro susceptibilities. J. Infect. Dis. 152:500-514. [DOI] [PubMed] [Google Scholar]
- 204.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, and R. C. Good. 1982. Disk diffusion testing with polymyxin and amikacin for differentiation of Mycobacterium fortuitum and Mycobacterium chelonei. J. Clin. Microbiol. 16:1003-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, R. C. Good, J. A. Tschen, and M. S. Stone. 1983. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5:657-679. [DOI] [PubMed] [Google Scholar]
- 206.Wallace, R. J., Jr., D. Tanner, P. J. Brennan, and B. A. Brown. 1993. Clinical trial of clarithromycin for cutaneous (disseminated) infection due to Mycobacterium chelonae. Ann. Intern. Med. 119:482-486. [DOI] [PubMed] [Google Scholar]
- 207.Wallace, R. J., Jr., Y. Zhang, B. A. Brown, V. Fraser, G. H. Mazurek, and S. Maloney. 1993. DNA large restriction fragment patterns of sporadic and epidemic nosocomial strains of Mycobacterium chelonae and Mycobacterium abscessus. J. Clin. Microbiol. 31:2697-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wayne, L. G., and G. P. Kubica. 1986. Mycobacteria, p. 1435-1457. In P. H. A. Sneath (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 209.Wayne, L. G., and H. A. Sramek. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin. Microbiol. Rev. 5:1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wenger, J. D., J. S. Spika, R. W. Smithwick, V. Pryor, D. W. Dodson, G. A. Carden, and K. C. Klontz. 1990. Outbreak of Mycobacterium chelonae infection associated with use of jet injectors. JAMA 264:373-376. [PubMed] [Google Scholar]
- 211.Wheeler, P. W., D. Lancaster, and A. B. Kaiser. 1989. Bronchopulmonary cross-colonization and infection related to mycobacterial contamination of suction valves of bronchoscopes. J. Infect. Dis. 159:954-58. [DOI] [PubMed] [Google Scholar]
- 212.Wilson, R. W., V. A. Steingrube, E. C. Böttger, B. Springer, B. A. Brown- Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks, and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. E vol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 213.Wolinsky, E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107-159. [DOI] [PubMed] [Google Scholar]
- 214.Wolinsky, E. 1992. Mycobacterial diseases other than tuberculosis. Clin. Infect. Dis. 15:1-12. [DOI] [PubMed] [Google Scholar]
- 215.Wolinsky, E., and T. K. Rynearson. 1968. Mycobacteria in soil and their relation to disease-associated strains. Am. Rev. Respir. Dis. 97:1032-1037. [DOI] [PubMed] [Google Scholar]
- 216.Woods, G. L., J. S. Bergmann, F. G. Witebsky, G. A. Fahle, B. Boulet, M. Plaunt, B. A. Brown, R. J. Wallace, Jr., and A. Wanger. 2000. Multisite reproducibility of Etest for susceptibility testing of Mycobacterium abscessus, Mycobacterium chelonae, and Mycobacterium fortuitum. J. Clin. Microbiol. 38:656-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Woods, G. L., J. S. Bergmann, F. G. Witebsky, G. A. Fahle, A. Wanger, B. Boulet, M. Plaunt, B. A. Brown, and R. J. Wallace, Jr. 1999. Multisite reproducibility of results obtained by the broth microdilution method for susceptibility testing of Mycobacterium abscessus, Mycobacterium chelonae, and Mycobacterium fortuitum. J. Clin. Microbiol. 37:1676-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Woods, G. L., B. A. Brown-Elliott, E. P. Desmond, G. S. Hall, L. Heifets, G. E. Pfyffer, M. R. Plaunt, J. C. Ridderhof, R. J. Wallace, Jr., N. G. Warren, and F. G. Witebsky. 2000. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; tentative standard 20:M24-T2, 2nd ed. NCCLS, Wayne, Pa. [PubMed]
- 219.Yakrus, M. A., S. M. Hernandez, M. M. Floyd, D. Sikes, W. R. Butler, and B. Metchock. 2001. Comparison of methods for identification of Mycobacterium abscessus and M. chelonae isolates. J. Clin. Microbiol. 39:4103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yew, W. W., S. Y. L. Kwan, P. C. Wong, and J. Lee. 1990. Ofloxacin and imipenem in the treatment of Mycobacterium fortuitum and Mycobacterium chelonae lung infections. Tubercle 71:131-133. [DOI] [PubMed] [Google Scholar]
- 221.Yew, W., P. Wong, H. Woo, C. Yip, C. Chan, and F. Cheng. 1993. Characterization of Mycobacterium fortuitum isolates from sternotomy wounds by antimicrobial susceptibilities, plasmid profiles, and ribosomal ribonucleic acid gene restriction patterns. Diagn. Microbiol. Infect. Dis. 17:111-117. [DOI] [PubMed] [Google Scholar]
- 222.Zahid, M. A., S. A. Klotz, E. Goldstein, and W. Bartholomew. 1994. Mycobacterium chelonae (M. chelonae subspecies chelonae): report of a patient with a sporotrichoid presentation who was successfully treated with clarithromycin and ciprofloxacin. Clin. Infect. Dis. 18:999-1001. [DOI] [PubMed] [Google Scholar]
- 223.Zenone, T., A. Boibieux, S. Tigaud, J.-F. Fredenucci, V. Vincent, and D. Peyramond. 1998. Nontuberculous mycobacterial tenosynovitis: report of two cases. Clin. Infect. Dis. 26:1467-1468. [DOI] [PubMed] [Google Scholar]
- 224.Zhang, Y., M. Rajagopalan, B. A. Brown, and R. J. Wallace, Jr. 1997. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J. Clin. Microbiol. 35:3132-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang, Y., R. J. Wallace Jr., V. A. Steingrube, B. A. Brown, D. R. Nash, A. Silcox, and M. Tsukamura. 1992. Isoelectric focusing patterns of β-lactamases in the rapidly growing mycobacteria. Tubercle Lung Dis. 73:337-344. [DOI] [PubMed] [Google Scholar]
- 226.Zolg, J. W., and S. P. Schulz. 1994. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J. Clin. Microbiol. 32:2801-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]


