Abstract
Francisella tularensis is the etiological agent of tularemia, a serious and occasionally fatal disease of humans and animals. In humans, ulceroglandular tularemia is the most common form of the disease and is usually a consequence of a bite from an arthropod vector which has previously fed on an infected animal. The pneumonic form of the disease occurs rarely but is the likely form of the disease should this bacterium be used as a bioterrorism agent. The diagnosis of disease is not straightforward. F. tularensis is difficult to culture, and the handling of this bacterium poses a significant risk of infection to laboratory personnel. Enzyme-linked immunosorbent assay- and PCR-based methods have been used to detect bacteria in clinical samples, but these methods have not been adequately evaluated for the diagnosis of pneumonic tularemia. Little is known about the virulence mechanisms of F. tularensis, though there is a large body of evidence indicating that it is an intracellular pathogen, surviving mainly in macrophages. An unlicensed live attenuated vaccine is available, which does appear to offer protection against ulceroglandular and pneumonic tularemia. Although an improved vaccine against tularemia is highly desirable, attempts to devise such a vaccine have been limited by the inability to construct defined allelic replacement mutants and by the lack of information on the mechanisms of virulence of F. tularensis. In the absence of a licensed vaccine, aminoglycoside antibiotics play a key role in the prevention and treatment of tularemia.
INTRODUCTION
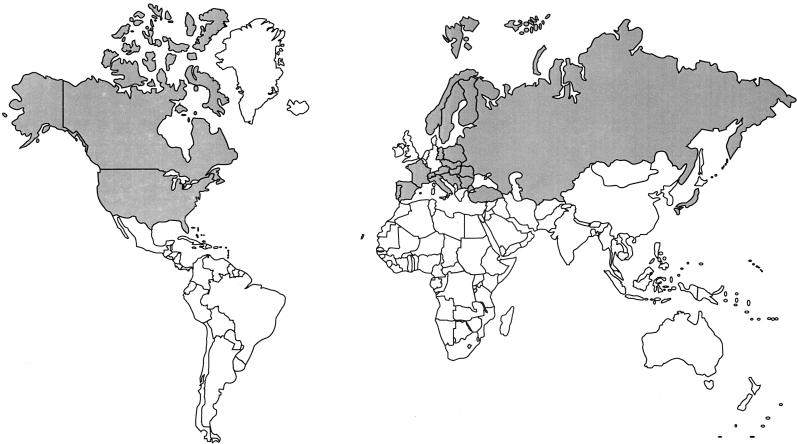
In 1911, an outbreak of a plague-like disease in rodents in Tulare County, California, provided the first isolation of a small gram-negative bacterium which was named Bacterium tularensis (115). Subsequently, natural infections with Francisella tularensis have been reported in a range of vertebrates including mammals, birds, amphibians, and fish, and even in invertebrates (122). Tularemia occurs only in the northern hemisphere and most frequently in Scandinavia, northern America, Japan, and Russia (15, 17, 126, 166, 175). However, tularemia has recently been reported from Turkey, Yugoslavia, Spain, Kosovo, and Switzerland (4, 11, 143, 183), indicating that tularemia is even more widely distributed than was previously thought (Fig. 1).
FIG. 1.
World map showing areas where F. tularensis is endemic (shaded).
In many parts of the world, colloquial names such as rabbit fever, hare fever, deerfly fever, and lemming fever have been used to describe the disease (122). Tularemia is not a disease which is notifiable to the World Health Organization, and the worldwide incidence of disease is not known. Nevertheless, some data on the incidence of disease are available. In Sweden, the annual number of reported cases of disease in humans over the period from 1973 to 1985 ranged from less than 5 cases to over 500 (122). In Japan 1,355 cases of tularemia were reported over the period 1924 to 1987 (126). In Turkey, 205 cases were reported over the period 1988 to 1998 (75), while in Slovakia 126 cases of disease were reported during the period 1985 to 1994 (73).
In some parts of the world, the number of cases of tularemia has declined markedly during the twentieth century: in the United States, the annual number of reported cases of tularemia declined from several thousand in the 1930s to several hundred in the 1980s (17, 65). It is likely that these figures are a gross underrepresentation of the true incidence of tularemia, because many infections are not diagnosed as a consequence of the relatively benign nature of disease caused by some strains of F. tularensis and by some difficulties in laboratory testing.
F. tularensis has been an organism of concern as a biological threat agent since the large state-funded biological weapons programs of the 1950s, when the United States first evaluated the organism as a biological weapon, and it was subsequently incorporated into weapons by the U.S.S.R. (reviewed in reference 113). Now that the emphasis has shifted towards defending against biological terrorism, public health and medical management protocols following a release of tularemia have been reviewed (5, 31).
EPIDEMIOLOGY
F. tularensis is thought to be maintained in the environment principally by various terrestrial and aquatic mammals such as ground squirrels, rabbits, hares, voles, muskrats, water rats, and other rodents (15, 17, 122, 175). In regions where tularemia is endemic, antibodies to F. tularensis are frequently detected in the sera of trapped wild animals (63, 106, 107, 121). Outbreaks of disease in humans often parallel outbreaks of tularemia in wild animals. For example, in Sweden a clear correlation between peaks in vole and hare populations and outbreaks of tularemia in humans have been reported (175), and outbreaks of tularemia in humans in the former Soviet Union have been linked to epizootics of disease in ground voles (122). However, it is not clear whether these animal species are the true reservoir of the bacterium in the environment.
A wide range of arthropod vectors have also been implicated in the transmission of tularemia between mammalian hosts. In central Europe, the ticks Dermacentor reticulatus and Ixodes ricinus are important vectors. In areas of the Czech Republic and Austria where natural foci of tularemia occur, between 2.1 and 2.8% of D. reticulatus ticks analyzed contained F. tularensis (84). In the United States, biting flies are the most common vectors in Utah, Nevada, and California (17, 122), while ticks are the most important vectors east of the Rocky Mountains. In the former Soviet Union, the bacterium is transmitted by both mosquitoes (Aedes, Culex, and Anopheles species) and the Ixodes species of tick (122). Such arthropod vectors play a role both in the transmission of the disease within wild animal populations and in the transmission of disease to humans. It therefore follows that rural populations, especially individuals who spend time in endemic areas such as farmers, hunters, walkers, and forest workers, are most at risk of contracting tularemia (76, 108, 170).
There is evidence that the bacterium can persist in watercourses, possibly in association with amoebae (15). Beavers and muskrats in North America and lemmings and beavers in Scandinavia might also play a role in the maintenance of the bacterium in watercourses (16, 121, 122). During a prolonged period in water (140 days in the study reported), the number of directly culturable bacterial cells declined to below detectable levels (49). However, on the basis of rhodamine 123 staining, which is indicative of metabolic activity, at least 30% of these cells were judged to be viable. Although this finding suggests that F. tularensis can persist in a viable but nonculturable form in water, the bacteria were not able to cause disease when injected into mice (49). These findings might have implications for techniques which rely on the direct culture of F. tularensis to identify bacteria in watercourses.
TAXONOMY OF F. TULARENSIS
The taxonomic position of F. tularensis is complex and has changed frequently. F. tularensis was originally included in the genus Bacterium, later in the Pasteurella genus, and subsequently provisionally placed in the Brucella genus (131, 184). In 1947, the proposal was made that the bacterium should be the sole member of a new genus called Francisella (35).
The most recent issue of Bergey's Manual of Systematic Bacteriology (40) indicates that F. tularensis and F. novicida are the two species in the genus Francisella. F. novicida has been differentiated on the basis of inability to produce acid from sucrose, the relative ease of culture, and the lack of virulence for humans and rabbits (40). However, several workers have since questioned whether F. novicida should be considered a separate species. F. novicida is now known to be capable of causing a tularemia-like illness in humans, and on the basis of DNA hybridization, F. novicida was not distinguishable from F. tularensis (80). More recently, the 16S ribosomal DNA (rDNA) sequences of F. tularensis and F. novicida have been shown to have a high degree of sequence similarity (99.6%) (50). On the basis of these studies, both Hollis et al. (80) and Forsman et al. (50) proposed that F. novicida should be considered a subspecies of F. tularensis.
More recently, a detailed analysis of several strains of Yersinia philomiragia revealed their fatty acid compositions to be typical of the genus Francisella (80). In addition, the DNA from these strains showed a high degree of relatedness to F. tularensis DNA (80). These observations indicated that Y. philomiragia should be included in the Francisella genus and renamed Francisella philomiragia (80). This proposal is supported by the finding that the 16S rDNA sequences of F. tularensis and F. philomiragia confirm both its placement in the Francisella genus and its identity as a separate species (50). F. philomiragia strains can also be distinguished from F. tularensis because the former are oxidase positive in the Kovacs modification of this test, are often able to hydrolyze gelatin, and are relatively easily cultured. In addition, F. philomiragia is considered virulent only in immunocompromised individuals or in those who have recently had a near-drowning experience (80, 180).
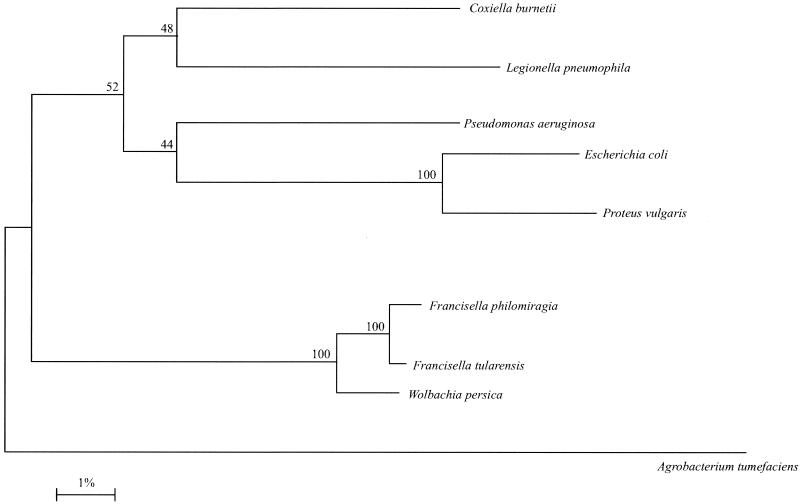
Therefore, the current consensus position appears to be that F. tularensis and F. philomiragia are the sole members of the Francisella genus within the Francisellaceae family. The 16S rDNA sequences of the members of this family contain the consensus sequence for members of the γ-subclass of Proteobacteria (Fig. 2). This family also includes Wolbachia persica (50), a parasite of arthropods and worms, the Dermacentor andersoni symbiont, which was originally isolated from Rocky Mountain wood ticks (124), and Ornithodorous moubata symbiont B (125). Most of these arthropod endosymbionts are poorly characterized. However, W. persica is known to exist in the Malpighian tubules of the soft tick Argas arboreus (125) and is not considered a pathogen of mammals.
FIG. 2.
Evolutionary distance tree, based on 16S rDNA sequences, showing the relationship of F. tularensis with other putative members of the Francisellaceae and with other closely related members of the γ subclass of the Proteobacteria. Agrobacterium tumefaciens was included as an outgroup. Reproduced from Forsman et al. (50) with the kind permission of the International Union of Microbiological Societies.
For F. tularensis, several subspecies other than novicida have been proposed (Table 1). Originally, F. tularensis strains were identified as belonging to either subsp. tularensis (also known as type A or subspecies nearctica) or subspecies palaearctica (also know as type B or subspecies holarctica) principally on the basis of virulence, citrulline ureidase activity (conversion of l-citrulline to ornithine), and acid production from glycerol (40, 128, 130). Subspecies holarctica is now the most widely used terminology in place of subspecies palaearctica (129). A more detailed description of the properties of subsp. tularensis and subspecies holarctica can be found in Olsufjev and Mesheryakova (130). Three biovars of F. tularensis subspecies holarctica have been suggested (130); biovar I (erythromycin sensitive) (100), biovar II (erythromycin resistant) (100), and biovar japonica (146).
TABLE 1.
Taxonomy and characteristics of the subspecies of F. tularensis
| Current terminology | Previous names | Biovara | 16s rRNA genotypeb | Geographic locationa | Citrulline ureidase activity | Glycerol fermentation | Glucose fermentation | Sensitivity to erythromycin |
|---|---|---|---|---|---|---|---|---|
| Francisella tularensis subsp. tularensis | Francisella tularensis A; Francisella tularensis subsp. nearartica | NA | A | Primarily N. America | +c | + | + | + |
| Francisella tularensis subsp. holarctica | Francisella tularensis B; Francisella tularensis subsp. palaearctica | I | B | Primarily Europe, Siberia, Far East, Kazakhstan, and N. America | −c | − | + | + |
| II | B | Primarily Eurasia | − | − | + | − | ||
| Japonica | A | Japan | − | + | + | + | ||
| Francisella tularensis subsp. mediaasiatica | NA | A | Primarily central Asia and some parts of the former USSR | + | + | −d | + | |
| Francisella novicida | Francisella tularensis subsp. novicida | NA | NR | Primarily N. America | NR | NR | + | NR |
Strains of F. tularensis subsp. tularensis are considered the most virulent for humans, with an infectious dose of less than 10 CFU and a mortality of 5 to 6% in untreated cases of cutaneous disease (36, 46, 130). F. tularensis strain Schu S4 is the proposed subsp. tularensis type strain (130). Strains of subspecies holarctica do not cause disease in rabbits, and the mortality rate associated with cutaneous disease in humans is less than 0.5% (40, 130). Until recently, F. tularensis subsp. tularensis was thought to be found only in North America. However, the recent isolation of this subspecies in Europe suggests that it may be more widely distributed than was previously thought (74). F. tularensis subspecies holarctica is found mainly in North America and in Eurasia (40, 123, 130). A fourth subspecies, F. tularensis subsp. mediaasiatica, is found predominantly in the central Asian republics of the former USSR (2, 40). These strains possess citrulline ureidase activity and are able to ferment glycerol, but are less virulent than strains of F. tularensis subsp. tularensis in rabbits (130, 151).
GENOMIC ANALYSIS OF F. TULARENSIS
The genetic makeup of F. tularensis is poorly understood, with sequences of only 34 genes deposited with GenBank. Two different cryptic plasmids (pOM1 and pNFL10) are reportedly found in the live vaccine strain (LVS) and in F. novicida, respectively (132) (GenBank accession AFO55345). This paucity of information has prompted two projects to sequence the F. tularensis genome. The genome sequence of a strain of F. tularensis subsp. tularensis (strain Schu S4) is currently being determined by a consortium of laboratories in Europe and the United States (94, 138). A project to sequence the genome of LVS, originally derived from a virulent F. tularensis subsp. holarctica strain, has recently commenced in the United States.
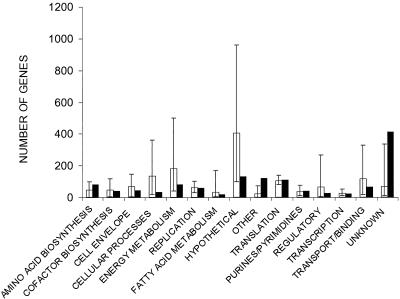
Preliminary results from the strain Schu S4 genome sequencing project suggest a total genome size of <2 Mbp, making this one of the smaller bacterial genomes (94). Strain Schu S4 does not appear to possess either plasmid pOM1 or pNFL10 (94). The G+C content of the genome is approximately 34% (94, 138). A preliminary annotation has identified 1,804 candidate open reading frames which are characterized by a slight shift in G+C content. Of these, 1,289 are thought to encode proteins, of which 413 had no database match (138), meaning that in comparison with many other bacterial genomes, F. tularensis contains a high proportion of unique genes (Fig. 3). On the basis of database matches with known proteins, genes encoding putative transport/binding, gene regulation, energy metabolism, and cellular processes appeared to be underrepresented in the F. tularensis genome (Fig. 3) (138).
FIG. 3.
Preliminary annotation of the F. tularensis strain Schu S4 genome sequence; comparison of the number of F. tularensis genes in 15 functional categories (solid bars). The mean number of genes in these categories in 20 other bacterial species is shown as open bars, with error bars indicating the lowest and highest numbers of genes in each category. Reproduced from Prior et al. (138) with the kind permission of Blackwell Scientific Limited.
DETECTION AND DIAGNOSIS
F. tularensis is a fastidious organism which requires enriched medium for growth. Traditionally, cysteine glucose blood agar has been the growth medium of choice. However, enriched chocolate agar (cysteine heart agar supplemented with 9% heated sheep red blood cells [CHAB]) and nonselective buffered charcoal yeast extract agar also support the growth of the organism and may be used for isolation (5, 65). The Centers for Disease Control and Prevention guidelines recommend the use of CHAB once growth on the general microbiological agars, such as sheep blood agar, chocolate agar, and Thayer-Martin agar, which are routinely used in U.S. laboratories for the isolation of bacteria from clinical specimens, indicates the pathogen to be present (5).
A heavy inoculum on appropriate medium will yield visible growth in 18 h, but the appearance of individual colonies may require 2 to 4 days of incubation (33). F. tularensis grows slowly at 37°C and poorly at 28°C, and this can be exploited to distinguish F. tularensis from Yersinia pestis, F. philomiragia, and F. tularensis subsp. novicida, all of which grow well at 28°C (5). On CHAB, colonies are 2 to 4 mm in size, greenish-white, round, smooth, and slightly mucoid, while on media containing whole blood there is usually a small zone of alpha-hemolysis surrounding colonies (5, 65). Gram staining of cultured material reveals small (0.2 to 0.5 μm by 0.7 to 1.0 μm), mainly single gram-negative coccobacilli which stain weakly. Chemically defined media capable of supporting the growth of F. tularensis do exist, but growth on such media is slower and the colonies are smaller than on traditional rich agars.
F. tularensis does not grow well in liquid media even when the medium is supplemented with cysteine and requires a large inoculum to obtain visible growth within 24 h. Clinical laboratories routinely inoculate clinical specimens into broth, usually brain heart infusion or Trypticase soy broth, for the recovery of bacteria. However, F. tularensis requires enriched media for growth, supplemented with cysteine. The organism has been cultivated in modified Mueller-Hinton broth and thioglycollate broth. In Mueller-Hinton broth, the addition of 0.025% ferric pyrophosphate appeared to enhance the growth of F. tularensis. Growth in liquid media is slow, requiring up 3 to 7 days of incubation if the broth is shaken or a minimum of 10 days in unshaken broth to produce visible growth. In static thioglycollate broth, growth is seen first as a dense band near the top which diffuses throughout the broth as growth progresses (5, 12, 181). The most widely used synthetic medium used for the growth of tularemia was devised by Chamberlain (22). However, defined media are not used for routine diagnosis of tularemia.
Direct isolation of bacteria is frequently achieved from ulcer scrapings, lymph node biopsies, or sputum (65). The organism is rarely cultured directly from blood, although this is becoming feasible with the development of sensitive blood culture systems (139, 141, 174). The isolation of bacteria from urine or feces is not frequently done (65), but antigen detection in urine and RNA hybridization of wound specimens have been reported (72, 172). In stained tissue sections, bacteria may be found both intra- and extracellularly (5, 65). Enzyme-linked immunosorbent assay (ELISA)-based tests can also be used to detect the bacteria in clinical samples. For example, a capture ELISA using monoclonal antibodies against F. tularensis lipopolysaccharide (LPS) recognized all strains of F. tularensis tested other than those of subspecies novicida, with no cross-reactivity with other bacterial species tested. The sensitivity was 103 CFU/ml in phosphate-buffered saline (PBS) and 104 CFU/ml in spiked human sera (72).
An immunochromatographic hand-held assay has also been developed based on a polyclonal and a monoclonal antibody to LPS of F. tularensis LVS; the detection limits of this assay were 106 CFU/ml PBS and 106 to 107 CFU/ml in spiked human sera. This assay is principally designed for field use, being compact and easy to use and giving results in 15 min, but the relatively low sensitivity means that a negative result does not exclude tularemia (72).
Because of the difficulty in culturing F. tularensis, most cases of tularemia are diagnosed on the basis of clinical picture and/or serology (23, 103). Serological tests for the diagnosis of F. tularensis infection are attractive because diagnostic work involving culture procedures carries a risk of infection for nonvaccinated laboratory staff (20, 88). The diagnosis of human cases of tularemia is usually confirmed by the demonstration of an antibody response to F. tularensis, which occurs about 2 weeks after the onset of the disease (96). The detection of serum antibodies is most frequently achieved by agglutination or an ELISA (21, 96, 171).
A latex agglutination test commercially available from BBL (Becton Dickinson, Franklin Lakes, N.J.) has been used by some workers to identify individuals with antibodies to F. tularensis: reactions at dilutions greater than 1:20 are considered specific and significant. Using this test, 2% of trappers in Quebec were found to be seropositive (at serum dilutions of 1:20 to 1:2,048) for F. tularensis (108). The only association between this population and the development of antibodies appeared to be the trapping of muskrats. Commercially available antigens can also be used with standard tube agglutination tests. A fourfold increase during illness or a single titer of 1:160 or greater is considered diagnostic (18, 65). However, while serological tests are frequently used for diagnosis, strains of F. tularensis have occasionally been isolated which fail to agglutinate commercially available F. tularensis antigens (23).
A range of PCR-based assays have been reported for the detection of F. tularensis or for the diagnosis of tularemia. These PCR assays have all used primers directed against genes encoding outer membrane proteins such as fopA (60) or the 17-kDa outer membrane lipoprotein (72, 91). These PCR assays offer high specificity, failing to generate an amplicon using template DNA from a range of other bacterial pathogens (60, 162). When used with pure cultures of F. tularensis as the source of template DNA, the assays also offer high levels of sensitivity. The PCR assay reported by Grunow et al. allowed the detection of 102 CFU/ml in PBS (72), and the use of a nested PCR assay allowed the detection of 1 CFU of F. tularensis (60). However, samples such as blood contain compounds capable of inhibiting the PCR, making detection of such low levels of bacteria in clinical samples impossible. In addition, blood samples invariably require some form of processing to allow high-sensitivity PCR tests to be carried out (60, 72, 162). Nevertheless, the PCR assay reported by Grunow et al. was shown to detect 103 to 104 CFU/ml of spiked serum (72), while the nested PCR assay reported by Fulop et al. was able to detect 102 CFU/ml of spiked blood (60).
Even in their current forms, these assays do appear to offer advantages over the direct culture of bacteria. In mice, 83% of blood samples taken 24 h after experimental infection were positive by PCR, while bacteria could be cultured from only 48% of the samples (91). This advantage appears to extend to clinical samples: in a study with swab samples taken from the lesion site in 40 human cases of ulceroglandular tularemia, a PCR assay was positive for 73% of the samples, whereas bacteria were cultured from only 25% of the samples (162). Moreover, PCR identified F. tularensis in one sample which had not been identified by culture or serology in a parallel study (162).
Subsequently, a number of other workers have reported the use of PCR assays for the diagnosis of tularemia (34, 93) or for the analysis of environmental samples (16). These PCR-based tests might also be safer than tests which involve the culture of bacteria (162). Improved methods for the isolation and processing of tissue or blood samples and for the transport of samples to the laboratory might further improve the sensitivity and utility of the PCR test (88, 162). Indeed, a specially formulated filter paper designed for sample collection has been proposed for the rapid preparation of template DNA from clinical or field-collected tick vectors (72). When linked with a TaqMan 5′ nuclease assay or a PCR-enzyme immunoassay, sensitivities of <100 CFU were reported (77).
While PCR assays have allowed the rapid detection of a range of F. tularensis strains, other workers have developed molecular methods to discriminate between Francisella strains (50, 85). PCR-based methods have been evaluated for their potential to identify F. tularensis and discriminate between the different subspecies. Long random sequence oligonucleotide primers and primers specific for repetitive extragenic palindromic (REP) sequences and enterobacterial repetitive intragenic consensus (ERIC) sequences were evaluated (140).
REP-PCR has been applied to specifically identifying strains of F. tularensis subsp. novicida, but patterns from subspecies holarctica and tularensis were found to be similar (89). A one-base difference in the 16S rRNA sequences of F. tularensis subsp. tularensis and F. tularensis subsp. holarctica has been demonstrated, and on this basis a PCR has been developed which can differentiate the two subspecies (50). In one study, PCR analyses based on the use of ERIC, REP or long random sequence primers yielded reproducible banding patterns of similar complexity and allowed differentiation of strains at the subspecies level, but the methods investigated do not meet the criteria for typing of individual isolates (89). More recent studies suggest that PCR amplification of tandem repeat regions may be highly discriminatory and a useful tool in strain typing (48, 90). A specific PCR has been developed which produces amplicons of different lengths (target unknown), which has been used in combination with the 17-kDa lipoprotein PCR and can distinguish F. tularensis subsp. holarctica from strains of other F. tularensis subspecies (89).
CLINICAL DISEASE
Tularemia in humans can occur in several forms, depending to a large extent on the route of entry of the bacterium into the body. Although tularemia can be a severely debilitating disease, especially when caused by F. tularensis subsp. tularensis, many cases of disease caused by lower-virulence strains are undiagnosed. The most common form of the disease is ulceroglandular tularemia, which usually occurs as a consequence of a bite from an arthropod vector which has previously fed on an infected animal (127, 176). Some cases of ulceroglandular tularemia occur in hunters and trappers as a consequence of the handling of infected meat, with infection via cuts or abrasions.
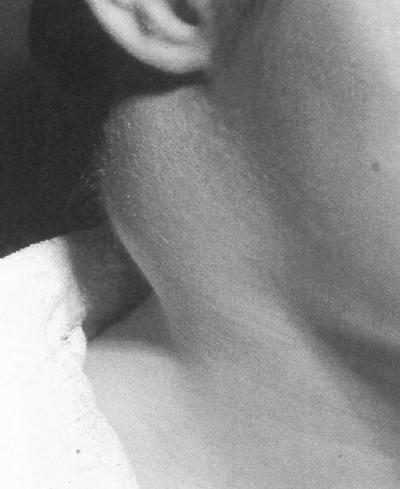
After an incubation period of typically 3 to 6 days (47), the patient experiences the sudden onset of flu-like symptoms, especially chills, fever, headache, and generalized aches (126). An ulcer, which can persist for several months, forms at the site of infection, usually on the lower limbs in the cases of tick-borne disease (a disease with similar symptoms but without the appearance of an ulcer is termed glandular tularemia). Bacteria are disseminated from this site via the lymphatic system to regional lymph nodes. The enlargement of these lymph nodes (Fig. 4) often resembles the classical bubo associated with bubonic plague. From this site, bacteria may be disseminated to other tissues such as the spleen, liver, lungs, kidneys, intestine, central nervous system, and skeletal muscles. However, the bacteremic phase of the infection is transient and occurs relatively early in the infection process.
FIG. 4.
Enlarged lymph node in a tularemia patient.
Recovery from the disease can be protracted, but the ulceroglandular form of tularemia is rarely fatal (the mortality rate from ulceroglandular tularemia is typically less than 3% [47]). Even without treatment, ulceroglandular and glandular tularemia are rarely fatal, but may take a significant length of time to resolve. On the other hand, an acute form of the disease produced by F. tularensis subsp. tularensis, typhoidal tularemia, is typified as a septicemia without lymphadenopathy or the appearance of an ulcer and carries a mortality rate of 30 to 60% (47, 64). In addition to the symptoms described above, the patient may be delirious and shock may develop.
A rare variation of ulceroglandular disease is oculoglandular tularemia, where the conjunctiva is the initial site of infection, usually as a result of the transfer of bacteria on the fingertips (165). The disease is marked by the appearance of ulcers and nodules on the conjunctiva, and without treatment the infection spreads to the local lymph nodes
The ingestion of infected foodstuffs (14, 166) or of bacteria in drinking water (14, 16, 75, 175) can result in oropharyngeal or gastrointestinal tularemia, depending on the site of colonization of host tissues. The former is often described as a painful sore throat with enlargement of the tonsils and the formation of a yellow-white pseudomembrane. This is most often accompanied by swollen cervical lymph nodes (143). Depending on the infecting dose, gastrointestinal tularemia ranges from mild but persistent diarrhea to an acute fatal disease with extensive ulceration of the bowel.
Probably the most acute form is disease is associated with the inhalation of bacteria, although pneumonic disease can also occur as a complication of ulceroglandular, glandular, oculoglandular, or oropharyngeal tularemia (64). The clinical and roentgenographic features of pneumonic tularemia are quite variable, making diagnosis difficult (64). On occasion, pneumonic tularemia may even occur without any overt signs of pneumonia. The occasional naturally occurring cases of inhalation tularemia often arise from farming activities which involve the handling (and the subsequent generation of dusts) of hay which has previously been the site of residence of infected rodents. (79, 166, 170, 175).
ANIMAL MODELS
The pathogenesis of the LVS strain in mice has been used as a model to study the behavior of intracellular pathogens. A study published in 1946 compared the sensitivities of various animal models to strain Schu S4. It was shown that Schu S4 was fully virulent in the mouse, guinea pig, rabbit, hamster, and cotton rat. The last was shown to have a high degree of individual variation in response to Schu S4, and accordingly this model has received little attention. Strains from all of the subspecies of F. tularensis are reported to be virulent in the murine and guinea pig models of disease, but only strains of F. tularensis subsp. tularensis are considered virulent in the rabbit model of disease (Table 2).
TABLE 2.
Virulence of F. tularensis biotypesa
| Biotype | 50% lethal dose, CFU (reference[s])
|
|||
|---|---|---|---|---|
| Humans | Mice | Guinea pigs | Rabbits | |
| Francisella tularensis subsp. tularensis | <10 (74) | <10 (100) | <10 (100) | 1-10 (100, 130, 151) |
| Francisella tularensis subsp. holarctica | <103 (40) | <1 (100) | <10 (100) | >106 (100, 130) |
| Francisella tularensis subsp. mediaasiatica | NR | NR | NR | >106 (151) |
| Francisella tularensis subsp. novicida | >103 (40) | <103 (40) | <103 (40) | >103 (40) |
All doses were given subcutaneously. NR, not reported.
Most studies on the behavior of F. tularensis in vivo rely on the infection of inbred mice, and this murine tularemia system has become a model to study the behavior of intracellular pathogens. There is some evidence that strains with differing virulence can be differentiated by using this model. For example, strain Schu S4 is fully virulent regardless of the route of administration. However, the virulence of LVS has been shown to be dependent on the route of delivery; LVS is fully virulent in the mouse, with a median lethal dose of <10 CFU when delivered intraperitoneally, but is attenuated in the mouse when delivered intradermally (42).
PATHOGENESIS AND HOST RESPONSES TO INFECTION
In typical human ulceroglandular tularemia, a skin lesion first appears at the site of infection 3 to 5 days after infective exposure (126). During this initial phase, T cells appear to play little role in combating infection. The systemic administration of tumor necrosis factor or gamma interferon (IFN-γ) is capable of reducing the severity of tularemia (52), indicating the key role these cytokines play in response to infection (41). During the early stages of disease, studies with other pathogens have suggested that keratinocytes are a likely source of tumor necrosis factor, while natural killer (NK) cells may produce IFN-γ (101, 182). A transient bacteremia occurs, during which the pathogen must resist lysis by complement. Protection appears to be due to the presence of capsule, as a nonencapsulated mutant was susceptible to killing by nonimmune serum (150).
The bacteremic phase allows the organism to be seeded throughout the body, infecting all reticuloendothelial tissues. Within days, a wide range of cytokines are expressed in the reticuloendothelial tissues. In the liver, for example, the bacteria are able to invade and multiply within hepatocytes (24), and tumor necrosis factor alpha, interleukin-10, interleukin-12, and IFN-γ are produced within 48 h (66). However, although this cytokine response, assumed to evoke the expansion of a protective Th1 response, was capable of mounting a defense against the LVS strain, it was unable to prevent a more virulent strain from mounting a lethal infection in mice (66, 68). Infection also induces a stress response in infected host cells; hsp72 was induced in peritoneal exudate cells of infected mice which were susceptible to tularemia for 3 days following infection (167).
Later during infection, T cells appear to play a major role in protection. Mice depleted of CD4+ and/or CD8+ T cells were capable of controlling a primary LVS infection, but were not able to resolve it (25), indicating a scenario of complex interactions of different cell types in primary infection. Studies with knockout mice infected with strain LVS confirm these observations, but CD4−, β2-microglobulin-deficient/CD8−, and γδ T-cell receptor-negative mice were all able to resolve an infection (185). However, αβ T-cell receptor-negative knockout mice succumbed to infection, indicating that while either CD4 or CD8 T cells are individually sufficient to resolve infection with strain LVS, αβ T-cell receptor cells are required for protection.
In mice challenged with a high-virulence strain (Schu S4), both CD4 and CD8 T cells appeared to play key roles in controlling disease (62). Circulating γδ T cells are well known for controlling bacterial intracellular infections; for example, they are essential in controlling Listeria monocytogenes infections (78). A significant expansion in the Vγ9Vδ2 T-cell subpopulation was observed in a Japanese patient with tularemia (168). This effect was subsequently shown to occur within a week of infection, but could persist for over a year (99, 137). This response appears to be due to exposure to phosphoantigens expressed by F. tularensis in vivo (137). Phosphoantigens are powerful stimuli for Vγ9Vδ2 cells, as has been shown for Mycobacterium tuberculosis and Plasmodium falciparum (57). Interestingly, vaccination with strain LVS did not result in an expansion of the Vγ9Vδ2 T cells similar to that observed in tularemia patients. Neutrophils also appear to play a role in defense by ingesting and killing microorganisms, lysing infected hepatocytes and acting as a source of cytokines (25, 161).
Different strains of mice show different degrees of susceptibility to infection, indicating that resistance involves multiple genetic loci (6). One locus which appears to play a role in the natural resistance to primary infection is Bcg (Nramp1) (98). Expression of this allele has many pleiotropic effects associated with activation of macrophages by IFN-γ or LPS, and mutation of the allele can confer susceptibility to infection by a range of intracellular pathogens (112), including F. tularensis (98).
The pathogenicity of intracellular bacteria depends on their ability to survive within macrophages, although other cell types such as hepatocytes may play an important but as yet poorly defined role (24). As such, most of the work on the cellular interaction of F. tularensis has concentrated on the macrophage.
INTERACTION WITH HOST CELLS
Although a facultative intracellular organism in vitro, F. tularensis has been described as an “obligate intracellular pathogen of macrophages in vivo” (52). Early work showed the organism multiplied in murine macrophages (7), guinea pig hepatic cells, endothelium (28), and gut endothelial cells isolated from ticks (58). In various artificial in vitro culture systems, such as chick embryos (19), HeLa cells (156), and mouse fibroblasts (119), intracellular multiplication has also been demonstrated. That the organism is an intracellular pathogen in vivo as well as capable of intracellular growth in vitro is supported by the similarity of the histopathology of tularemia to that seen in infections produced other intracellular pathogens such as M. tuberculosis (reviewed in reference 173). In addition, resistance to highly virulent strains of F. tularensis is dependent on lymphoid tissue rather than serum (97), further supporting the case that F. tularensis is an intracellular pathogen.
F. tularensis enters macrophages using a cytochalasin B-insensitive pathway without triggering the respiratory burst (52). However, opsonized F. tularensis has been shown to be actively phagocytosed by polymorphonuclear leukocytes, which are capable of killing the bacteria by oxidative killing mechanisms (110, 111). A protein, AcpA, has been identified in F. tularensis which has an acid phosphatase function (142). AcpA is capable of inhibiting the respiratory burst more efficiently than previously described acid phosphatases of other intracellular pathogens, such as Leishmania donovani and Legionella micdadei (144, 148). Bacteria live within the macrophage in a phagosome which does not fuse with lysosomes (7), but acidification of the phagosome does occur and is essential for growth of F. tularensis and acquisition of iron (53).
In some animals, nitric oxide (NO) production is produced by macrophages to limit infections by intracellular bacterial pathogens (reviewed in reference 29). NO production by the infected host appears to have a nonspecific role in protection against F. tularensis infection (71). Phase variation of F. tularensis LPS has been observed, and different forms of the LPS appeared to affect NO induction, thus modulating the innate immune response (29). Unlike peritoneal macrophages (9, 51), NO has been shown not to be involved in killing of F. tularensis by alveolar macrophages (136), which can behave differently from other resident macrophage populations (reviewed in reference 136). Alveolar macrophages activated by IFN-γ were able to kill F. tularensis, and this killing was resistant to inhibitors of NO production, although the inhibition of growth correlated with nitrite production by the cell. Cytokines known to regulate the effector functions of activated macrophages (tumor necrosis factor alpha, interleukin-10, transforming growth factor beta 1, and IFN-α) also did not affect the IFN-γ-induced killing by alveolar macrophages (136), indicating that the IFN-γ-induced responses of alveolar and peritoneal macrophages are fundamentally different.
Bacteria must modify gene expression to survive in hostile environments.
F. tularensis was shown to upregulate the expression of four proteins during growth in a macrophage cell line (69). Compared to the stress response induced in other intracellular pathogens such as Salmonella enterica serovar Typhimurium (1), Legionella pneumophila (102), and M. tuberculosis (105), F. tularensis demonstrates a very low level of response on encountering an adverse environment. Although DnaK, GroEL, and GroES have all been shown to be upregulated in vitro in response to exposure to heat and hydrogen peroxide stress (45), these were not upregulated in the macrophage (69).
The four proteins which showed increased levels of expression relative to broth-grown cells had molecular masses of 20, 23, 55, and 70 kDa. Subsequent separation of protein extracts by two-dimensional gels revealed the 23-kDa protein to have a pI of 5.8, but the other three proteins of interest appeared to have pI values outside the range of 4 to7 and thus were not visible on the gels (69). The 23-kDa protein had no amino acid sequence homology with any known protein, and its function has not been elucidated to date, although it appears to play a role in response to stress, as it is upregulated under conditions of oxidative stress but not heat shock (69).
One operon shown to be essential for growth of F. novicida inside macrophages is mglAB (13), which is also present in F. tularensis (94). MglAB shows high similarity to the SspAB of Escherichia coli and is therefore likely to be a transcriptional regulator. In E. coli, SspAB regulates the expression of a range of proteins in response to nutritional stress, and in F. novicida inactivation of the operon resulted in a change in expression of a number of proteins; most notably, four proteins of 33, 38, 20, and 70 kDa were absent in the mutant compared to the parent strain (13). Phenotypic assays also indicated that MglAB may regulate expression of at least one exported phosphatase, a group of proteins which includes AcpA.
Another locus described as necessary for survival in macrophages is minD (10). MinD is a 29-kDa protein and thus is not one of those described above that are upregulated intracellularly. Two roles for MinD have been proposed based on possible actions of the first-described minD in E. coli. Initially, the E. coli MinD was suggested to have a role in septum formation during cell division, but subsequently homology with a heavy-metal ion pump indicated an alternative role. Thus, Anthony et al. proposed that the MinD in F. tularensis may be essential for intracellular growth either because it acts as a pump for toxic or radical ions or because abnormal septum formation in the minD mutant results in loss of cell wall integrity, allowing bactericidal agents into the cell (10).
After multiplication within the macrophage, F. tularensis induces cell death by apoptosis (104). This releases the bacteria from the cell, allowing infection of fresh cells. Interestingly, relatively large numbers of bacteria were required to be present within the macrophage before apoptosis could occur and required a longer time from infection to induction of apoptosis compared to Salmonella, Shigella, Yersinia, and Legionella species. This probably reflects the slow growth rate and the obligate intracellular lifestyle of F. tularensis in vivo.
VIRULENCE DETERMINANTS
Few virulence factors have been identified for F. tularensis. However, studies with F. novicida and the availability of genome sequence data for F. tularensis (94, 138) may help to dissect the pathogenic mechanisms of this enigmatic organism. One of the problems in analyzing the contribution of a specific gene to virulence is that isogenic mutants in F. tularensis have not yet been produced, although methods have been described for producing mutants in F. novicida (8). Transposon mutagenesis has been employed successfully and has been used to identify the genes mentioned above involved in macrophage survival and growth, but so far this technology has not been used to specifically target virulence factors.
The capsule, although essential for serum resistance (150, 164), is not required for survival following phagocytosis by polymorphonuclear leukocytes (150). Noncapsular mutants possess higher neuraminidase activity than capsular wild-type strains (133). The reason for this is not known, but the authors proposed a role for neuraminidase in colonization, as the enzyme was active in degrading natural mucins but not glycoproteins.
The LPS of F. tularensis does not exhibit the properties of a classical endotoxin. It fails to induce interleukin-1 from mononuclear cells and only poorly induces tumor necrosis factor and NO production from macrophages (2, 152). The inability of F. tularensis LPS to antagonize a range of endotoxin-induced cellular responses seen with most LPS molecules indicates that F. tularensis LPS does not interact with LPS receptors (2). LPS from F. tularensis has been shown to undergo phase variation, which affects both antigenicity due to variations in the O antigen and the NO response of macrophages due to variation in the lipid A moiety. The phase variation of the lipid A has been demonstrated to affect the organism's ability to grow intracellularly. In one phase, reduced NO induction results in bacterial growth, while in another phase, increased NO production suppresses growth (29). This growth restriction was observed only in rat macrophages and not in mouse macrophages (29).
Studies on F. novicida have also shown a role for LPS in macrophage growth. The valA gene encoded an ABC transporter possibly required for transport of LPS to the outer membrane (117), and mutants defective in this gene were unable to grow in macrophages and showed an increased susceptibility to serum killing (118).
As can be seen, very few classical virulence factors have been identified for this pathogen. Some of these, such as secreted toxins, are not produced by the organism (163), while others await discovery. For example, tularemia can be contracted by drinking contaminated water (14), but how the organisms invade from the gut to produce infection is not known and no invasin has been identified for this pathogen. Probing the genome sequence data will help to identify putative virulence factors such as adhesins, but the inability to create specific allelic replacement mutants will delay research into their role until this problem is overcome.
DEVELOPMENT OF LIVE TULAREMIA VACCINES
Initial efforts to develop a live attenuated tularemia vaccine began in the former Soviet Union prior to the Second World War. Attenuation of strains was achieved either by repeatedly subculturing fully virulent strains in media supplemented with antiserum or by drying the strains (95). In 1934, El'bert et al. inoculated animals with a weakly virulent tularemia culture. Protection was demonstrated when these immunized animals were challenged with a virulent culture, and it was suggested that the same immunization procedure might be applicable to humans (178). Strain Moscow was reported to show weakened virulence and high immunogenicity and was used as a live vaccine in humans in 1942. The effectiveness of vaccination was successfully demonstrated in volunteers, and several thousand individuals were reportedly vaccinated before the strain was apparently lost (178). Strain 15 was later identified and shown to have reduced virulence in guinea pigs while retaining virulence for mice. These strains were administered subcutaneously into humans in clinical trials, and it was concluded at this time that vaccination of humans with attenuated F. tularensis strains was harmless (178). In subsequent years, a number of mass vaccination programs using strain 15 were carried out in areas of the former Soviet Union where tularemia outbreaks were prevalent. As many as 60 million individuals were immunized with live vaccine preparations in the former Soviet Union until 1960 (160).
In subsequent years it was shown that strain 15 had become so attenuated that it was no longer virulent in mice (135). The strain was passaged in animals and a variant, strain 15 restored, was derived. Another attenuated vaccine, strain 155, was also developed at this time, and both strains were produced as live vaccines at the Gamaleya Institute in Moscow. These strains were transferred to the United States in 1956 (153). However, cultures grown from reconstituted ampoules showed that both vaccine strains segregated into the two colony types, designated blue colony variant or grey colony variant depending on their appearance when viewed microscopically under oblique light. The blue colony variant was shown to be more virulent and immunogenic in small animals than the grey colony variant (36).
Mice immunized with the blue colony variant vaccine were protected against subsequent challenge with the fully virulent strain Schu S4. Guinea pigs immunized with the blue colony variant showed increased resistance to challenge. Lyophilized preparations of the blue colony variant were prepared, and a live vaccine strain (LVS) was derived after five passages through mice. The LVS appeared to be an effective vaccine, protecting immunized mice and guinea pigs against an inhalation challenge with F. tularensis strain Schu S4 (36).
These studies were sufficiently encouraging to warrant an extension of studies to humans. It was shown that clinical tularemia could be induced in nonvaccinated individuals by inhalation of approximately 10 to 50 CFU (116). Treatment of infected volunteers occurred at the earliest indication of systemic disease, employed either streptomycin or tetracycline, and resulted in the complete recovery of infected individuals. In a subsequent study, 18 volunteers were vaccinated with LVS prior to an inhalation challenge with strain Schu S4. Whereas 8 of 10 controls showed evidence of infection, only 3 of 18 LVS-immunized individuals showed evidence of infection (154). It was concluded from this study that immunization with LVS induced significant protection against respiratory challenge with F. tularensis (154, 155).
A subsequent investigation involved volunteers who had been immunized with LVS approximately 1 year before challenge by the aerosol route with strain Schu S4. The challenge dose ranged from 200 to 20,000 CFU. Results showed that the majority of vaccinees challenged with up to 2,000 organisms escaped clinical illness. Immunized volunteers challenged with 20,000 organisms showed modified disease symptoms compared to nonimmunized volunteers infected with a similar dose of Schu S4 (39).
Several routes of immunization with F. tularensis LVS have been evaluated over the past 40 years. Airborne administration of LVS was investigated in nonhuman primates and guinea pigs in order to enhance the immunity provided by the live tularemia vaccine (37, 38) and was sufficiently encouraging for aerogenic vaccination of humans to be initiated. Initial studies demonstrated that aerogenic immunization of animals gave at least comparable protection against tularemia infection as dermal immunization. When these studies were repeated in humans, there was an apparent greater level of protection after respiratory immunization compared to the conventional intradermal method of administration (82). Similarly, oral administration of high doses of F. tularensis LVS was reported to induce protection against aerosol challenge (83).
In the United States, the initial batches of the LVS vaccine were produced by the National Drug Company in 1959. In 1977, a retrospective study was published that showed the effectiveness of the LVS vaccine in the prevention of laboratory-acquired tularemia (20). The period from 1950 to 1959, prior to LVS vaccination, was compared with the period 1960 to 1969, during which LVS vaccination was used routinely. Figures showed that the incidence of typhoidal tularemia fell from 5.7 to 0.27 cases per 1,000 at-risk employees. The incidence of ulceroglandular tularemia remained unchanged in the two periods, but the clinical signs and symptoms of this form of the disease were moderated in vaccinated individuals.
An application to license the LVS vaccine was submitted to the Federal Drug Administration. In this instance, LVS was derived from NDBR 101, lot 9, and grown under fermentation conditions. The new vaccine contained fewer of the immunogenic blue colony variant types than the parent strain and also had a higher residual moisture content (153). The immunogenicity of the new vaccine lot was evaluated in 19 human volunteers after administration by scarification (179). Tests were conducted to evaluate humoral and cell-mediated immune responses in volunteers between 7 and 63 days after immunization. Immune responses were assayed by ELISA and lymphocyte proliferation techniques. Results showed that by day 63, a positive IgG, IgA, and IgM response towards an ether-extracted antigen was evident in 100% of volunteers. Lymphocyte proliferation assays using the same antigen demonstrated a positive response in 40% of volunteers 7 days after immunization, and 80% of volunteers gave a positive result by day 63.
In the early 1960s, the vaccine was approved by the Food and Drug Administration only for use in clinical trials under investigational new drug status. However, some properties of the LVS vaccine may give cause for concern and make licensing difficult. For example, the protective response induced by the vaccine has not been characterized. In addition, the basis of attenuation of the LVS strain is not known, and studies to examine the virulence of LVS in the mouse model have shown that the strain is fully virulent when delivered intraperitoneally, with a median lethal dose of less than 10 CFU. However, when delivered intradermally, the strain is avirulent in the mouse. The median lethal dose for LVS after intravenous and subcutaneous delivery is reported to be approximately 103 and 105 CFU, respectively. However, these results were derived after repeated passage through mice to increase the virulence of the strain. Studies in mice with ampoule-derived LVS that has not been animal passaged have shown the median lethal doses to be 105 CFU after intravenous administration and 107 CFU when delivered subcutaneously.
The LVS vaccine remains the only effective vaccine against tularemia developed to date. However, this vaccine is not currently available, though work to fully license this vaccine is under way in the United States. The finding that an attenuated mutant of F. tularensis can induce protective immunity suggests that this approach to vaccine development is feasible. In a range of other pathogens, the introduction of defined mutations into genes required for growth of the pathogen in vivo has yielded safe and effective vaccines. The construction of a defined attenuated mutant of F. tularensis could provide a safe, effective, and licensable tularemia vaccine. The aromatic amino acid and purine biosynthesis pathways have already been identified from genome sequence information as targets for the construction of a defined attenuated mutant (94, 138). However, the utility of this approach is limited because, as outlined in a previous section of this review, work to date has failed to devise methods for the construction of allelic replacement mutants of F. tularensis.
DEVELOPMENT OF NONLIVING TULAREMIA VACCINES
Prior to the development of the LVS vaccine, immunologically based therapies against tularemia were reported in the 1930s by Lee Foshay, who suggested that immune serum could be administered to favorably modify the clinical course of tularemia in humans. This finding stimulated Foshay to work to develop a killed tularemia vaccine that induced humoral immunity (54, 55). A number of techniques were employed to prepare the killed bacterial cells, including heating, acetone, and phenol treatment, and the Foshay vaccines were administered to human volunteers with variable results. There were reports of lesions at the site of administration and that the killed vaccine caused severe local reactions when administered to immune humans. These reactions were reduced somewhat if the vaccine was prepared with acetone rather than phenol killing of bacterial cells. Subsequently, vaccination was preceded by a simple skin test to determine whether immunity was present, and the vaccine dose regimen was modified accordingly to limit severe reactions (56).
The efficacy of such killed whole-cell vaccines appears to be questionable. The phenol-killed vaccine was able to protect nonhuman primates against very low systemic challenges with F. tularensis strain Schu S4, although immunized nonhuman primates were as readily infected as controls (27). When tested in mice, the Foshay vaccine afforded a similarly low level of protection against virulent strains. However, although these animal studies suggest that killed whole-cell vaccines induced only low levels of protection against disease, studies in humans indicated that immunization with these vaccines reduced the number of infections and considerably modified the course of the disease (92).
Administration of the LVS vaccine has been demonstrated to induce a variable cell-mediated immune response in humans (173). The nature of the protective response to tularemia is generally believed to be T-cell mediated (173). Previous studies with killed vaccines generated a predominantly humoral immune response that was nonprotective and was believed to have failed to generate a sufficient cell-mediated immune response.
An ongoing strategy towards subunit vaccine development has been to identify those antigens of LVS that are capable of inducing a protective immune response. Up to 23 cytoplasmic and envelope antigens have been identified in F. tularensis LVS (81). The next step was to identify whether any of the identified antigens could individually elicit the activation of T cells (81).
Cell-mediated immune reactions can be readily demonstrated by the lymphocyte stimulation test, and in 1987 a study was published that investigated T-lymphocyte stimulation by membrane proteins from F. tularensis (149). It had previously been shown that in LVS-vaccinated individuals, lymphocytes reacted with protein antigens and antibodies were produced against carbohydrate antigens present on the capsule. Protein antigens that elicited T-cell reactivity in LVS-immunized humans were investigated by using the capsule-deficient mutant of LVS. Several polypeptide antigens were identified, and four major ones were purified, estimated at 61, 37, 32, and 17.5 kDa. It was believed that the latter two proteins were present on the bacterial surface. All four polypeptides caused lymphocyte proliferation in cells from vaccinated individuals.
Subsequently, the cell-mediated immune response to LVS of individuals immunized by natural infection with F. tularensis was examined (157). Several membrane polypeptides of LVS were recognized, including the 40-kDa protein as well as the four polypeptide antigens identified in 1987. Among the conclusions of this study was that the polypeptides relevant to the inducement of cell-mediated immune are well conserved in the live vaccine strain. There was high immunological specificity of these proteins, because T cells from nonimmunized and uninfected individuals did not show evidence of proliferation against them.
Additionally, Surcel and coworkers (169) identified two heat-modifiable proteins of 17 and 40 kDa. The 17-kDa protein showed strong T-cell proliferative activity in subjects immunized with LVS, whereas the 40-kDa protein did not induce proliferation of T cells, although a strong antibody response to it was evident. A similarity between this 40-kDa protein and the OmpA-like outer membrane proteins in other bacterial species was also suggested. The significance of the lack of T-cell proliferation was explained because OmpA proteins have very little alpha-helical structure in contrast to major T-cell epitopes, which are alpha-helical peptides. A further study investigated whether immunization with another protein, FopA, could induce a protective response in the mouse model of infection (61). The protein was delivered in Salmonella cells to enhance the cell-mediated immune response, and although a strong immune response was generated, there was no evidence of protection against an LVS challenge.
In addition to FopA, another membrane component has been the focus of a number of studies. LPS from F. tularensis is reported to have low endotoxicity in the Limulus amoebocyte lysate assay in comparison to LPS from other bacterial species (152). Studies in mice have demonstrated that immunization with LPS purified from LVS offers protection against both LVS and Schu S4 challenge (41, 59, 62). This protection has been shown to be antibody mediated, as demonstrated by serum passive transfer experiments. This protective role for LPS identifies it as one possible component of a subunit vaccine.
Analyses of immune responses after LVS immunization have identified the 17-kDa membrane protein as a suitable subunit candidate because of its strong T-cell proliferative activity. A DNA fragment containing two genes, one of which encoded the 17-kDa protein, was cloned into an attenuated S. enterica serovar Typhimurium strain (158). After oral immunization with the construct, mice were challenged with LVS. Immunized mice showed lower viable counts of LVS in tissues after challenge compared to controls, and it was suggested that this might involve a T-cell-mediated mechanism. Further studies with this construct showed that the 17-kDa protein-mediated protection was not as high as LVS-mediated protection in the mouse model (159). Incorporation of the 17-kDa protein into immunostimulatory complexes again elicited a strong immune response but did not confer any protection against LVS infection (67).
Although a subunit tularemia vaccine that offers protection against challenge with a fully virulent strain has not yet been identified, progress towards identifying protective antigens has been made. Sequencing of the F. tularensis strain Schu S4 genome will facilitate the identification of protective antigens through bioinformatics. Analysis of the immune response to the LVS vaccine has shown a heterogeneity of immunogenic epitopes recognized in humans, and this indicates it is likely that a subunit vaccine will be composed of a number of protective antigens to provide protection against virulent strains.
CHEMOTHERAPY
The aminoglycosides streptomycin and gentamicin are bactericidal against F. tularensis and are currently the drugs of choice for the treatment of tularemia infections (31, 44, 177). Alternative therapies have been proposed, although generally there is a lack of supporting clinical data (120, 134). The fluoroquinolones have been shown to have good bactericidal activity against F. tularensis in in vitro systems (86, 109, 114), and both ciprofloxacin and doxycycline have been found to be effective in treating F. tularensis infection in mice (26, 32, 147).
In a recent human epidemic outbreak in Spain, ciprofloxacin was the antibiotic with the lowest level of therapeutic failure and with the fewest side effects (134). Ciprofloxacin was also shown to be suitable for the treatment of tularemia in children (87) and in a case where relapse was evident after initial gentamicin therapy (145). The efficacy of levofloxacin has also been demonstrated in the treatment of two immunocompromised patients diagnosed with tularemia (109).
Tetracycline and chloramphenicol are bacteriostatic against F. tularensis and have been used to treat tularemia. However, treatment failures have been associated with these antibiotics, although the chances of relapse are reduced with longer treatment regimens (31). In a study of in vitro susceptibility of F. tularensis strains isolated from humans and animals, all isolates tested were found to be resistant to beta-lactams and azithromycin (86). Also, evidence of in vitro activity for ceftriaxone against F. tularensis did not correlate to successful treatment when used clinically (30, 44). The ketolide telithromycin has been shown to be bactericidal for F. tularensis both in axenic medium and within a cell culture system and is known to be highly active against other intracellular pathogens, including Chlamydia and Legionella spp. (114)
Since there is no effective control of this disease in nature, public awareness of the ubiquitous presence of this organism and the potential for human infection should be maintained. In areas where tularemia is endemic, the handling of dead or moribund animals should be avoided, and the possibility of insect bites should be reduced. The chlorination of municipal drinking water has virtually eliminated epidemics from that source, but untreated water should be considered when other routes are not evident (65).
CONCLUSIONS
In spite of the clinical significance of F. tularensis, little is known about the genetic makeup of the bacterium or its mechanisms of pathogenicity. While it is known that the bacterium can invade a range of cell types, and it is clear that the bacterium is primarily an intracellular pathogen, the mechanisms allowing invasion and growth within host cells are not known. This lack of information is hindering research aimed at developing improved vaccines against tularemia.
One obvious starting point for these investigations would be to determine the mechanisms of attachment to host cells. The availability of genome sequence information should facilitate the identification of candidate adhesins and pili. Second, the ability of F. tularensis to survive and grow within the endocytic vacuoles of macrophages appears to be critical for its lifestyle. The mechanisms by which this pathogen prevents phagosome-lysosome fusion should be addressed. The completion of the ongoing genome sequencing projects will certainly provide a wealth of information, and the challenge for the future will be to interpret this information. Microarrays should allow this information to be fully exploited, for example, by identifying genes which are upregulated in macrophages. However, it is unlikely that significant progress will be made in understanding the molecular basis of virulence unless efficient methods for the construction of defined allelic replacement mutants can be devised. The development of such methods therefore remains one of the highest priorities for workers in this field.
Equally intriguing is the relationship between Francisella strains of differing virulence and between F. tularensis and the related arthropod endosymbionts such as W. persica. Are these arthropod endosymbionts, with a reduced host range and limited metabolic ability, possible ancestors of F. tularensis, which has acquired additional DNA sequences? Alternatively, the arthropod endosymbionts may have evolved from F. tularensis and their constrained lifestyles might reflect genome downsizing. The ongoing genome sequencing projects and the use of microarrays to probe the genetic makeup of F. tularensis strains belonging to different subspecies should allow these questions to be answered. However, again it will be difficult to prove the relationship between genetic makeup and virulence in these different subspecies without the ability to generate defined allelic replacement mutants.
Although only limited genetic information is available, it has been very effectively exploited to provide modern methods for the detection of F. tularensis and for the diagnosis of tularemia. However, in spite of the availability of these tools, we are still not sure of the life cycle of the bacterium or the true reservoirs of the bacterium in the environment. In this respect, the association with amoebae is intriguing and certainly merits further attention. Similarly, the finding that the bacterium can enter a viable but nonculturable state should be investigated further. For example, can such bacteria be recovered after multiple passages in animals? The availability of genome sequence information is now likely to provide methods for strain typing and for detailed epidemiological analyses which might support these studies.
The LVS vaccine is known to be effective in preventing tularemia in humans, and since an improved vaccine is likely to be at least a decade away, additional studies should be undertaken to characterize this vaccine, with a view towards licensure. In parallel, work should continue to attempt to identify protective subunits (this work will also serve to support licensing if the LVS vaccine). In the absence of an effective vaccine, antibiotics are the only available treatment or therapy, and the need to monitor for the appearance of antibiotic-resistant variants strains remains critical.
Acknowledgments
The image shown in Fig. 4 was kindly provided by Arne Tarnvik, University of Umea, Umea, Sweden.
REFERENCES
- 1.Abshire, K. Z., and F. C. Neidhardt. 1993. Analysis of protein synthesised by Salmonella typhimurium during growth within a host macrophage. J. Bacteriol. 175:3734-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aikimbaev, M. A. 1966. Taxonomy of genus Francisella. Rep. Acad. Sci. Kaz. SSR Ser. Biol. 5:42-44. [Google Scholar]
- 3.Ancuta, P., T. Pedron, R. Girard, G. Sandstrom, and R. Chaby. 1996. Inability of the Francisella tularensis lipopolysaccharide to mimic or to antagonize the induction of cell activation by endotoxins. Infect. Immun. 64:2041-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2000. Tularemia, Kosovo. Wkly. Epidemiol. Rec. 75:133-134. [PubMed] [Google Scholar]
- 5.Anonymous. 2001. Basic laboratory protocols for the presumptive identification of Francisella tularensis. Centers for Disease Control and Prevention, Atlanta, Ga.
- 6.Anthony, L. S. D., E. Skamene, and P. A. L. Kongshaven. 1988. Influence of genetic background on host resistance to experimental murine tularemia. Infect. Immun. 56:2089-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony, L. S. D., R. D. Burke, and F. E. Nano. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59:3291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anthony, L. S. D., M. Gu, S. C. Cowley, W. W. S. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 137:2697-2703. [DOI] [PubMed] [Google Scholar]
- 9.Anthony, L. S. D., P. J. Morrisey, and F. E. Nano. 1992. Growth inhibition of Francisella tularensis live vaccine strain by IFN-gamma-activated macrophages is mediated by reactive nitrogen intermediates derived from l-arginine metabolism. J. Immunol. 148:1829-1832. [PubMed] [Google Scholar]
- 10.Anthony, L. S. D., S. C. Cowley, K. E. Mdluli, and F. E. Nano. 1994. Isolation of a Francisella tularensis mutant that is sensitive to serum and oxidative killing and is avirulent in mice: correlation with the loss of MinD homologue expression. FEMS Microbiol. Lett. 124:157-166. [DOI] [PubMed] [Google Scholar]
- 11.Bachiller Luque, P., J. L. Perez Castrillon, M. Martin Luquero, F. J. Mena Martin, J. de la Lama Lopez-Areal, P. Perez Pascual, M. A. Mazon, and V. Herreros Guilarte. 1998. Preliminary report of an epidemic tularemia outbreak in Valladolid. Rev. Clin. Esp. 198:789-793. [PubMed] [Google Scholar]
- 12.Baker, C. N., D. G. Hollis, and C. Thornsberry. 1985. Antimicrobial susceptibility testing of Francisella tularensis with a modified Mueller-Hinton broth. J. Clin. Microbiol. 22:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29:247-259. [DOI] [PubMed] [Google Scholar]
- 14.Bell, J. F. 1977. Tularemia — a review. CRC Handbook Series in Zoonoses, Section A, p. 161. CRC Press, Boca Raton, Fla.
- 15.Berdal, B. P., R. Mehl, N. K. Meidell, A.-M. Lorentzen-Styr, and O. Scheel. 1996. Field investigations of tularemia in Norway. FEMS Immunol. Med. Microbiol. 13:191-195. [DOI] [PubMed] [Google Scholar]
- 16.Berdal, B. P., R. Mehl, H. Haaheim, M. Loksa, R. Grunow, J. Burans, C. Morgan, and H. Meyer. 2000. Field detection of Francisella tularensis. Scand. J. Infect. Dis. 32:287-291. [DOI] [PubMed] [Google Scholar]
- 17.Boyce, J. M. 1975. Recent trends in the epidemiology of tularemia in the United States. J. Infect. Dis. 131:197-199. [DOI] [PubMed] [Google Scholar]
- 18.Brown, S. L., F. T. McKinney, G. C. Klein, and W. L. Jones. 1980. Evaluation of a safranin-O-stained antigen microagglutination test for Francisella tularensis antibodies. J. Clin. Microbiol. 11:146-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buddingh, G. J., F. C. Womack. 1941. Observations on the infection of chick embryos with Bacterium tularense, Brucella, and Pasteurella pestis. J. Exp. Med. 74:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burke, D. S. 1977. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J. Infect. Dis. 135:55-60. [DOI] [PubMed] [Google Scholar]
- 21.Carlsson, H. E., A. A. Lindberg, G. Lindberg, B. Hederstedt, K. A. Karlsson, and B. O. Agell. 1979. Enzyme-linked immunosorbent assay for immunological diagnosis of human tularemia. J. Clin. Microbiol. 10:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chamberlain, R. E. 1965. Evaluation of a live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarridge, J. E. I., T. J. Raich, A. Sjostedt, G. Sandstrom, R. O. Darouiche, R. M. Shawar, P. R. Georghiou, C. Osting, and L. Vo. 1996. Characterization of two unusual clinically significant Francisella strains. J. Clin. Microbiol. 34:1995-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conlan, J. W., and R. J. North. 1992. Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect. Immun. 60:5164-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conlan, J. W., A. Sjostedt, and R. J. North. 1994. CD4+ and CD8+ T-cell-dependent and -independent host defense mechanisms can operate to control and resolve primary and secondary Francisella tularensis LVS infection in mice. Infect. Immun. 62:5603-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conley, J., H. M. Yang, T. Wilson, K. Blasetti, V. DiNinno, G. Schnell, and J. P. Wong. 1997. Aerosol delivery of liposome-encapsulated ciprofloxacin: aerosol characterization and efficacy against Francisella tularensis infection in mice. Antimicrob. Agents Chemother. 41:1288-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coriell, L. L., E. O. King, and M. G. Smith. 1948. Studies on tularemia IV. Observations on tularemia in control and vaccinated monkeys. J. Immunol. 58:183-202. [PubMed] [Google Scholar]
- 28.Councilman, W. T., and R. P. Strong. 1921. Plague-like infections in rodents. Trans. Assoc. Am. Physicians 36:135-143. [Google Scholar]
- 29.Cowley, S. C., S. V. Myltseva, and F. E. Nano. 1996. Phase variation in Francisella tularensis affecting intracellular growth, lipopolysaccharide antigenicity and nitric oxide production. Mol. Microbiol. 20:867-874. [DOI] [PubMed] [Google Scholar]
- 30.Cross, J. T., and R. F. Jacobs. 1993. Tularemia — treatment failures with outpatient use of ceftriaxone. Clin. Infect. Dis. 17:976-980. [DOI] [PubMed] [Google Scholar]
- 31.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, A. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon-medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 32.D'iakov, S. I., V. P. Bubnov, I. K. Lebedeva, S. V. Sidorenko, and V. I. Moskalenko. 2000. The chemotherapeutic efficacy of ciprofloxacin and lomefloxacin in the inhalation model infecting white mice with tularemia. Antibiot. Khimioter. 45:17-20. [PubMed] [Google Scholar]
- 33.Doern, G. V. 2000. Detection of selected fastidious bacteria. Clin. Infect. Dis. 30:166-173. [DOI] [PubMed] [Google Scholar]
- 34.Dolan, S. A., C. B. Dommaraju, and G. B. DeGuzman. 1998. Detection of Francisella tularensis in clinical specimens by use of polymerase chain reaction. Clin. Infect. Dis. 26:764-765. [DOI] [PubMed] [Google Scholar]
- 35.Dorofe'ev, K. A. 1947. Classification of the causative agent of tularemia. Symp. Res. Works Inst. Epidemiol. Mikrobiol. Chita. 1:170-180. [Google Scholar]
- 36.Eigelsbach, H. T., and C. M. Downs. 1961. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87:415-425. [PubMed] [Google Scholar]
- 37.Eigelsbach, H. T., and J. J. Tulis. 1961. Aerogenic immunization of the monkey and guinea pig with live tularemia vaccine. Proc. Soc. Exp. Biol. Med. 108:732-734. [DOI] [PubMed] [Google Scholar]
- 38.Eigelsbach, H. T., J. J. Tulis, M. H. McGavran, and J. D. White. 1962. Live tularemia vaccine. I. Host-parasite relationship in monkeys vaccinated intracutaneously or aerogenically. J. Bacteriol. 84:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eigelsbach, H. T., R. B. Hornick, and J. J. Tulis. 1967. Recent studies on live tularemia vaccine. Med. Ann. Dist. Columbia 36:282-286. [PubMed] [Google Scholar]
- 40.Eigelsbach, H. T., and V. G. McGann. 1984. Genus Francisella Dorofe'ev 1947, 176AL. p. 394-399. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Williams and Wilkins Co., Baltimore, Md.
- 41.Elkins, K. L., T. Rhinehart-Jones, C. A. Nacy, R. K. Winegar, and A. H. Fortier. 1993. T-cell-independent resistance to infection and generation of immunity to Francisella tularensis. Infect. Immun. 61:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elkins, K. L., T. R. Rhinehart-Jones, S. J. Culkin, D. Yee, and R. K. Winegar. 1996. Minimal requirements for murine resistance to infection with Francisella tularensis LVS. Infect. Immun. 64:3288-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Enderlin, G., L. Morales, R. F. Jacobs, and J. T. Cross. 1994. Streptomycin and alternative agents for the treatment of tularemia: review of the literature. Clin. Infect. Dis. 19:42-47. [DOI] [PubMed] [Google Scholar]
- 45.Ericsson, M., A. Tarnvik, K. Kuoppa, G. Sandstrom, and A. Sjostedt. 1994. Increased synthesis of DnaK, GroEL, and GroES homologs by Francisella tularensis LVS in response to heat and hydrogen peroxide. Infect. Immun. 62:178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans, M. E. 1985. Francisella tularensis. Infect. Control 6:381-383. [PubMed] [Google Scholar]
- 47.Evans, M. E., D. W. Gregory, W. Schaffner, and Z. A. McGee. 1985. Tularemia: a 30 year experience with 88 cases. Medicine (Baltimore) 64:251-269. [PubMed] [Google Scholar]
- 48.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forsman, M., E. W. Henningson, E. Larsson, T. Johansson, and G. Sandstrom. 2000. Francisella tularensis does not manifest virulence in viable but nonculturable state. FEMS Microbiol. Ecol. 31:217-224. [DOI] [PubMed] [Google Scholar]
- 50.Forsman, M., G. Sandstrom, and A. Sjostedt. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38-46. [DOI] [PubMed] [Google Scholar]
- 51.Fortier, A. H., T. Polsinelli, S. J. Green, and C. A. Nacy. 1992. Activation of macrophages for destruction of Francisella tularensis: identification of cytokines, effector cells and effector molecules. Infect. Immun. 60:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fortier, A. H., S. J. Green, T. Polsinelli, T. R. Jones, R. M. Crawford, D. A. Leiby, K. L. Elkins, M. S. Meltzer, and C. A. Nacy. 1994. Life and death of an intracellular pathogen: Francisella tularensis and the macrophage. Immunol. Ser. 60:349-361. [PubMed] [Google Scholar]
- 53.Fortier, A. H., D. A. Leiby, R. B. Narayanan, E. Asafoadjei, R. M. Crawford, C. A. Nacy, and M. S. Meltzer. 1995. Growth of Francisella tularensis LVS in macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect. Immun. 63:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foshay, L., W. H. Hesselbrock, H. J. Wittenberg, and A. H. Rodenberg. 1942. Vaccine prophylaxis against tularemia in man. Am. J. Public Health 32:1131-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foshay, L. 1946. A comparative study of the treatment of tularemia with immune serum, hyperimmune serum and streptomycin. Am. J. Med. 1:180-188. [DOI] [PubMed] [Google Scholar]
- 56.Foshay, L. 1950. Tularemia. Annu. Rev. Microbiol. 4:313-330. [DOI] [PubMed] [Google Scholar]
- 57.Fournie, J. J., and M. Bonneville. 1996. Stimulation of gamma-delta cells by phosphoantigens. Res. Immunol. 147:338-347. [DOI] [PubMed] [Google Scholar]
- 58.Francis, E. 1927. Microscopic changes of tularemia in the tick Dermacentor andersoni and the bedbug Cimex lectularius. Public Health Rep. 42:2763-2772. [Google Scholar]
- 59.Fulop, M., R. Manchee, and R. W. Titball. 1995. Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine 13:1220-1225. [DOI] [PubMed] [Google Scholar]
- 60.Fulop, M., D. Leslie, and R. Titball. 1996. A rapid, highly sensitive method for the detection of Francisella tularensis in clinical samples using the polymerase chain reaction. Amer. J. Trop. Med. Hyg. 54:364-366. [DOI] [PubMed] [Google Scholar]
- 61.Fulop, M., R. Manchee, and R. Titball. 1996. Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS Immunol. Med. Microbiol. 13:245-247. [DOI] [PubMed] [Google Scholar]
- 62.Fulop, M., P. Mastroeni, M. Green, and R. W. Titball. 2001. Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of F. tularensis. Vaccine 19:4465-4472. [DOI] [PubMed] [Google Scholar]
- 63.Gese, E. M., R. D. Schultz, M. R. Johnson, E. S. Williams, R. L. Crabtree, and R. L. Ruff. 1997. Serological survey for diseases in free-ranging coyotes (Canis latrans) in Yellowstone National Park, Wyoming. J. Wildl. Dis. 33:47-56. [DOI] [PubMed] [Google Scholar]
- 64.Gill, V., and B. A. Cunha. 1997. Tularemia pneumonia. Semin. Respir. Infect. 12:61-67. [PubMed] [Google Scholar]
- 65.Gilligan, P. H. 1995. Pseudomonas and Burkholderia, p. 509-519. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology Press, Washington, D.C.
- 66.Golovliov, I., G. Sandstrom, M. Ericsson, A. Sjostedt, and A. Tarnvik. 1995. Cytokine expression in the liver during the early phase of murine tularemia. Infect. Immun. 63:534-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golovliov, I., M. Ericsson, L. Akerblom, G. Sandstrom, A. Tarnvik, and A. Sjostedt. 1995. Adjuvanticity of ISCOMS incorporating a T-cell reactive lipoprotein of the facultative intracellular pathogen Francisella tularensis. Vaccine 13:261-267. [DOI] [PubMed] [Google Scholar]
- 68.Golovliov, I., K. Kuoppa, A. Sjostedt, A. Tarnvik, and G. Sandstrom. 1996. Cytokine expression in the liver of mice infected with a highly virulent strain of Francisella tularensis. FEMS Immunol. Med. Microbiol. 13:239-244. [DOI] [PubMed] [Google Scholar]
- 69.Golovliov, I., M. Ericsson, G. Sandstrom, A. Tarnvik, and A. Sjostedt. 1997. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65:2183-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reference deleted.
- 71.Green, S. J., L. F. Scheller, M. A. Marletta, M. C. Seguin, F. W. Klotz, M. Slayter, B. J. Nelson, and C. A. Nacy. 1994. Nitric oxide: cytokine-regulation of nitric oxide in host resistance to intracellular pathogens. Immunol. Lett. 43:87-94. [DOI] [PubMed] [Google Scholar]
- 72.Grunow, R., W. Splettstoesser, S. McDonald, C. Otterbein, T. O'Brien, C. Morgan, J. Aldrich, E. Hoffer, E. Finke, and H. Meyer. 2000. Detection of Francisella tularensis in biological specimens using a capture enzyme-linked immunosorbent assay, an immunochromatographic handheld assay, and a PCR. Clin. Diagn. Lab. Immunol. 7:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurycova, D. 1997. Analysis of the incidence and routes of transmission of tularemia in Slovakia. Epidemiol. Mikrobiol. Immunol. 46:67-72. [PubMed] [Google Scholar]
- 74.Gurycova, D. 1998. First isolation of Francisella tularensis subsp. tularensis in Europe. Eur. J. Epidemiol. 14:797-802. [DOI] [PubMed] [Google Scholar]
- 75.Helvaci, S., S. Gedikoglu, H. Akalin, and H. B. Oral. 2000. Tularemia in Bursa, Turkey: 205 cases in ten years. Eur. J. Epidemiol. 16:271-276. [DOI] [PubMed] [Google Scholar]
- 76.Hermanowska-Szpakowicz, T., and S. A. Pancewicz. 1996. The presence of antibodies against Francisella tularensis among inhabitants of North-Eastern Poland. Przegl. Epidemiol. 50:55-59. [PubMed] [Google Scholar]
- 77.Higgins, J. A., Z. Hubalek, J. Halouzka, K. L. Elkins, A. Sjostedt, M. Shipley, and M. S. Ibrahim. 2000. Detection of Francisella tularensis in infected mammals and vectors using a probe-based polymerase chain reaction. Am. J. Trop. Med. 62:310-318. [DOI] [PubMed] [Google Scholar]
- 78.Hiromatsu, K., Y. Yoshikai, G. Matsuzaki, S. Ohga, K. Muramori, J. Bluestone, and K. Nomoto. 1992. A protective role of γδ T cells in primary infection with Listeria monocytogenes in mice. J. Exp. Med. 175:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoel, T., O. Scheel, S. H. Nordahl, and T. Sandvik. 1991. Water- and airborne Francisella tularensis biovar palaearctica isolated from human blood. Infection 19:348-350. [DOI] [PubMed] [Google Scholar]
- 80.Hollis, D. G., R. E. Weaver, A. G. Steigerwalt, J. D. Wenger, C. W. Moss, and D. J. Brenner. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holm, S. E., A. Tarnvik, and G. Sandstrom. 1980. Antigenic composition of a vaccine strain of Francisella tularensis. Int. Arch. Allergy Appl. Immunol. 61:136-144. [DOI] [PubMed] [Google Scholar]
- 82.Hornick, R. B., and H. T. Eigelsbach. 1966. Aerogenic immunization of man with live tularemia vaccine. Bacteriol. Rev. 30:532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hornick, R. B., A. T. Dawkins, H. T. Eigelsbach, and J. J. Tulis. 1966. Oral tularemia vaccine in man. Antimicrob. Agents Chemother. 6:11-14. [DOI] [PubMed] [Google Scholar]
- 84.Hubalek, Z., W. Sixl, and J. Halouzka. 1998. Francisella tularensis in Dermacentor reticulatus ticks from the Czech Republic and Austria. Wien. Klin. Wochenschr. 110:909-910. [PubMed] [Google Scholar]
- 85.Ibrahim, A., P. Gerner-Smidt, and A. Sjostedt. 1996. Amplification and restriction endonuclease digestion of a large fragment of genes coding for rRNA as a rapid method for discrimination of closely related pathogenic bacteria. J. Clin. Microbiol. 34:2894-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ikaheimo, I., H. Syrjala, J. Karhukorpi, R. Schildt, and M. Koskela. 2000. In vitro antibiotic susceptibility of Francisella tularensis isolated from humans and animals. J. Antimicrob. Chemother. 46:287-290. [DOI] [PubMed] [Google Scholar]
- 87.Johansson, A., L. Berglund, L. Gothefors, A. Sjostedt, and A. Tarnvik. 2000. Ciprofloxacin for treatment of tularemia in children. Pediatr. Infect. Dis. J. 19:449-453. [DOI] [PubMed] [Google Scholar]
- 88.Johansson, A., L. Berglund, U. Eriksson, I. Goransson, R. Wollin, M. Forsman, A. Tarnvik, and A. Sjostedt. 2000. Comparative analysis of PCR versus culture for diagnosis of ulceroglandular tularemia. J. Clin. Microbiol. 38:22-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johansson, A., A. Ibrahim, I. Goransson, U. Eriksson, D. Gurycova, J. E. Clarridge, and A. Sjostedt. 2000. Evaluation of PCR-based methods for discrimination of Francisella species and subspecies and development of a specific PCR that distinguishes the two major subspecies of Francisella tularensis. J. Clin. Microbiol. 38:4180-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johansson, A., I. Goransson, P. Larsson, and A. Sjostedt. 2001. Extensive allelic variation among Francisella tularensis strains in a short-sequence tandem repeat region. J. Clin. Microbiol. 39:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Junhui, Z., Y. Ruifu, L. Jianchun, Z. Songle, C. Meiling, C. Fengxiang, and C. Hong. 1996. Detection of Francisella tularensis by the polymerase chain reaction. J. Med. Microbiol. 45:477-482. [DOI] [PubMed] [Google Scholar]
- 92.Kadull, P. J., H. R. Reames, L. L. Coriell, and L. Foshay. 1950. Studies on tularemia. V. Immunization of man. J. Immunol. 65:425-435. [PubMed] [Google Scholar]
- 93.Karhukorpi, E. K., and J. Karhukorpi. 2001. Rapid laboratory diagnosis of ulceroglandular tularemia with polymerase chain reaction. Scand. J. Infect. Dis. 33:383-385. [DOI] [PubMed] [Google Scholar]
- 94.Karlsson, J., R. G. Prior, K. Williams, L. Lindler, K. A. Brown, N. Chatwell, K. Hjalmarsson, N. Loman, K. A. Mack, M. Pallen, M. Popek, G. Sandstom, A. Sjostedt, T. Svensson, I. Tamas, S. G. E. Andersson, B. W. Wren, P. C. F. Oyston, and R. W. Titball. 2000. Sequencing of the Francisella tularensis strain Schu 4 genome reveals the shikimate and purine metabolic pathways, targets for the construction of a rationally attenuated auxotrophic vaccine. Microbiol. Comp. Gen. 5:25-39. [DOI] [PubMed] [Google Scholar]
- 95.Khatenever, L. M. 1943. The allergic diagnosis, specific prophylaxis and vaccine therapy of tularemia. p. 62-79. In E. B. Balsky, I. G. Kochergin, and V. V. Porin (ed.), Microbiology and epidemiology. Medical Publications Ltd,, London, United Kingdom.
- 96.Koskela, P., and A. Salminen. 1985. Humoral immunity against Francisella tularensis after natural infection. J. Clin. Microbiol. 22:973-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kostiala, A. A. I., D. D. McGregor, and P. S. Logie. 1975. Tularemia in the rat. I. The cellular basis of host resistance to infection. Immunology 28:855-869. [PMC free article] [PubMed] [Google Scholar]
- 98.Kovarova, H., L. Hernychova, M. Hajduch, M. Sirova, and A. Macela. 2000. Influence of the Bcg locus on natural resistance to primary infection with the facultative intracellular bacterium Francisella tularensis in mice. Infect. Immun. 68:1480-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kroca, M., A. Tarnvik, and A. Sjostedt. 2000. The proportion of circulating gamma delta T cells increases after the first week of onset of tularemia and remains elevated for more than one year. Clin. Exp. Immunol. 120:280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kudelina, R. I., and N. G. Olfsufjev. 1980. Sensitivity to macrolide antibiotics and lincomycin in Francisella holarctica. J. Hyg. Epidemiol. Microbiol. Immunol. 24:84-91. [PubMed] [Google Scholar]
- 101.Kupper, T. S. 1990. The activated keratinocyte: a model for inducible cytokine production by non-bone marrow-derived cells in cutaneous inflammatory and immune responses. J. Investig. Dermatol. 94:146S-150S. [DOI] [PubMed] [Google Scholar]
- 102.Kwaik, Y. A., B. I. Eisenstein, and N. C. Engelberg. 1993. Phenotypic modification by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Labayru, C., A. Palop, L. Lopez-Urrutia, C. Avellaneda, M. A. Mazon, A. Alberte, and P. P. Pascual. 1999. Francisella tularensis: update on microbiological diagnosis after epidemic outbreak. Enferm. Infecc. Microbiol. Clin. 17:458-462. [PubMed] [Google Scholar]
- 104.Lai, X. H., I. Golovliov, and A. Sjostedt. 2001. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69:4691-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee, B. Y., and M. A. Horowitz. 1995. Identification of macrophage and stress-induced proteins by Mycobacterium tuberculosis. J. Clin. Investig. 96:245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leighton, F. A., H. A. Artsob, M. C. Chu, and J. G. Olson. 2001. A serological survey of rural dogs and cats on the southwestern Canadian prairie for zoonotic pathogens. Can. J. Public Health 92:67-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lepitzki, D. A., A. Woolf, and M. Cooper. 1990. Serological prevalence of tularemia in cottontail rabbits of southern Illinois. J. Wildl. Dis. 26:279-282. [DOI] [PubMed] [Google Scholar]
- 108.Levesque, B., G. de Serres, R. Higgins, M. A. D'Halewyn, H. Artsob, J. Grondin, M. Major, M. Garvie, and B. Duval. 1995. Seroepidemiologic study of three zoonoses (leptospirosis, Q fever and tularemia) among trappers in Quebec, Canada. Clin. Diagn. Lab. Immunol. 2:496-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Limaye, A. P., and C. J. Hooper. 1999. Treatment of tularemia with fluoroquinolones: two cases and review. Clin. Infect. Dis. 29:922-924. [DOI] [PubMed] [Google Scholar]
- 110.Lofgren, S., A. Tarnvik, G. D. Bloom, and W. Sjoberg. 1983. Phagocytosis and killing of Francisella tularensis by human polymorphonuclear leukocytes. Infect. Immun. 39:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lofgren, S., T. A., M. Thore, and J. Carlsson. 1984. A wild and susceptible strain of Francisella tularensis differs in susceptibility to hypochlorous acid: a possible explanation to their different handling by polymorphonuclear leukocytes. Infect. Immun. 43:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malo, D., K. Vogan, S. Vidal, J. Hu, M. Cellier, E. Schurr, A. Fuks, N. Bumstead, K. Morgan, and P. Gros. 1994. Halotype mapping and sequence analysis of the mouse Nramp gene predicts susceptibility to infection with intracellular parasites. Genomics 23:51-61. [DOI] [PubMed] [Google Scholar]
- 113.Mangold, T., and J. Goldberg. 1999. Plague wars: a true story of biological warfare. Macmillan Publishers Ltd., London, United Kingdom.
- 114.Maurin, M., N. F. Mersali, and D. Raoult. 2000. Bactericidal activities of antibiotics against intracellular Francisella tularensis. Antimicrob. Agents Chemother. 44:3428-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCoy, G. W., and C. W. Chapin. 1912. Further observations on a plague-like disease of rodents with a preliminary note on the causative agent, Bacterium tularense. J. Infect. Dis. 10:61-72. [Google Scholar]
- 116.McCrumb, F. R. 1961. Aerosol infection of man with Pasteurella tularensis. Bacteriol. Rev. 25:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McDonald, M. K., S. C. Cowley, and F. E. Nano. 1997. Temperature-sensitive lesions in the Francisella novicida valA gene cloned into an Escherichia coli msbA lpxK mutant affecting deoxycholate resistance and lipopolysaccharide assembly at the restrictive temperature. J. Bacteriol. 179:7638-7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mdluli, K. E., L. S. Anthony, G. S. Baron, M. K. McDonald, S. V. Myltseva, and F. E. Nano. 1994. Serum sensitive mutation of F. novicida: association with an ABC transporter gene. Microbiology 140:3309-3318. [DOI] [PubMed] [Google Scholar]
- 119.Merriott, J., A. Shoemaker, and C. M. Downs. 1961. Growth of Pasteurella tularense in cultured cells. J. Infect. Dis. 108:136-150. [DOI] [PubMed] [Google Scholar]
- 120.Montejo, M., J. Perez-Irezabal, P. G. de Zarate, K. Aguirrebengoa, J. M. Vicente, E. Martinez, S. Ibarra, E. Bereciartua, and C. Castell. 1998. Tularemia: report of 16 cases in the Castilla-Leon Community. Rev. Clin. Esp. 198:794-798. [PubMed] [Google Scholar]
- 121.Morner, T., and K. Sandstedt. 1983. A serological survey of antibodies against Francisella tularensis in some Swedish mammals. Nord. Vet. Med. 35:82-85. [PubMed] [Google Scholar]
- 122.Morner, T. 1992. The ecology of tularemia. Rev. Sci. Tech. Off. Int. Epiz. 11:1123-1130. [PubMed] [Google Scholar]
- 123.Morner, T., R. Mattsson, M. Forsman, K. E. Johansson, and G. Sandstrom. 1993. Identification and classification of different isolates of Francisella tularensis. Zentralbl. Veterinarmed B 40:613-620. [DOI] [PubMed] [Google Scholar]
- 124.Niebylski, M. L., M. G. Peacock, E. R. Fischer, S. F. Porcella, and T. G. Schwan. 1997. Characterization of an endosymbiont infecting wood ticks, Dermacentor andersoni, as a member of the genus Francisella. Appl. Environ. Microbiol. 63:3933-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Noda, H., U. G. Munderloh, and T. J. Kurtti. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ohara, Y., T. Sato, H. Fujita, T. Ueno, and M. Homma. 1991. Clinical manifestations of tularemia in Japan — analysis of 1355 cases observed between 1924 and 1987. Infection 19:14-17. [DOI] [PubMed] [Google Scholar]
- 127.Ohara, Y., T. Sato, and M. Homma. 1998. Arthropod-borne tularemia in Japan: clinical analysis of 1374 cases observed between 1924 and 1996. J. Med. Entomol. 35:471-473. [DOI] [PubMed] [Google Scholar]
- 128.Olsufjev, N. G., O. S. Emelyanova., and T. N. Dunaeva. 1959. Comparative study of strains of B. tularense in the Old and New World and their taxonomy. J. Hyg. Epidemiol. Microbiol. Immunol. 3:138-149. [PubMed] [Google Scholar]
- 129.Olsufjev, N. G. 1970. Taxonomy and characteristic of the genus Francisella Dorofeev, 1947. J. Hyg. Epidemiol. Microbiol. Immunol. 14:67-74. [PubMed] [Google Scholar]
- 130.Olsufjev, N. G., and I. S. Meshcheryakova. 1983. Subspecific taxonomy of Francisella tularensis McCoy and Chapin 1912. Int. J. Syst. Bacteriol. 33:872-874. [Google Scholar]
- 131.Owen, C. R. 1974. Genus Francisella, p. 283-285. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology. The Williams and Wilkins Co., Baltimore, Md.
- 132.Pavlov, V. M., A. N. Mokrievich, and K. Volkovoy. 1996. Cryptic plasmid pFNL10 from Francisella novicida-like F6168: the base of plasmid vectors for Francisella tularensis. FEMS Immunol. Med. Microbiol. 13:253-256. [DOI] [PubMed] [Google Scholar]
- 133.Pavlovich, N. V., N. I. Shimaniuk, and B. N. Mishan'kin. 1992. Neuraminidase activity in representatives of the genus Francisella. Zh. Mikrobiol. Epidemiol. Immunobiol. 9-10:8-10. [PubMed] [Google Scholar]
- 134.Perez-Castrillon, J. L., P. Bachiller-Luque, M. Martin-Luquero, F. J. Mena-Martin, and V. Herreros. 2001. Tularemia epidemic in northwestern Spain: Clinical description and therapeutic response. Clin. Infect. Dis. 33:573-576. [DOI] [PubMed] [Google Scholar]
- 135.Pollitzer, R. 1967. History and incidence of tularemia in the Soviet Union. A review. The Institute of Contemporary Russian Studies, Fordham University, New York, N.Y.
- 136.Polsinelli, T., M. S. Meltzer, and A. H. Fortier. 1994. Nitric oxide-independent killing of Francisella tularensis by IFN-γ-stimulated murine alveolar macrophages. J. Immunol. 153:1238-1245. [PubMed] [Google Scholar]
- 137.Poquet, Y., M. Kroca, F. Halary, S. Stenmark, M. Peyrat, M. Bonneville, J. J. Fournie, and A. Sjostedt. 1998. Expansion of Vγ9Vδ2 T cells is triggered by Francisella tularensis-derived phosphoantigens in tularemia but not after tularemia vaccination. Infect. Immun. 66:2107-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Prior, R. G., L. Klasson, P. Larsson, K. Williams, L. Lindler, A. Sjostedt, T. Svensson, I. Tamas, B. W. Wren, P. C. F. Oyston, S. G. E. Andersson, and R. W. Titball. 2001. Preliminary analysis and annotation of the partial genome sequence of Francisella tularensis strain Schu 4. J. Appl. Microbiol. 91:1-7. [DOI] [PubMed] [Google Scholar]
- 139.Provenza, J. M., S. A. Klotz, and R. L. Penn. 1986. Isolation of Francisella tularensis from blood. J. Clin. Microbiol. 24:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Puente-Redondo, V. A. de la, N. Garcia del Blanco, C. B. Gutierrez-Martin, F. J. Garcia-Pena, and, E. F. Rodriguez Ferri. 2000. Comparison of different PCR approaches for typing of Francisella tularensis strains. J. Clin. Microbiol. 38:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reary, B. W., and S. A. Klotz. 1988. Enhancing recovery of Francisella tularensis from blood. Diagn. Microbiol. Infect. Dis. 11:117-119. [DOI] [PubMed] [Google Scholar]
- 142.Reilly, T. J., G. S. Baron, F. E. Nano, and M. S. Kuhlenschmidt. 1996. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 271:10973-10983. [DOI] [PubMed] [Google Scholar]
- 143.Reintjes, R., I. Dedushaj, A. Gjini, T. R. Jorgensen, B. Cotter, A. Lieftucht, F. D'Ancona, D. T. Dennis, M. A. Kosoy, G. Mulliqi-Osmani, R. Grunow, A. Kalaveshi, L. Gashi, and I. Humolli. 2002. Tularemia outbreak investigation in Kosovo: case control and environmental studies. Emerg. Infect. Dis. 8:69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Remaley, A. T., C. Das, P. I. Campbell, G. M. LaRocca, M. T. Pope, and R. H. Glew. 1985. Characterisation of Leishmania donovani acid phosphatases. J. Biol. Chem. 260:880-886. [PubMed] [Google Scholar]
- 145.Risi, G. F., and D. J. Pombo. 1995. Relapse of tularemia after aminoglycoside therapy — case-report and discussion of therapeutic options. Clin. Infect. Dis. 20:174-175. [DOI] [PubMed] [Google Scholar]
- 146.Rodionova, I. V. 1967. Respiration of geographic variants of Francisella tularensis in the presence of glycerin. Zh. Mikrobiol. Epidemiol. Immunobiol. 44:21-25. [PubMed] [Google Scholar]
- 147.Russell, P., S. M. Eley, M. J. Fulop, D. L. Bell, and R. W. Titball. 1998. The efficacy of ciprofloxacin and doxycycline against experimental tularemia. J. Antimicrob. Chemother. 41:461-465. [DOI] [PubMed] [Google Scholar]
- 148.Saha, A. K., J. N. Dowling, K. L. LaMarco, S. Das, A. T. Remaley, N. Olomu, M. T. Pope, and R. H. Glew. 1985. Properties of an acid phosphatase from Legionella micdadei which blocks superoxide anion production by human neutrophils. Arch. Biochem. Biophys. 243:150-160. [DOI] [PubMed] [Google Scholar]
- 149.Sandstrom, G., and A. Tarnvik. 1987. Immunospecific T-lymphocyte stimulation by membrane proteins from Francisella tularensis. J. Clin. Microbiol. 25:641-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sandstrom, G., S. Lofgren, and A. Tarnvik. 1988. A capsule-deficient mutant of Francisella tularensis LVS exhibits enhanced sensitivity to killing by serum but diminished sensitivity to killing by polymorphonuclear leukocytes. Infect. Immun. 56:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sandstrom, G., A. Sjostedt, M. Forsman, N. V. Pavlovich, and B. N. Mishan'kin. 1992. Characterization and classification of strains of Francisella tularensis isolated in the central Asian focus of the Soviet Union and in Japan. J. Clin. Microbiol. 30:172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sandstrom, G., A. Sjostedt, T. Johansson, K. Kuoppa, and J. C. Williams. 1992. Immunogenicity and toxicity of lipopolysaccharide from Francisella tularensis LVS. FEMS Microbiol. Immunol. 5:201-210. [DOI] [PubMed] [Google Scholar]
- 153.Sandstrom, G. 1994. The tularemia vaccine. J. Chem. Tech. Biotech. 59:315-320. [DOI] [PubMed] [Google Scholar]
- 154.Saslaw, S., H. T. Eigelsbach, J. A. Prior, H. E. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:702-714. [DOI] [PubMed] [Google Scholar]
- 155.Saslaw, S., H. T. Eigelsbach, H. E. Wilson, J. A. Prior, and S. Carhart. 1961. Tularemia vaccine study. I. Intracutaneous challenge. Arch. Intern. Med. 107:689-701. [DOI] [PubMed] [Google Scholar]
- 156.Shepard, C. C. 1959. Non-acid-fast bacteria and HeLa cells: their uptake and subsequent intracellular growth. J. Bacteriol. 77:701-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sjostedt, A., G. Sandstrom, and A. Tarnvik. 1990. Several membrane polypeptides of the live vaccine strain Francisella tularensis LVS stimulate T cells from naturallly infected individuals. J. Clin. Microbiol. 28:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sjostedt, A., G. Sandstrom, and A. Tarnvik. 1990. Immunization of mice with an attenuated Salmonella typhimurium strain expressing a membrane protein of Francisella tularensis. A model for identification of bacterial determinants relevant to the host defence against tularemia. Res. Microbiol. 141:887-891. [DOI] [PubMed] [Google Scholar]
- 159.Sjostedt, A., G. Sandstrom, and A. Tarnvik. 1992. Humoral and cell-mediated immunity in mice to a 17-kilodalton lipoprotein of Francisella tularensis expressed by Salmonella typhimurium. Infect. Immun. 60:2855-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sjostedt, A., A. Tarnvik, and G. Sandstrom. 1996. Francisella tularensis: host-parasite interaction. FEMS Immunol. Med. Microbiol. 13:181-184. [DOI] [PubMed] [Google Scholar]
- 161.Sjostedt, A., J. W. Conlan, and R. J. North. 1994. Neutrophils are critical for host defense against primary infection with the facultative intracellular bacterium Francisella tularensis in mice and participate in defense against reinfection. Infect. Immun. 62:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sjostedt, A., U. Eriksson, L. Berglund, and A. Tarnvik. 1997. Detection of Francisella tularensis in ulcers of patients with tularemia by PCR. J. Clin. Microbiol. 35:1045-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Skrodzki, E. 1968. Investigations on the pathogenesis of tularemia. VII. Attempts to discover F. tularensis toxins. Biol. Inst. Med. Morsk. Gdansk 19:69-76. [PubMed] [Google Scholar]
- 164.Sorokin, V. M., N. V. Pavlovich, and L. A. Prozorova. 1996. Francisella tularensis resistance to bactericidal action of normal human serum. FEMS Immunol. Med. Microbiol. 13:249-252. [DOI] [PubMed] [Google Scholar]
- 165.Steinemann, T. L., M. R. Sheikholeslami, H. H. Brown, and R. W. Bradsher. 1999. Oculoglandular tularemia. Arch. Opthalmol. 117:132-133. [DOI] [PubMed] [Google Scholar]
- 166.Stewart, S. J. 1996. Tularemia: association with hunting and farming. FEMS Immunol. Med. Microbiol. 13:197-199. [DOI] [PubMed] [Google Scholar]
- 167.Stulik, J., L. Hernychova, A. Macela, Z. Krocova, and M. Kroca. 1999. Production of stress-inducible form of heat-shock protein 70 in mouse peritoneal adherent cells after in vivo infection by Francisella tularensis. Folia Microbiol. (Prague) 44:306-310. [DOI] [PubMed] [Google Scholar]
- 168.Sumida, T., T. Maeda, H. Takahashi, S. Yoshida, F. Yonaha, A. Sakamoto, H. Tomioka, T. Koike, and S. Yoshida. 1992. Predominant expansion of V γ9/Vδ2 T cells in a tularemia patient. Infect. Immun. 60:2554-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Surcel, H. M., M. Sarvas, I. M. Helander, and E. Herva. 1989. Membrane proteins of Francisella tularensis LVS differ in ability to induce proliferation of lymphocytes from tularemia-vaccinated individuals. Microb. Pathog. 7:411-419. [DOI] [PubMed] [Google Scholar]
- 170.Syrjala, H., P. Kujala, V. Myllyla, and A. Salminen. 1985. Airborne transmission of tularemia in farmers. Scand. J. Infect. Dis. 17:371-375. [DOI] [PubMed] [Google Scholar]
- 171.Syrjala, H., P. Koskela, T. Ripatti, A. Salminen, and E. Herva. 1986. Agglutination and ELISA methods in the diagnosis of tularemia in different clinical severities of the disease. J. Infect. Dis. 153:142-145. [DOI] [PubMed] [Google Scholar]
- 172.Tarnvik, A., S. Lofgren, L. Ohlund, and G. Sandstrom. 1987. Detection of antigen in urine of a patient with tularemia. Eur. J. Clin. Microbiol. 6:318-319. [DOI] [PubMed] [Google Scholar]
- 173.Tarnvik, A. 1989. Nature of protective immunity to Francisella tularensis. Rev. Infect. Dis. 11:440-451. [PubMed] [Google Scholar]
- 174.Tarnvik, A., C. Henning, E. Falsen, and G. Sandstrom. 1989. Isolation of Francisella tularensis biovar palaeartica from human blood. Eur. J. Clin. Microbiol. Infect. Dis. 8:146-150. [DOI] [PubMed] [Google Scholar]
- 175.Tarnvik, A., G. Sandstrom, and A. Sjostedt. 1996. Epidemiological analysis of tularemia in Sweden, 1931-1993. FEMS Immunol. Med. Microbiol. 13:201-204. [DOI] [PubMed] [Google Scholar]
- 176.Tarnvik, A., M. Ericsson, I. Golovliov, G. Sandstrom, and A. Sjostedt. 1996. Orchestration of the protective immune response to intracellular bacteria: Francisella tularensis as a model organism. FEMS Immunol. Med. Microbiol. 13:221-225. [DOI] [PubMed] [Google Scholar]
- 177.Thompson, S., L. Omphroy, and T. Oetting. 2001. Parinaud's oculoglandular syndrome attributable to an encounter with a wild rabbit. Am. J. Ophthalmol. 131:283-284. [DOI] [PubMed] [Google Scholar]
- 178.Tigertt, W. D. 1962. Soviet viable Pasteurella tularensis vaccines: a review of selected articles. Bacteriol. Rev. 26:354-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Waag, D. M., G. Sandstrom, M. J. England, and J. C. Williams. 1996. Immunogenicity of a new lot of Francisella tularensis live vaccine strain in human volunteers. FEMS Immunol. Med. Microbiol. 13:205-209. [DOI] [PubMed] [Google Scholar]
- 180.Wenger, J. D., D. G. Hollis, R. E. Weaver, C. N. Baker, G. R. Brown, D. J. Brenner, and C. V. Broome. 1989. Infection caused by Francisella philomiragia (formerly Yersinia philomiragia). A newly recognised human pathogen. Ann. Intern. Med. 110:888-892. [DOI] [PubMed] [Google Scholar]
- 181.Westerman, E. L., and J. McDonald. 1983. Tularemia pnuemonia mimicking Legionnaires' disease: isolation of organism on CYE agar and successful treatment with erythromycin. South. Med. J. 76:1169-1170. [PubMed] [Google Scholar]
- 182.Wherry, J. C., R. D. Schreiber, and E. R. Unanue. 1991. Regulation of gamma interferon production by natural killer cells in SCID mice: roles of tumor necrosis factor and bacterial stimuli. Infect. Immun. 59:1709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wicki, R., P. Sauter, C. Mettler, A. Natsch, T. Enzler, N. Pusterla, P. Kuhnert, G. Egli, M. Bernasconi, R. Lienhard, H. Lutz, and C. M. Leutenegger. 2000. Swiss Army survey in Switzerland to determine the prevalence of Francisella tularensis, members of the Ehrlichia phagocytophila genogroup, Borrelia burgdorferi sensu lato, and tick-borne encephalitis virus in ticks. Eur. J. Clin. Microbiol. Infect. Dis. 19:427-432. [DOI] [PubMed] [Google Scholar]
- 184.Wilson, G. S., and A. A. Miles. 1964. Brucella tularensis, p. 1013-1014 In Topley and Wilson's principles of bacteriology and immunity. The Williams and Wilkins Co., Baltimore. Md.
- 185.Yee, D., T. R. Rhinehart-Jones, and K. L. Elkins. 1996. Loss of either CD4+ or CD8+ T cells does not affect the magnitude of protective immunity to an intracellular pathogen. Francisella tularensis strain LVS. J. Immunol. 157:5042-5048. [PubMed] [Google Scholar]