Abstract
Recently, the role of the environment and climate in disease dynamics has become a subject of increasing interest to microbiologists, clinicians, epidemiologists, and ecologists. Much of the interest has been stimulated by the growing problems of antibiotic resistance among pathogens, emergence and/or reemergence of infectious diseases worldwide, the potential of bioterrorism, and the debate concerning climate change. Cholera, caused by Vibrio cholerae, lends itself to analyses of the role of climate in infectious disease, coupled to population dynamics of pathogenic microorganisms, for several reasons. First, the disease has a historical context linking it to specific seasons and biogeographical zones. In addition, the population dynamics of V. cholerae in the environment are strongly controlled by environmental factors, such as water temperature, salinity, and the presence of copepods, which are, in turn, controlled by larger-scale climate variability. In this review, the association between plankton and V. cholerae that has been documented over the last 20 years is discussed in support of the hypothesis that cholera shares properties of a vector-borne disease. In addition, a model for environmental transmission of cholera to humans in the context of climate variability is presented. The cholera model provides a template for future research on climate-sensitive diseases, allowing definition of critical parameters and offering a means of developing more sophisticated methods for prediction of disease outbreaks.
INTRODUCTION
The relationship between human health and climate is not a new concept, nor is it a new subject. In fact at least as far back as Hippocrates, many believed that human health was intricately linked to the seasons, local weather patterns, and other environmental factors (130). While preceding the advent of Pasteur's germ theory in the mid-1800s, these notions described certain patterns of disease, and often therapies were based directly on changes in the environment. Proponents of miasma, i.e., a poisonous atmosphere arising from swamps and putrid matters, as a source of disease noted that fevers and intestinal ailments were common in warm seasons and were often associated with wet, poorly drained, and humid areas, such as swamps (130). This association motivated draining of swamps in rural areas and the installation of the first central sewer systems in urban areas in North America (130). Of course, the therapeutic benefits of these practices had little to do with ridding the environment of miasma, but these practices did reduce the size and number of habitats breeding mosquitoes (disease vectors) and removed raw waste (hence, waterborne pathogens) from immediate contact with humans. In spite of the developments in the mid- to late 1800s demonstrating the role of microorganisms in disease, the theory of weather-borne diseases held firm in early medicine until the late 1800s (130).
Clearly, the relationship between climate and health has been a subject of study for a very long time. However, during the past 2 decades, development of modern tools and technologies has led to fascinating observations sparking new interest in the role of the environment, including weather and climate, in infectious disease dynamics. Scientific interest has been further stimulated by the growing problems of antibiotic resistance among pathogens, emergence and reemergence of infectious diseases worldwide, the potential threat of bioterrorism, and the debate concerning climate change. During the past few years, scientists and several agencies, including the World Health Organization, American Academy of Microbiology, Intergovernmental Panel on Climate Change (IPCC), and U.S. Global Change Research Program, among others, have published papers and issued reports highlighting the topic of climate and health (e.g., see references 8, 25, 35, 56, 58, 135, 137, 162, and 163). Concerns over the impact of anthropogenic alterations to both terrestrial and aquatic habitats, coupled with a changing global climate, have helped to spawn an expanding cross-disciplinary effort to understand how such changes might affect human health. Progress is under way in using climate factors in predictive models for certain diseases, notably cholera.
CLIMATE AND HEALTH
Part of the growing interest in the effects of climate on health is due to concerns about global climate change and variability. In concurrently appearing papers in Science, Barnett et al. (4) and Levitus et al. (99) independently reported warming in the top 3,000 m of the global ocean since 1950. Furthermore, models incorporating anthropogenic forces (e.g., gases) match the observed changes in oceanic heat content (4). These studies provide more evidence of human-induced climate change than those models that rely on near-surface temperatures, largely because the heat capacity of the oceans gives a more stable parameter than the surface temperatures (99). Superimposed on global trends or long-term changes in climate are periodic signals in the ocean-atmosphere that result in climate variability at interannual to interdecadal scales. In particular, the El Niño-southern oscillation (ENSO) has been the focus of much attention in the popular press and scientific communities. This phenomenon originates in the eastern equatorial Pacific, offshore from Peru, where every few years near Christmastime (hence, “El Niño,” the Christ child) the normally cool waters of this region are suppressed by a layer of warm water arriving from the west. This event triggers changes in weather patterns across the globe and is considered to be among the most-important factors in global climate variability (135). Climatologists have also begun to look at global impacts of other modes of atmospheric variability associated with large ocean basins, e.g., the North Atlantic oscillation (NAO), where decadal oscillations in the atmospheric pressure difference between Iceland and the Azores affect wintertime weather in Europe and the Atlantic coast of the United States. Research is under way to improve prediction of such climate signals, which is poor in comparison to the better-understood ENSO (J. Marshall, Y. Kushnir, D. Battisti, P. Change, J. Hurrell, M. McCartney, and M. Visbeck, unpublished data [http://geoid.mit.edu/accp/avehtml.html]). Although controversy remains over how human activities might be driving climate variability and change, most scientists in the field agree that our climate is changing and there is an increasing need to understand the potential outcomes of such changes on human health.
MODES OF DISEASE TRANSMISSION
Vector Borne
Vector-borne diseases are those that require a host (most often arthropods or rodents) to transmit a pathogen. Variability in climate and weather can affect both the vector population and the amplification of the pathogen within the host. Malaria and dengue fever continue to be the most-important vector-borne diseases worldwide (163). Unlike yellow fever and rift valley fever, which affect humans only incidentally, malaria and dengue are transmitted directly by infected mosquitoes that commonly bite humans. Other mosquito-borne infections, including rift valley fever, yellow fever, and encephalitis, are transmitted between the mosquito vector and a nonhuman animal host under normal circumstances and affect humans only incidentally. Plague and Lyme disease, among others, are transmitted to animals by fleas and ticks, respectively, and hantavirus is transmitted by rodents (58). While humans are not a normal host for some of these agents, they can all pose a threat when the pathogen uses a vector that bites or cohabitates with humans.
Vector-borne diseases remain the most-studied group of diseases in the context of climate variability. The geographical range of the vector population is indicative of ecological and climatic conditions that limit the distribution of both the host and the pathogen. Often, ecological disturbances, including weather-related events, can cause a shift in host vectors or change in habitat that results in a greater likelihood of the pathogen coming in contact with humans, thus increasing the risk of human infection (163). In border areas where conditions are not optimal for the pathogen and/or vector, small ecological disturbances or changes in weather may also alter conditions for disease transmission by creating a more favorable environment (163). It is often in such border regions that the most-dramatic results of climate change and/or variability are recognized. In the recent literature, many researchers have documented relationships between changes in weather due to the ENSO and certain vector-borne disease outbreaks (e.g., see references 60, 61, 65, and 100).
In this review, we have adopted a more inclusive definition of vector-borne disease that encompasses pathogens that do not require an obligate host (vector) for transmission but which are likely to be transmitted to humans by such a route. Below we present how this definition applies to Vibrio cholerae.
Airborne
As a whole, little is known about airborne infectious diseases and how they are affected by climate. While there is a growing body of work focused on health impacts associated with air pollution and climate change, these studies largely focus on noninfectious agents (ozone and particulate matter, etc.) or allergens (8). Influenza- and rhinoviruses, which cause flu and colds, respectively, are recognized as infectious airborne pathogens. Indeed, there does seem to be a regular season for flu and colds, and climatic factors are suspected to contribute in part to these cycles. In the Northern Hemisphere, flu cases peak in the late fall and winter, but there is no significant correlation with mean temperature (98). Humidity, however, may affect survival of aerosolized virus particles (140). In the southwestern United States, Coccidioides immitis, a soil fungus responsible for valley fever, causes higher infection rates during dry periods that follow a rainy season, when wind can distribute the pathogen (37).
Dust-borne agents distributed in the air are also affected by large atmospheric phenomena. Relative to normal conditions, large numbers of viable microorganisms have been isolated in Caribbean air samples during “African dust events” (57). Periodically, massive plumes of dust are transported across the tropical Atlantic from the sub-Saharan region of Africa. Such events have been exacerbated by drought in the Sahel region, overgrazing, and the drying of Lake Chad and may be related to the NAO (Marshall et al., unpublished data [http://geoid.mit.edu/accp/avehtml.html]). To date, microorganisms found in the dust particles include Aspergillus spp., Pseudomonas spp., and Sphingomonas spp., among others (57); however, it remains unknown if any human infections can be related to the African dust events. Similar phenomena are associated with the Gobi Desert and wind-swept dust from this region of the world.
Waterborne
The quality and quantity of drinking water, irrigation water, and environmental and/or recreational waters can be associated with changes in environmental conditions including weather- or climate-related variables. Floods may cause the overflow of wastewater treatment plants, failure of septic systems, or combined sewer overflows, which could contaminate nearby surface waters or wells. Furthermore, there is increasing concern about pathogens in storm water runoff (121). Maintaining sanitary water conditions is also an issue during drought conditions, when contaminants may become concentrated in available water. Additionally, the likelihood of multiple uses in a water body may increase (e.g., for cleaning, bathing, and drinking) during droughts and consequently enhance the risk of contamination and exposure.
Increasingly, the importance of the effect that weather and climate have on water quality, and not simply quantity, is being realized. Severe weather events appear to be correlated with enteric diseases, such as outbreaks of cryptosporidiosis related to excessive demand placed on sewage treatment plants from heavy rains and flooding (136). A retrospective study of drinking-water-related outbreaks of acute gastrointestinal illness in the United States by Rose et al. (136) revealed that 20 and 40% of groundwater and surface water outbreaks, respectively, between 1971 and 1994 were statistically associated with extreme precipitation. Other work has also demonstrated an ENSO connection to rates of enteric illness (18, 59) in South America, levels of enteric microorganisms in coastal areas of south Florida (United States) (101, 103), and cholera in Bangladesh (25, 104, 124).
In addition to enteric pathogens, climate also influences the abundance and ecology of nonenteric and other pathogens which are naturally present in the environment. The best examples of this include the pathogenic Vibrio spp., which are autochthonous in estuarine ecosystems. Vibrio parahaemolyticus and Vibrio vulnificus are responsible for a majority of the nonviral infections related to shellfish consumption in the United States (159) and also result in infections of open wounds during recreational exposure, such as swimming and fishing. Along with V. cholerae, these bacteria thrive in warm waters of moderate salinity (102, 147) and are closely associated with aquatic invertebrates (88, 89, 145). Therefore, with a changing climate, the geographic range of these pathogens may also change, potentially resulting in increased exposure and risk of infection for humans. Furthermore, changes in plankton populations, and other hosts for which vibrios are commensals or symbionts, would similarly alter the ecology of these pathogens that are autochthonous to the aquatic environment.
CHOLERA AS A MODEL FOR CLIMATE-RELATED INFECTIOUS DISEASE
Cholera, caused by V. cholerae, lends itself to the study of the role of climate in infectious disease. This disease has a historical context linking it to specific seasons and biogeographical zones. Although cholera is an ancient disease and had disappeared from most of the developed world during the second half of the 20th century, it persists in many parts of the world with serious epidemics, most often centered in tropical areas. The association between the disease and water has been long held in folk tradition but was first established epidemiologically in the 1850s when John Snow made the link between the disease and a shallow well in London, United Kingdom (138). Subsequently, extensive studies demonstrated that V. cholerae is, in fact, native to coastal ecosystems (34). Vibrios, including V. cholerae, can be found in virtually any coastal water body, especially in the tropics and subtropics, when appropriate techniques are used. Recently, ecologically based models have been developed which define the role of environmentally, weather-, and climate-related variables in outbreaks of the disease (32, 72, 104, 124). Data on the association between plankton and V. cholerae have been gathered over the last 20 years and strongly support the hypothesis of a commensal or symbiotic relationship between zooplankton and this bacterium. Because V. cholerae is highly concentrated on zooplankton carapaces and in their gut, the risk of ingesting an infectious dose increases when untreated surface waters are used for consumption. In this way, cholera shares some properties of a vector-borne disease. This hypothesis is currently under study in our laboratory at the Center of Marine Biotechnology and elsewhere (E. Whitcomb, personal communication, 2001).
Cholera and V. cholerae offer an excellent example of the effect of climate and weather on infectious diseases and pathogens. Here we discuss some of the recent research on the subjects of V. cholerae, climate, and health and describe a framework for future studies using the cholera model.
VIBRIOS AND V. CHOLERAE
During a cholera epidemic in 1854 in Florence, Italy, Pacini (122) first described the comma-shaped gram-negative Vibrio, the “comma bacillus,” responsible for cholera, which was subsequently named V. cholerae by Robert Koch. Pacini carried out detailed studies on the etiology of the disease. Pacini studied cholera for about 20 years and published several articles (51) in which he described the destruction of the intestinal mucosa during cholera, ascribing it to the bacterium. Furthermore, Pacini clearly stated that the Vibrio was the “specific” causative agent of the disease and insisted the disease was contagious. However, at this time, the idea that cholera was contagious was under dispute. His data were ignored, and the etiological agent of cholera was rediscovered by Robert Koch in May 1884 (30 years after Pacini's description and 1 year after Pacini's death). Microscope slides from Pacini's studies on cholera are on exhibit in the Department of Anatomy and Histology of the University of Florence (7).
During the same period of time that Pacini did his research, cholera ravaged Western Europe, and John Snow made the link between cholera and a drinking water source in London, establishing cholera as a waterborne disease. Although V. cholerae is the cause of a devastating diarrheal disease, it is also a natural member of the aquatic microbial community, a finding not clearly understood until more than 100 years later (34).
Vibrio, a diverse genus, is the most intensely studied of the Vibrionaceae family and has engendered a great deal of interest in the taxonomy and systematics of Vibrio spp. (42, 67). Taxonomic debates aside, Vibrio spp. are among the most commonly isolated bacteria in marine and estuarine waters (23, 64, 96). They significantly affect nutrient cycling in these habitats and often comprise a major portion of the natural flora. Furthermore, several of the members of the genus are pathogenic either for humans or marine animals.
To date, ca. 200 serogroups of V. cholerae have been recorded, of which only two (O1 and O139) have been associated with major epidemics. Furthermore, genes for cholera toxin (CT) production are rarely found in serogroups other than O1 or O139. Prior to the seventh pandemic, which started in 1961, outbreaks were traced to the classical biotype of V. cholerae O1. V. cholerae O1 El Tor strains largely replaced the classical biotype by 1961 (90). Toxigenic V. cholerae O139 first emerged as a pandemic threat in southeastern India and the Bay of Bengal in 1992 and appears to have arisen by genetic exchange with V. cholerae O1 El Tor (131, 157). Recent evidence also suggests that O139 isolates may have arisen by genetic exchange with non-O1 V. cholerae strains as well as clinical strains of O1 (49; S. M. Faruque, M. S. Islam, G. B. Nair, and R. R. Colwell, unpublished data).
Before the seventh pandemic, V. cholerae O1 El Tor was considered to be a different species, referred to as Vibrio eltor (127). Similarly, V. cholerae isolated from the environment and clinical strains that did not agglutinate with O1 antiserum were designated nonagglutinating vibrios and were believed to be a different species from clinical isolates of V. cholerae O1 (24). Over the last 30 years, biochemical and molecular tests and polyphasic numerical taxonomy have proven that O1 and non-O1 strains of V. cholerae are the same species (20, 26). Recent work using 16S rRNA gene sequencing has also shown no differences at the species level between V. cholerae O1 El Tor and classical strains or between V. cholerae O1 and non-O1 strains.
While environmental and clinical strains of V. cholerae are now known to represent a single species, there is significant genetic diversity among environmental and non-O1, non-O139 strains of V. cholerae (6, 15, 84, 132). Molecular tools have been used to fingerprint and track toxigenic strains of V. cholerae (6, 15, 84, 85, 132). These types of analyses have demonstrated that O1 and O139 isolates group closely with one another, forming a tight cluster, while environmental non-O1, non-O139 isolates are generally diverse (6, 15, 84, 132), as would be expected, given that ca. 200 serotypes have been described. Detailed investigation of the V. cholerae genome and of specific housekeeping genes revealed that recombination may be the major source of this variability, a common feature in the evolution of V. cholerae (15, 64). Furthermore, the three main pathogenic clones of V. cholerae O1 (sixth pandemic [classical], seventh pandemic [El Tor], and U.S. Gulf Coast isolates) appear to have evolved independently (15, 85). Evidence indicates that toxigenic strains may arise from environmental, nontoxigenic progenitors in coastal areas (17, 85). The emergence of toxigenic O139 strains may have a similar history, with confirming evidence now being developed (S. M. Faruque, M. S. Islam, R. R. Colwell, and G. B. Nair, personal communication, 2002).
Most V. cholerae strains, especially those from the environment, lack the genes required to produce CT, but the possibility of genetic exchange in the environment allows the potential emergence of new toxigenic clones. While horizontal gene transfer is common among V. cholerae, as well as between and among other bacteria, the significance of this phenomenon was not recognized until widespread patterns of antibiotic resistance in bacteria began to emerge in the 1950s (118). Since then, similar rapid evolution by acquisition of “foreign” DNA has also been implicated in the transfer of metabolic properties and virulence genes (12, 13, 48, 64, 91, 118, 157). In particular, horizontal transfer and genetic reassortment have been a key mechanism for the emergence of new toxigenic strains of V. cholerae as well as clonal diversity (12, 13, 46, 48, 64, 118, 157).
The best example of horizontal gene transfer in V. cholerae is the transduction of the elements that code for CT, the CTX genetic element, and the V. cholerae pathogenicity island (VPI). The CTX element includes six genes in its core region: ctxAB, zot, ace, cep, and orfU. The VPI codes for 15 separate open reading frames, including the tcp (toxin-coregulated pilus) gene (45). Evidence now indicates that the CTX element is actually the genome of a distinct filamentous phage that lysogenizes V. cholerae (157). VPI may also be part of a phage genome, but the evidence is not as clear. The presence of tcp (part of the VPI) is required as a receptor for CTXΦ. Furthermore, this phage-mediated transfer occurs not only among strains of V. cholerae but also with the very closely related V. mimicus, an occasional source of diarrheal disease in humans (13).
In addition to acquiring toxin-encoding genes, V. cholerae and other gram-negative bacteria have a selective advantage in their ability to enter a dormant stage, termed viable but nonculturable (VBNC), when environmental conditions are unfavorable for active growth and cell division. In 1982, Xu et al. (164) first demonstrated this phenomenon in Escherichia coli and V. cholerae using direct viable count and fluorescent-antibody staining methods. In V. cholerae and other vibrios, this state, in which metabolically active cells cannot be cultured on microbiological media, is induced by changes in environmental conditions, including temperature (reduction) and salinity (32). Microcosm studies demonstrated that these cells could remain viable in the environment for years (32, 115) and continue to be capable of causing disease (28, 29).
The existence of VBNC cells continues to be debated in the scientific community, as the definition of living, as opposed to dead, bacteria is complex and difficult to articulate. Some maintain that bacterial cells not growing on culture plates are dead. Yet, Gonzalez et al. (55) suggested bacterial cells should be considered dead only when they lose both culturability and cellular integrity. Early studies by Valentine and Bradfield (155), Postgate (128), Jannasch (83), Stevenson (150), Kurath and Morita (97), and others provide a framework for the concept of a resting stage for nonsporulating bacteria. Xu et al. (164) concluded that cells that were unable to grow on conventional culture media but that responded to nalidixic acid treatment were VBNC. This phenomenon represents a state of dormancy and allows survival and persistence of bacterial cells in the natural or host environment. To date, at least 18 species undergoing a VBNC-type shift have been documented (77), and more-sophisticated methods are being employed that confirm continued viability (39). The ability to adapt to unfavorable conditions may be a key factor in the success of V. cholerae as an opportunistic pathogen and is most likely explained by its commensal existence with zooplankton, which themselves undergo diapause.
CLIMATE, V. CHOLERAE, AND CHOLERA
Seasonality
Analysis of the frequency of specific disease cases throughout the year is a logical first-order assessment of the potential role of climate in disease transmission. For many diseases (vector-, air-, and waterborne) there is a clear seasonal trend in the detection or isolation of a pathogen and prevalence of disease. For cholera, a distinct seasonal pattern is evident, particularly in regions of endemicity. In the following sections we discuss environmental and climatic factors that drive the seasonality and occurrence of V. cholerae and the disease cholera.
Endemicity.
For most of its known history, V. cholerae was believed by physicians and clinical workers to be only coincidentally associated with environmental waters and detectable only if there was contamination by an infected individual. While we now know that V. cholerae is autochthonous in riverine, estuarine, and coastal waters, as first proposed in 1977 by Colwell et al. (34), Cockburn and Cassanos (22) reported that as early as the 1800s there was discussion about environmental connections of the disease. By the late 1800s, areas of endemicity had been defined, along with relevant descriptions of the environment, and an association of cholera with monsoons had been noted (127). V. cholerae was shown to withstand high-pH conditions, and the notion that ponds may be a potential source of infection had been put forward (22). In retrospective studies of cholera in India during the 19th century, there is clearly a series of epidemic years associated with monsoons and other weather events (Whitcomb, personal communication, 2001).
Today, geographical areas once known to have experienced cholera epidemics can be characterized into three levels. Cholera-free communities are defined as having no locally acquired infections. In areas of cholera epidemicity, the disease diminishes after an outbreak. In regions of cholera endemicity, the disease does not disappear after an epidemic peak and returns in successive waves (154). Of particular interest and relevance to identifying environmental or climate factors that may promote epidemics is the understanding of dynamics of the disease in areas of endemicity. For example, what causes periodic oscillations in cholera outbreaks, and why are some areas more prone to endemism? Torres Codeço (154) incorporated seasonality into a model of endemic cholera and showed that cholera infections occurred 2 to 4 months after the onset of V. cholerae growth in the environment; however, this predictive model was influenced by population size and rate of contact. Furthermore, only transient environmental reservoirs of V. cholerae were required to maintain endemism in poor communities (154).
(i) Ecology and biogeography of V. cholerae.
Detection of V. cholerae, both non-O1 and O1 strains, has been reported from almost every part of the world where there are rivers and coastal regions (78, 127). In regions no longer with endemic Asiatic cholera in North America, V. cholerae has been isolated from estuarine and coastal water collected along the Pacific, Atlantic, and Gulf coasts of the United States (e.g., see references 34 and 113), and it has also been isolated in Australia (40, 41), Southeast Asia, the island nations of the South Pacific, Africa, Asia, and throughout Europe. V. cholerae has been similarly isolated in areas of endemicity (India, Bangladesh, and Central and South America) (90). Despite the prevalence of the causative organism worldwide, only certain regions of the world, mainly in the tropics and subtropics, maintain endemicity for the disease. The nature of this phenomenon is not entirely understood but is most likely related to both environmental and socioeconomic factors. Following the sixth pandemic (∼1950), areas with endemic cholera had been reduced to southeastern India and Bangladesh, a significantly narrower geographic region, historically (22). During the current seventh pandemic, the range of endemicity has expanded and includes areas of Africa and South and Central America (11, 90).
Where cholera is endemic, cases tend to demonstrate distinct seasonal trends. These patterns are strongly related to the ecology of V. cholerae in the environment, where high numbers are observed during times of warm water temperatures and zooplankton blooms (104). Over the last 25 years, the major cholera epidemics, including the first outbreak of O139 disease, have originated in coastal areas (54; T. Ramamurthy et al., Letter, Lancet 341:703-704, 1993). Currently, the main regions of cholera endemicity include the coasts surrounding the Bay of Bengal, both Bangladesh and the Indian subcontinent, and coastal Latin America. In each of these three geographical regions, patterns of disease frequency follow similar trends and are most likely explained by the same physical or environmental drivers (see “Hierarchy in the cholera model” below). In Bangladesh, there is a bimodal distribution in the frequency of cases over an annual cycle. The first, and smaller, peak occurs in the spring. The larger peak, however, follows the monsoon season in the fall. The onset of epidemics coincides with dry weather and the warmest water temperatures of the year (August or September) (22, 90). The frequency of cases is reduced to background levels of a few cases as the temperature decreases in winter (22, 25, 104). In Calcutta, India, cholera cases tend to peak in April, May, and June (90). In South America, where the disease has become endemic since its reemergence in 1991, cases are concentrated in the austral summer months (January and February) (90). The geographic limits of endemicity are also consistent and suggest a biogeographical pattern of endemic disease that follows the environmental habitat of the causative organism. Although inland countries report cases of cholera (161) and freshwater plankton species harbor vibrios, case frequency and location of regions of endemicity increase near coastlines. Along the Indian subcontinent, there is a distinct boundary approximately 150 miles inland and to the northern part of the peninsula where the frequency of cholera cases rapidly diminishes (22, 127). In South America, outbreaks start along the coast (141), and epidemiological studies in Mexico demonstrate that people living in coastal states are at a high risk of contracting the disease (11; M. L. Lizarraga-Partida, I. Wong-Chang, G. Barrera-Escorcia, A. V. Botello, A. Flisser, L. Gutierrez, A. Huq, and R. R. Colwell, unpublished data).
V. cholerae O1 is also endemic in regions where there are now only sporadic cases of cholera. Such endemic clones are found along the U.S. Gulf Coast and in Australia and are unique, although closely related, based on phage typing and gene sequence analyses (1, 15, 92). In these regions, sanitation practices are well developed and the waterborne disease occurs rarely, although V. cholerae is often detected in estuaries and shellfish (116). Consumption of raw or improperly cooked shellfish harboring V. cholerae, and not contaminated drinking water, causes the sporadic cases of diarrhea and occasional septicemia that do occur (66). Yet, as in the case of the range of areas of cholera endemicity, the geographic distribution of these endemic strains tends to be limited to the subtropical and subtemperate climates. Similarly, along the U.S. Gulf Coast and in Australia, the frequency of isolation of V. cholerae from both the environment and patients peaks during the warmer months of the year (113). Non-O1 isolates also follow this pattern, with sporadic cases occurring mostly in the late summer in the United States and in the early summer in Bangladesh (prior to the annual fall epidemic) (112). Europe also reports detection of V. cholerae during the summer and early fall (3, 10, 53). Seasonality in detection of the presence of V. cholerae in water and sediment in the Chesapeake Bay of the east coast of the United States, a cholera-free region, has been abundantly demonstrated (62, 63, 85; V. Louis et al., submitted for publication).
(ii) Emergence of toxigenic strains from the environment.
When cholera reemerged in South America during the seventh pandemic in 1991, a new focus of endemicity was initiated. Many investigators have speculated on the reemergence of cholera in South America after its hundred-year absence, and several hypotheses have been put forth. One theory was that an isolated event led to the epidemics—that is, an exchange of ballast water by a ship previously traveling through areas of cholera endemicity. It has also been proposed that given the similarity between strains isolated in Latin American and Asia (O1 El Tor), cells in the VBNC state may have been transported from Southeast Asia to Peru with a transfer of water associated with the 1991 El Niño event (114). V. cholerae may have also already been a feature of the nearshore aquatic environment in South America, but some environmental cue triggered the bacterial population numbers past a threshold point, resulting in outbreaks of the disease (25, 115, 141). Following this, further contamination with infected stool and poor sanitary conditions led to outbreaks and epidemics. However, hospital records from the months prior to the official onset of the epidemic in Peru (January 1991) indicate the occurrence of cases matching the description of cholera as early as October of the previous year (141). Furthermore, cases were detected at about the same time in several geographically distant locales, suggesting that multiple strains contributed to the epidemic (141). This scenario supports the notion that a population of V. cholerae already existed along the coast of Peru and South America. Furthermore, data show that both O1 and non-O1 isolates had been detected in South and Central America prior to the onset of epidemics, as early as 1978 (108, 133, 141).
Recent molecular analyses also suggest that multiple strains may have been and continue to be involved in outbreaks or emergence of toxigenic strains. Prior to 1992, the origin of all cholera epidemics had been strains of V. cholerae O1; however, during that year cholera outbreaks were associated with a new toxigenic serogroup, V. cholerae O139, in India and for a short time this group replaced the V. cholerae O1 El Tor (19). Since the emergence of V. cholerae O139 strains in 1992, there has been much interest in determining the mechanisms by which this pathogenic serogroup could have arisen.
Various researchers have now demonstrated that O139 strains associated with epidemics arose via an homologous recombination event. Part of the sequence for O-antigen biosynthesis in an O1 El Tor strain was lost and replaced with sequences derived from a non-O1 strain (9, 44). This recombination event also led to the expression of a capsule in O139, a feature often associated with non-O1 strains (9, 44). Furthermore, ribotyping of both toxigenic and nontoxigenic strains of V. cholerae O139 revealed that isolates might have arisen from at least two different progenitors, including non-O1 serogroups; half of the nontoxigenic isolates produced distinctly different ribotype patterns than those produced by V. cholerae O1 El Tor (50). Moreover, analysis of housekeeping genes in the sixth (classical) and seventh (El Tor) pandemic and U.S. Gulf Coast strains suggests that these pathogenic clones were derived independently from nontoxigenic non-O1 progenitors (90). Thus, given the likelihood of more recombination events leading to new toxigenic strains, it becomes increasingly clear that there is a need to understand the occurrence of and monitor for V. cholerae non-O1 in the environment.
Interestingly, even in areas of endemicity it is often difficult to isolate V. cholerae O1 or O139 from the environment, particularly during interepidemic periods, while V. cholerae non-O1 is more readily detected (75, 94). The first study to demonstrate the presence of V. cholerae year-round in Bangladesh was conducted in the late 1990s using direct detection methods (75). In attachment studies using copepods, there was no difference observed between V. cholerae O1 and non-O1 strains. However, other studies have shown that non-O1 V. cholerae strains survive better in the environment and are more resistant than O1 strains to detergents and chelating agents, which may be discharged into the environment (111). There was early speculation about the possibility of conversion between O1 and non-O1 serogroups, and phage-mediated seroconversion between biovars (Inaba and Ogawa) has been reported (119). Later, serogroup conversion was demonstrated in laboratory microcosms (33, 111). Conversion from non-O1 to O1, and vice versa, was observed in all microcosms of both artificial seawater and natural water at various temperatures; however, seroconversion occurred earliest (within 5 days) at a salinity of ∼10‰ and temperatures near 35°C (111).
Environmental conditions affect both the overall abundance and, potentially, the serogroup of V. cholerae in the environment. Surprisingly, even in areas of cholera endemicity, O1 strains are detected infrequently in the environment compared to non-O1 strains using traditional culture methods. Evidence for spontaneous seroconversion, in part, may explain this observation (27). Furthermore, in both endemic and cholera-free areas (e.g., Chesapeake Bay), increased environmental water temperatures correspond with increased detection rates of V. cholerae, which suggests a triggering factor may be responsible for enhancing the number of organisms in a given environment and condition (V. Louis et al., submitted for publication).
Contributing to the apparent dichotomy between clinical (toxigenic) and environmental (nontoxigenic) strains is the issue of acquisition of virulence genes in the environment, as most environmental isolates harbor neither tcp nor ctxAB (146). Recombination and acquisition of foreign DNA appear to be common features among vibrios and V. cholerae (64). Genes for both TCP and CTX can be readily transduced into recipient strains via temperate phages. While this is a key issue in the emergence of toxigenic strains, how this occurs in the environment and/or intestine is still under investigation. Recent studies show that, in general, phage-mediated transduction may occur at higher rates in the marine and estuarine environment than originally thought. Jiang and Paul (86) showed transduction rates of 1013 transformants year−1 in Tampa Bay Estuary, Fla. Furthermore, in the same subtropical estuary, prophage induction was demonstrated in 52% of surface water samples, and at least 41% of the bacterial isolates were lysogens (carrying a prophage in the genome or as a self-replicating element) (21). Work to date on the CTXΦ reveals that while there is no induction in the intestine (48), exposure to sunlight is a key factor in the induction of CTX prophage from the host and subsequent propagation of the phage particle (47). Environmental conditions, such as pH and temperature, also influence V. cholerae phage infection and lysogenizing of the host or recipient cell (E. K. Lipp, I. N. G. Rivera, M. Talledo, A. Neale, D. Karaolis, A. Huq, and R. R. Colwell, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. Q427, 2001). Although continued research is needed, it appears that seasonal environmental factors may affect phage-host dynamics and acquisition of virulence genes to a significant degree.
Hierarchy in the cholera model.
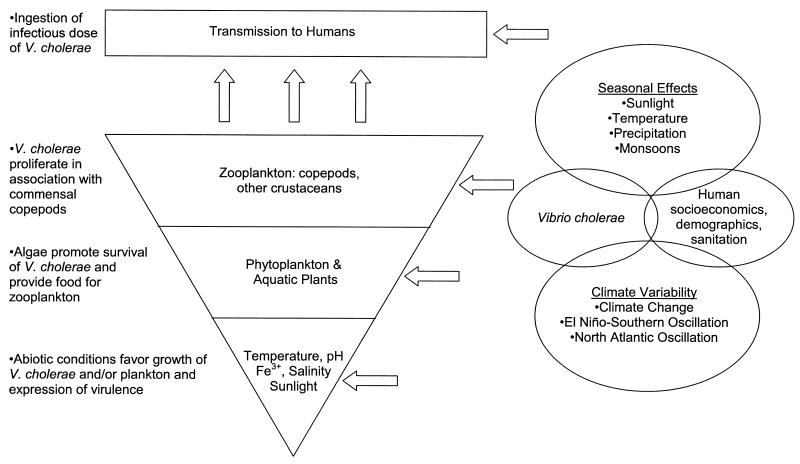
The spatial and temporal patterns of emerging and reemerging diseases have been changing throughout history due to a variety of factors. Their patterns as well as incidence or prevalence of disease are influenced by a complex interaction of direct and indirect factors. Among physical factors, temperature perhaps has the most direct and significant effect on the ecology of most bacteria. It is not any different for V. cholerae, because warmer temperatures in combination with elevated pH and plankton blooms can influence its attachment, growth, and multiplication in the aquatic environment, particularly in association with copepods (71). Here we revisit the model for environmental cholera transmission proposed by Colwell and Huq (30) and suggest a hierarchy to interpret the significance of climate and the environment in cholera and V. cholerae dynamics. A schematic representation of the model is presented in Fig. 1, and examples are detailed in Table 1.
FIG. 1.
Hierarchical model for environmental cholera transmission (modified from Colwell and Huq [30]).
TABLE 1.
Influence of environment, climate, and weather on cholera and V. cholerae dynamics
| Factor | Climate and/or weather drivers | Influence(s) | Representative reference(s) |
|---|---|---|---|
| Temperature | Seasons, interannual variability | Growth of V. cholerae, phytoplankton blooms, infection by temperate phages | 90; Lipp et al., Abstr. 101st Gen. Meet. Am. Soc. Microbiol. |
| Salinity | Seasons, monsoons, ENSO, sea level rise | Growth of V. cholerae, seroconversion, expression of cholera toxin | 111, 148, 151 |
| Sunlight | Seasons, monsoons, interannual variability | Survival of V. cholerae, phytoplankton blooms, induction of CTXφ | 47, 109 |
| pH | Seasons, interannual variability (phytoplankton growth) | Growth of V. cholerae | 22 |
| Fe3+ | Precipitation (runoff), atmospheric deposition (NAO) | Growth of V. cholerae, expression of cholera toxin | 125, 126 |
| Exogenous products of algal growth | Seasons, monsoons, interannual variability in light, nutrients | Survival of V. cholerae | 80, 144 |
| Chitin | Seasons, monsoons, zooplankton blooms (following phytoplankton) | Growth of V. cholerae, attachment to exoskeletons | 71, 117 |
(i) Abiotic factors.
Most Vibrio spp., including V. cholerae, are characterized by an increased growth rate at warm temperatures, which is evident in the higher rates of isolation in the environment during warm months. That is, between epidemics in areas of endemicity in the world it is exceedingly difficult to isolate V. cholerae O1 due to changes in salinity, lower water temperatures, and perhaps seroconversion; however, during periods of warmer water temperatures, success in isolation of V. cholerae O1 rises substantially. Although optimal salinity for growth is between 5 and 25‰, V. cholerae is one of the few Vibrio spp. that can withstand a salinity of 0‰, provided Na+ is available (16, 148). With sufficient dissolved organic matter present, V. cholerae can also grow well at salinities near 45‰ (148). Studies conducted by Miller et al. (110) suggested that strains of V. cholerae vary greatly in their survival in the culturable state under low-salinity conditions (0.05‰). Survival was not related to serogroup, source (clinical or environmental), or geographical origin of the strain (110). Because V. cholerae can withstand low-salinity conditions, it is well adapted to both fresh and brackish water of areas of endemicity such as Bangladesh. However, periodic intrusion of salt water appears to enhance survival in such environments (36). V. cholerae also thrives under high-pH conditions, a parameter that has been used to improve isolation of V. cholerae from environmental samples by enrichment (using alkaline peptone water at pH 8.0 to 8.6). Because of its low solubility in environmental water, iron is often a limiting micronutrient for both bacteria and algae. To deal with this, V. cholerae can produce iron-chelating siderophores to take up insoluble iron from the environment. This adaptive feature allows for prolonged survival when soluble iron is unavailable; survival is thus further improved when insoluble iron (Fe2O3) is available under alkaline pH conditions (126).
Environmental conditions may also affect expression of virulence genes in V. cholerae. As cited above, sunlight can induce propagation of the CTXΦ phage (48), and the viability, i.e., culturability, of V. cholerae remains stable in full sunlight compared to enteric bacteria such as E. coli, which may impart some selective advantage to vibrios at tropical latitudes (109). Expression of CT is optimal at salinities between 2 and 2.5‰, independent of cell concentration (151). Furthermore, moderate levels of introduced iron also increase the expression of CT (125). Therefore, environmental triggers may become epidemiologically important for prevalence of the organism and its virulence (potentially resulting in shorter onset times and lower infectious doses).
(ii) Phytoplankton.
Abiotic factors both affect and are affected by the biotic environment. Sunlight, temperature, and nutrients all influence the growth of phytoplankton and aquatic plants, which in turn alter the dissolved O2 and CO2 content of the water and, therefore, the pH of the surrounding water. Both direct and indirect effects of algal growth subsequently influence the population of V. cholerae in a given environment. Although this is a simplistic description, it does illustrate how environmental factors build upon one another to characterize cholera dynamics.
Cockburn and Cassanos (22) first proposed the theory that ponds in Bangladesh were the main source of infection to the community. They proposed that if the pH in ponds were sufficiently elevated, V. cholerae could outcompete other bacteria and reach infectious dose levels. Experimentally they showed a relationship between elevated pH and onset of cholera cases, which was also related to time of year, light, temperature, and precipitation. The driver of the temporal pH changes in their hypothesis was phytoplankton, which elevated the pH of the surrounding water by the sequestration of CO2 during photosynthesis (22), with the greatest changes occurring under conditions with elevated temperatures, low turbidity, and sufficient light. These insightful observations provided a foundation for the importance of environmental conditions in cholera dynamics but were still lacking critical information on other contributors influencing the ecology of V. cholerae.
Other authors have suggested that there may be a more direct relationship between algae, or aquatic plants, and V. cholerae. In general Vibrio spp. are found at high concentrations in zooplankton samples relative to the surrounding water column community. Simidu et al. (145) reported that nonfermenting bacteria such as pseudomonads are found in association with phytoplankton, whereas vibrios and aeromonads are more likely to be associated with zooplankton. Vibrios are also frequently isolated during red tides in Spain (134). V. cholerae produces mucinase, an enzyme that degrades mucin and mucin-like substances that are often encountered in the gut. Although clearly involved in intestinal colonization during the infection process, this enzyme can also degrade similar substances in plants and algae (139). Research has identified a close association between V. cholerae, algae (Volvox sp., Rhizoclonium fontanum, and green algae), and the mucilaginous sheath of Anabaena spp. (cyanobacteria) (36, 81, 82) and aquatic plants (143). Direct detection from the environment compared with plate counts showed that V. cholerae cells were often VBNC (143). Islam et al. (80, 81) also report that there may be some relationship between virulence and increased toxin expression when V. cholerae is associated with algae (R. fontanum) over time, although the observed relationship may be due to indirect effects such as pH change.
A few studies have shown that V. cholerae may increase its survival and persistence in the environment by association with aquatic plants or algae, and some research suggests that blooms of cyanobacteria, or other phytoplankton, may have a moderate effect on the multiplication of V. cholerae (149). However, epidemics of cholera in Bangladesh have two distinct peaks, one in late spring and one in late fall. Once an algal bloom disintegrates, the bacterial cells require another mechanism of survival and multiplication in the environment to cause later epidemics during a given year (144). Instead of any direct influence of phytoplankton on V. cholerae, it is more likely that the high nutrient levels arising from the breakdown of phytoplankton via algal viruses or other bacteria (142, 160) may have an indirect influence on bacteria other than those involved in phytoplankton disintegration, an interesting, but unproved hypothesis. High phytoplankton production does, however, produce food for zooplankton grazers, the tertiary stage in the cholera model.
(iii) Zooplankton.
In addition to favorably affecting water column pH, increased phytoplankton production also provides additional nutrients for the next level of the food chain, namely, zooplankton (95). Chitinous organisms, e.g., copepods, amphipods, and other crustaceans, are prevalent among zooplankton populations, and chitin is the second most abundant organic compound found in nature. Approximately 1011 metric tons of chitin year−1 is produced in aquatic environments; >109 metric tons is produced by copepods alone (93). If chitin were not degraded, high levels of carbon and nitrogen would remain insoluble and inaccessible to most organisms.
Using chitinase, marine bacteria including Vibrio spp. and V. cholerae play a significant role in chitin remineralization such that very little chitin can be detected in aquatic sediments (68, 88, 89, 93). In 1979, Nalin et al. (117), building on the observations of Kaneko and Colwell (87-89), demonstrated significant adhesion to chitin by V. cholerae and multiplication of cells in chitin suspensions. In addition to providing a food source for vibrios and enhancing survival under starvation conditions (38), chitin also offers protection to V. cholerae at low temperatures (2) and under acidic conditions (such as the human gut, which could be important if copepods carrying V. cholerae are ingested in drinking water) (117).
Other vibrios besides V. parahaemolyticus and V. cholerae have been studied with respect to chitin, as all vibrios tested to date are chitinolytic. Mechanisms of adhesion to chitin by Vibrio furnissi have been extensively studied. Yu et al. (165) found that populations of cells exhibited an adhesion-deadhesion behavior that allowed a number of free-swimming cells to relocate when a chitin source was depleted and, thereby, go in search of another source. Bassler et al. (5) subsequently demonstrated that Vibrio cells are also positively chemotactic to products of chitin hydrolysis. In V. cholerae, surface proteins have been identified in the attachment to chitin particles (152). While the exact mechanisms of chitin colonization in V. cholerae are not known, there is a distinct chitin recognition system (152). Together, these studies suggest a quorum-type response in the colonization of chitin particles and, therefore, copepods and other chitinous animals.
There may be a synergistic effect between phytoplankton and zooplankton in chitin colonization by V. cholerae. An alkaline pH of 8.5, often associated with algal blooms, was found to positively influence the attachment of V. cholerae to copepods (71). Huq et al. (79) proposed that once cells of V. cholerae attach to zooplankton, they are protected from the external environment and begin to proliferate, taking advantage of the increased surface area and improved conditions of nutrition, the latter derived from the disintegration of phytoplankton and release of nitrogenous products into the water (72). It was further suggested that during interepidemic monsoon seasons in Bangladesh, when there is a significant alteration of nutritional conditions arising from seasonal changes in the chemical parameters of the water (120), V. cholerae may become nonculturable (73).
The simple presence of crustacean copepods enhances the survival of V. cholerae O1, as demonstrated in laboratory microcosm experiments (79). In the field (Bangladesh), VBNC cells of V. cholerae O1 and O139, detected by microscopy using direct fluorescent antibodies, were found attached to chitin particles, an observation providing support for results obtained in laboratory experiments (A. Huq et al., Letter, Lancet 345:1249, 1995). Other investigators also confirmed results of previous studies showing V. cholerae O1 surviving for significantly longer periods of time when attached to copepods (43). Such protection afforded to VBNC cells by chitin or chitin-containing organisms in the environment and in the human digestive system surely may play a role in the epidemiology of cholera.
Scanning electron microscopy (SEM) showed that egg cases and the oral region of copepods were sites of significantly enhanced attachment by V. cholerae (43, 79). Attachment was found to be strong for V. cholerae but significantly less so for E. coli or Pseudomonas spp.; that is, preparation for SEM requires vigorous washing that did not separate cells of V. cholerae from sites where they were attached but did remove other bacteria. Dumontet et al. (43) found that bacterial cells attached to copepods in the natural environment detached when SEM was employed. This is a particularly important finding because both freely swimming and nonculturable V. cholerae attach to copepod eggs and multiply rapidly (79). Studies are currently under way in our laboratory to determine if attachment is reversible.
According to Oppenheimer et al. (120), during the Indian Ocean monsoon (rainy) season in June and July, zooplankton populations decrease because of reduced levels of nutrients. During August and September, after the monsoon season, levels of nutrients significantly increase and blooms of phytoplankton followed by zooplankton occur. Kiorboe and Neilson (95) published a report on an extensive study of copepod production, demonstrating two distinct seasons for the production of eggs by several species of copepods. One peak occurs in February through April, and the other occurs during the months of August and September. Interestingly, the timing correlates very well with incidence of diarrhea and cholera cases, particularly in India and Bangladesh. In Japan, a seasonal relationship between culturable V. cholerae and zooplankton was not detected (156), but Japan, like the United States, enjoys excellent sanitation and safe drinking water; in Japan, epidemics of cholera do not occur, and cases tend to be sporadic and few in number. Nonetheless, results of the research by Oppenheimer et al. (120) and Kiorboe and Neilson (95) in a region of cholera endemicity fit very well into the original hypothesis of Kaneko and Colwell (88) and Huq et al. (79) that copepods play an important role in the survival, multiplication, and transmission of vibrios, including V. cholerae, in the natural aquatic environment.
Cells of V. cholerae also colonize the gut of copepods. Singleton et al. (148) proposed that there might be an environmental role for CT production by V. cholerae in this type of association. In the human intestine, binding to the CT receptors of epithelial cells eventually results in an efflux of Na+ and other electrolytes and water, resulting in diarrhea. If CT affects the epithelial cells of crustaceans in a similar manner, it would provide an effective mechanism to regulate the concentration of Na+ when it is not optimal for the host. CT could, thereby, play a role in the osmoregulation of the copepod (25).
Evidence to date suggests that there may be a specific symbiosis between species of copepods and V. cholerae. A recent report stated that V. cholerae may also be associated with Chironomus sp. (midge) egg masses in freshwater (14), but V. cholerae is known to be more commonly found in association with zooplankton (25). Our preliminary analyses of data from Bangladesh ponds show only a very few chironomid egg masses, and none yielded V. cholerae cells. Furthermore, we detected no relationship of chironomid egg masses with cholera epidemics (S. Islam, A. Huq, G. B. Nair, and R. R. Colwell, unpublished data, 2002), whereas zooplankton samples from Chesapeake Bay, Peru, and Bangladesh all reveal the presence of V. cholerae when examined using direct staining and molecular genetic methods. Further evidence indicates that V. cholerae may preferentially associate with Acartia tonsa in coastal waters (M. L. Lizzaraga-Partida, personal communication, 2001).
(iv) Transmission to humans.
Cholera is primarily a waterborne disease, and an epidemic may be enhanced by secondary transmission. Yet, it has been demonstrated that V. cholerae can be transmitted to humans via the environment, in drinking and cooking water, irrigation water, and shellfish. With active environmental monitoring systems using epifluorescence microscopy and molecular methods for direct detection, the presence of V. cholerae, ctxAB-carrying bacteria, and vibriophages in sewage and surface waters has been shown to precede the onset of cholera outbreaks by 1 to 4 months (52, 104, 106, 154). Furthermore, cholera is dose dependent, with at least 104 cells required for infection (69). A single copepod may carry 104 to 106 cells of V. cholerae, and thus incidental ingestion of a few copepods in untreated drinking water could result in an infection (27). Although, V. cholerae can be transmitted directly in water, it also acts in a vector- or host-dependent manner because the presence of copepods substantially raises the dose of V. cholerae over that in copepod-free water. Preliminary studies show that Bangladeshi households which filter their pond water through sari cloth before drinking reduce their risk of cholera infections by approximately 50% (76).
Extreme Events
Cholera is often considered a reemerging disease, in part because infections are appearing in novel communities or communities where the disease had been absent for many years, and the range of areas of endemicity is expanding. This may be due to changes in the environment or climate that promote favorable conditions for the pathogens and/or its invertebrate host. Because V. cholerae clearly plays a role in coastal ecosystems in association with plankton communities, it is obvious that this opportunistic pathogen cannot be eradicated, but, certainly, the disease can be controlled (31). Ecological investigations that focus on relating V. cholerae temporally and spatially to easily measured predictors or proxies offer a sound method to prevent exposure and, therefore, cholera. Progress has been made along these lines via historical examination of cholera cases in areas of endemicity and by defining clear seasonal trends and anomalies associated with climate variability and change.
Climate variability and change.
Few would disagree that our climate is changing, although the drivers of climate change are a subject of continued controversy. Recent research has focused on the impact of such changes on various ecosystems and public health (137, 158). The effects of climate variability driven by natural cycles (e.g., ENSO, NAO, and others) or by anthropogenic activities (greenhouse gas emissions, etc.) will superimpose anomalous weather patterns onto typical seasonal trends.
Climate or weather may affect each of the levels in the cholera model presented here. Climatologists predict a 1.4 to 5.8°C rise in mean temperatures over the next 100 years (70). Increasing temperatures would be expected to expand the range and increase the prevalence of V. cholerae and cholera both geographically and temporally, if public health measures are not implemented. For example, temperature shifts would alter the latitudinal distribution of plankton species. Should sea level rise, as anticipated with higher temperatures, inland areas would experience greater saltwater intrusion and increased levels of marine and estuarine bacteria, including V. cholerae. Changes in sunlight and/or UV intensity could increase the rates of induction and propagation of the CTXΦ phage and, thereby, increase the potential for the emergence of new toxigenic V. cholerae strains. Introduction of insoluble iron into the environment by agricultural practices, industrial pollution, and/or dust events, such as the transport of African dust to the Caribbean and southeastern United States, may lead to increased survival and virulence in affected populations. The introduction of dust may be related to modes of climate variability such as the NAO.
In light of climatic and environmental drivers in cholera disease dynamics, it is necessary to take into account the capabilities of public health systems and improved sanitation systems. It cannot be assumed that the number of cholera cases will increase because the risk of cholera, especially in developing countries, may increase. The disease itself is preventable if sanitation is sufficient to protect public health and measures such as boiling of water can be implemented.
While it is impossible to predict precisely the outcome of cholera dynamics with climate change, analyses of cholera and V. cholerae in the context of known variability, e.g., ENSO events, have been undertaken that demonstrate anomalous patterns in disease dynamics superimposed on normal seasonal trends.
The ENSO story.
The ENSO is a normal mode of climate variability that, in simple terms, involves pressure shifts across the equatorial Pacific. During the El Niño phase, a zone of high pressure moves eastward, easterly trade winds slacken, and a slow-moving low-frequency wave moves from the west to the east, resulting in depression of the thermocline along the South American coast and warming in the eastern equatorial Pacific. During La Niña episodes the trend is somewhat reversed, and there is a cooling in the eastern equatorial Pacific. Teleconnections from this coupled ocean-atmosphere phenomenon result in anomalous weather patterns around the world. Because of the widespread effects of the ENSO and because it can be accurately forecast, it offers an attractive starting point for building predictive models for disease, such as cholera.
Though not necessarily directly related, the frequencies of both El Niño events and cholera outbreaks have been increasing since the 1970s (31). Cholera dynamics in areas of South America (Peru) and the Bay of Bengal seem to be particularly affected by ENSO-driven anomalies. It has been hypothesized that an El Niño event in 1991 may have been the trigger that resulted in the resurgence of cholera in Peru (25, 114). Warm waters along the coast, coupled with plankton blooms driven by El Niño rains, may have helped to amplify the population of V. cholerae already in the environment (25, 114). Warm temperatures in Peru during the 1997-1998 event were also associated with increased numbers of diarrheal cases, including cholera.
In Bangladesh, cholera outbreaks are related to the end of the monsoon season on an annual basis and to ENSO on an interannual frequency. Pascual et al. (124) used time series analyses to examine the historical association between cholera in Bangladesh and ENSO (1980 to1998). Both previous disease levels in the community and ENSO were significant predictors in the nonlinear models, and interannual climate variability associated with ENSO clearly plays a role in the year-to-year variability of cholera outbreaks (124). Because of the recognized connection between ENSO and weather around the Indian Ocean (153), it is tempting to use ENSO to predict variability in the monsoon season and thus variability in annual cholera outbreaks. However, the ENSO-monsoon connection appears to be bidirectional and therefore cannot be used in a predictive capacity (153).
CONCLUSIONS
V. cholerae has long been known as a fecal-oral pathogen, and indeed, infection rates are significantly greater in areas with poor sanitation; however, the evidence showing that V. cholerae is naturally present in warm, brackish environments is overwhelming. Evidence collected over the last 20 years indicates a close association of V. cholerae with copepods, such that the bacteria are concentrated on the exoskeleton and gut of the zooplankters relative to the surrounding water. When communities rely on untreated environmental water sources for bathing, cooking, and drinking water, the incidental ingestion of copepods, which carry a high “dose” of V. cholerae, can initiate an infection. Likewise, the likelihood of consuming an infectious dose is higher when a bloom of copepods occurs in the water. Based on this association, a simple and effective measure to filter plankton from water using sari material has been developed, and preliminary evidence shows a significant reduction in cholera cases in those households using this filtration procedure (R. R. Colwell, A. Huq, S. Islam, K. M. Aziz, M. Yunus, N. H. Khan, and E. Russek-Cohen, unpublished data, 2002).
Because of the aquatic and oceanic connection of V. cholerae and commensal copepods and the moderating effect of the aquatic environment, outbreaks theoretically follow more predictable patterns than most vector-borne diseases, because local microenvironments largely control the vector population. Drivers such as sea surface temperature, sea surface height, and plankton blooms can be remotely sensed and used to forecast outbreaks (104). While issues of sanitation and human behavioral adaptation cannot be ignored, we propose that the hierarchical model presented here (Fig. 1) and elsewhere (30) can serve as a template for future research on similar climate-sensitive diseases to define critical parameters and develop methods for prediction.
Acknowledgments
This work was supported by the National Institute of Nursing Research of the National Institutes of Health (grant ROINR042701A1).
REFERENCES
- 1.Almeida, R. J., D. N. Cameron, W. L. Cook, and I. K. Wachsmuth. 1992. Vibriophage VcA-3 as an epidemic strain marker for the U.S. Gulf Coast Vibrio cholerae O1 clone. J. Clin. Microbiol. 30:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amako, K., S. Shimodori, T. Imoto, S. Miake, and A. Umeda. 1987. Effects of chitin and its soluble derivatives on survival of Vibrio cholerae O1 at low temperature. Appl. Environ. Microbiol. 53:603-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbieri, E., L. Fladanzo, C. Fiorentini, A. Pianetti, W. Affone, A. Fabbri, P. Matarrese, A. Casiere, M. Katouli, I. Kuhn, R. Mollby, F. Bruscolini, and G. Donelli. 1999. Occurrence, diversity, and pathogenicity of halophilic Vibrio spp. and non-O1 Vibrio cholerae from estuarine waters along the Italian Adriatic coast. Appl. Environ. Microbiol. 65:2748-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, T. P., D. W. Pierce, and R. Schnur. 2001. Detection of anthropogenic climate change in the world's oceans. Science 292:270-274. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., P. J. Gibbons, C. Yu, and S. Roseman. 1991. Chitin utilization by marine bacteria: chemotaxis to chitin oligosaccharides by Vibrio furnissi. J. Biol. Chem. 266:24268-24275. [PubMed] [Google Scholar]
- 6.Beltran, P., G. Delgado, A. Navarro, F. Trujillo, R. K. Selander, and A. Cravioto. 1999. Genetic diversity and population structure of Vibrio cholerae. J. Clin. Microbiol. 37:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentivoglio, M., and P. Pacini. 1995. Filippo Pacini: a determined observer. Brain Res. Bull. 38:161-165. [DOI] [PubMed] [Google Scholar]
- 8.Bernard, S. M., A. G. J. M. Samet, K. L. Ebi, and I. Romieu. 2001. The potential impacts of climate variability and change on air pollution-related health effects in the United States. Environ. Health Perspect. 109:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bik, E. M., A. E. Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bockemuhl, J., K. Rock, B. Wohlers, V. Aleksic, and R. Wokatsch. 1986. Seasonal distribution of facultatively enteropathogenic vibrios (Vibrio cholerae, Vibrio mimicus and Vibrio parahaemolyticus) in the freshwater of the Elbe River at Hamburg. J. Appl. Bacteriol. 60:435-442. [DOI] [PubMed] [Google Scholar]
- 11.Borroto, R. J., and R. Martinez-Piedra. 2000. Geographical patterns of cholera in Mexico, 1991-1996. Int. J. Epidemiol. 29:764-772. [DOI] [PubMed] [Google Scholar]
- 12.Boyd, E. F., A. J. Heilpern, and M. K. Waldor. 2000. Molecular analyses of a putative CTXφ precursor and evidence for independent acquisition of distinct CTXφs by toxigenic Vibrio cholerae. J. Bacteriol. 182:5530-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd, E. F., K. E. Moyer, L. Shi, and M. K. Waldor. 2000. Infectious CTXΦ and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broza, M., and M. Halpern. 2001. Pathogen reservoirs. Chironomid egg masses and Vibrio cholerae. Nature 412:40. [DOI] [PubMed] [Google Scholar]
- 15.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavari, B. Z., and R. R. Colwell. 1981. Effect of salt on survival and growth of Vibrio cholerae. In H. Shuval (ed.), Dev. Arid Zone Ecol. Environ. Quality 31:227-231. [Google Scholar]
- 17.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Checkley, W., L. D. Epstein, R. H. Gilman, R. I. Figueroa, J. A. Cama, J. A. Patz, and R. E. Black. 2000. Effect of El Niño and ambient temperature on hospital admissions and diarrhoeal diseases in Peruvian children. Lancet 355:442-450. [DOI] [PubMed] [Google Scholar]
- 19.Cholera Working Group. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387-390. [PubMed] [Google Scholar]
- 20.Citarella, R. V., and R. R. Colwell. 1970. Polyphasic taxonomy of the genus Vibrio: polynucleotide sequence relationships among selected Vibrio species. J. Bacteriol. 104:434-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cochran, P. K., and J. H. Paul. 1998. Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl. Environ. Microbiol. 64:2308-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cockburn, T. A., and J. G. Cassanos. 1960. Epidemiology of endemic cholera. Public Health Rep. 75:791-803. [PMC free article] [PubMed] [Google Scholar]
- 23.Colwell, R. R. 1961. Commensal bacteria of marine animals. A study of their distribution, physiology and taxonomy. Ph.D. thesis. University of Washington, Seattle.
- 24.Colwell, R. R. 1973. Genetic and phenetic classification of bacteria. Adv. Appl. Microbiol. 16:137-176. [DOI] [PubMed] [Google Scholar]
- 25.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 26.Colwell, R. R. 1970. Polyphasic taxonomy of the genus Vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J. Bacteriol. 104:410-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colwell, R. R., P. Brayton, P. Harrington, B. Tall, A. Huq, and M. Levine. 1996. Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 12:28-31. [DOI] [PubMed] [Google Scholar]
- 28.Colwell, R. R., P. Brayton, M. Tamplin, S. Huq, M. Voll, D. Roszack, S. Muralidhar, C. Somerville, and J. Grimes. 1988. Pathogenic potential, including production of cholera toxin, in viable but not recoverable Vibrio cholerae O1 in environmental microcosms, p. 29-39. In S. Kuwahara and N. F. Pierce (ed.), Advances in research on cholera and related diarrheas, vol. 5. KTK Scientific Publishers, Tokyo, Japan.
- 29.Colwell, R. R., P. R. Brayton, D. J. Grimes, D. R. Roszak, S. A. Huq, and L. M. Palmer. 1985. Viable but non-culturable Vibrio cholerae and related environmental pathogens in the environment: implication for release of genetically engineered microorganisms. Bio/Technology 3:817-820. [Google Scholar]
- 30.Colwell, R. R., and A. Huq. 1994. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann. N. Y. Acad. Sci. 740:44-54. [DOI] [PubMed] [Google Scholar]
- 31.Colwell, R. R., and A. Huq. 1999. Global microbial ecology: biogeography and diversity of Vibrios as a model. J. Appl. Microbiol. Symp. Suppl. 85:134S-137S. [DOI] [PubMed] [Google Scholar]
- 32.Colwell, R. R., and A. Huq. 1994. Vibrios in the environment: viable but nonculturable Vibrio cholerae, p. 117-133. In K. Wachsmuth, P. A. Blake, and R. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 33.Colwell, R. R., A. Huq, M. A. R. Chowdhury, P. R. Brayton, and B. Xu. 1995. Serogroup conversion of Vibrio cholerae. Can. J. Microbiol. 41:946-950. [DOI] [PubMed] [Google Scholar]
- 34.Colwell, R. R., J. Kaper, and S. W. Joseph. 1977. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science 198:394-396. [PubMed] [Google Scholar]
- 35.Colwell, R. R., and J. A. Patz. 1998. Climate, infectious disease and health: an interdisciplinary perspective. American Academy of Microbiology, Washington, D.C. [PubMed]
- 36.Colwell, R. R., M. L. Tamplin, P. R. Brayton, A. L. Gauzens, B. D. Tall, D. Harrington, M. M. Levine, S. Hall, A. Huq, and D. A. Sack. 1990. Environmental aspects of Vibrio cholerae in transmission of cholera, p. 327-343. In R. B. Sack and Y. Zinnaka (ed.), Advances in research on cholera and related diarrhoeas. K.T.K. Scientific Publishers, Tokyo, Japan.
- 37.Comrie, A. C., K. N. Kolivras, N. Ampel, M. Bultman, P. Johnson, S. Rogan, K. Smith, and S. Yool. 1999. Exploring climate variability and valley fever, p. 393. In Proceedings of the 24th Annual Climate Diagnostics and Prediction Workshop. U.S. Department of Commerce, Springfield, Va.
- 38.Dawson, M. P., B. A. Humphrey, and K. C. Marshall. 1981. Adhesion: a tactic in the survival strategy of a marine vibrio during starvation. Curr. Microbiol. 6:195-199. [Google Scholar]
- 39.del Mar Lleo, M., S. Pierobon, M. C. Tufi, C. Signoretto, and P. Canepari. 2000. mRNA detection over time by reverse transcription-PCR for monitoring viability over time in Enterococcus faecalis viable but nonculturable populations maintained in a laboratory microcosm. Appl. Environ. Microbiol. 66:4564-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desmarchelier, P. M., and C. R. Senn. 1989. A molecular epidemiological study of Vibrio cholerae in Australia. Med. J. Aust. 150:631-634. [DOI] [PubMed] [Google Scholar]
- 41.Desmarchelier, P. M., F. Y. K. Wong, and K. Mallard. 1995. An epidemiological study of V. cholerae O1 in the Australian environment based on the RNA gene polymorphism. Epidemiol. Infect. 115:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorsch, M., D. Lane, E. Stack, and E. Brandt. 1992. Towards a phylogeny of the genus Vibrio based on 16S rRNA sequences. Int. J. Syst. Bacteriol. 42:58-63. [DOI] [PubMed] [Google Scholar]
- 43.Dumontet, S., K. Krovacek, S. B. Baloda, R. Grottoli, V. Pascuale, and S. Vanucci. 1996. Ecological relationship between Aeromonas and Vibrio spp. and planktonic copepods in the coastal marine environment in southern Italy. Comp. Immunol. Microbiol. Infect. Dis. 19:245-254. [DOI] [PubMed] [Google Scholar]
- 44.Dumontier, S., and P. Berche. 1998. Vibrio cholerae O22 might be a putative source of exogenous DNA resulting in the emergence of the new strain of Vibrio cholerae O139. FEMS Microbiol. Lett. 164:91-98. [DOI] [PubMed] [Google Scholar]
- 45.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faruque, S. M., Asadulghani, M. N. Saha, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faruque, S. M., Asadulghani, M. M. Rahman, M. K. Waldor, and D. A. Sack. 2000. Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect. Immun. 68:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faruque, S. M., Asadulghani, A. R. M. Abdul Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. N. Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXΦ. Infect. Immun. 67:5723-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faruque, S. M., M. N. Saha, Asadulghani, D. A. Sack, R. B. Sack, Y. Takeda, and G. B. Nair. 2000. The O139 serogroup of Vibrio cholerae comprises diverse clones of epidemic and nonepidemic strains derived from multiple V. cholerae O1 or non-O1 progenitors. J. Infect. Dis. 182:1161-1168. [DOI] [PubMed] [Google Scholar]
- 51.Franceschini, P. 1971. Filippo Pacini e il cholera. Phisis 13:324-332. [PubMed] [Google Scholar]
- 52.Franco, A., A. D. Fix, A. Prada, E. Paredes, J. C. Palomino, A. C. Wright, J. A. Johnson, R. McCarter, H. Guerra, and J. G. Morris. 1997. Cholera in Lima, Peru correlates with prior isolation of Vibrio cholerae from the environment. Am. J. Epidemiol. 146:1067-1075. [DOI] [PubMed] [Google Scholar]
- 53.Garay, E., A. Arnau, and C. Amaro. 1985. Incidence of Vibrio cholerae and related vibrios in a coastal lagoon and seawater influenced by lake discharges along an annual cycle. Appl. Environ. Microbiol. 50:426-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glass, R. I., and R. E. Black. 1992. The epidemiology of cholera, p. 129-154. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum Medical Book Company, New York, N.Y.
- 55.Gonzalez, J. M., J. Iriberri, L. Egea, and I. Barcina. 1992. Characterization of culturability, protistan grazing, and death of enteric bacteria in aquatic ecosystems. Appl. Environ. Microbiol. 58:998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenough, G., M. McGeehin, S. M. Bernard, J. Trtanj, J. Riad, and D. Engleberg. 2001. The potential impacts of climate variability and change on health impacts of extreme weather events in the United States. Environ. Health Perspect. 109:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Griffin, D. W., V. Garrison, and E. A. Shinn. 2001. African dust in the Caribbean atmosphere: microbiology and public health. Aerobiologia 17:203-213. [Google Scholar]
- 58.Gubler, D. J., P. Reiter, K. L. Ebi, W. Yap, R. Nasci, and J. A. Patz. 2001. Climate variability and change in the United States: potential impacts on vector- and rodent-borne diseases. Environ. Health Perspect. 109:223-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gueri, M., C. Gonzalez, and V. Morin. 1986. The effect of the floods caused by “El Niño” on health. Disasters 10:118-124. [Google Scholar]
- 60.Hales, S., P. Weinstein, Y. Souares, and A. Woodward. 1999. El Niño and the dynamics of vectorborne disease transmission. Environ. Health Perspect. 107:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hay, S. A., M. F. Myers, D. S. Burke, D. W. Vaughn, T. Endy, N. Ananda, G. D. Shanks, R. W. Snow, and D. J. Rogers. 2000. Etiology of interepidemic periods of mosquito-borne disease. Proc. Natl. Acad. Sci. USA 97:9335-9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heidelberg, J. F. 1997. Seasonal abundance of bacterioplankton populations of bacteria, gamma proteobacteria, Vibrio/Photobacterium, Vibrio vulnificus, Vibrio cholerae/Vibrio mimicus, and Vibrio cincinnatiensis associated with zooplankton in the Choptank River, Maryland. Ph.D. thesis. University of Maryland, College Park.
- 63.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 64.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishman, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Ventor, and C. M. Frasier. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hjelle, B., and G. E. Glass. 2000. Outbreak of hantavirus infections in the Four Corners region of the United States in the wake of the 1997-1998 El Niño-southern oscillation. J. Infect. Dis. 181:1569-1573. [DOI] [PubMed] [Google Scholar]
- 66.Hlady, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176-1183. [DOI] [PubMed] [Google Scholar]
- 67.Holt, J. G., N. R. Krieg, P. H. A. Sneath, and D. Bergey. 1994. Bergey's manual of determinative bacteriology, 9th ed. Lippincott, Williams & Wilkins, East Lansing, Mich.
- 68.Hood, M. A., and S. P. Meyers. 1977. Microbiological and chitinoclastic activities associated with Penaeus stiferus. J. Oceanogr. Soc. Jpn. 33:235-241. [Google Scholar]
- 69.Hornick, R. B., S. I. Music, and R. Wenzel. 1971. The Broad Street pump revisited: response of volunteers to ingested cholera Vibrio. Bull. N. Y. Acad. Med. 47:1181-1191. [PMC free article] [PubMed] [Google Scholar]
- 70.Houghton, J. T., Y. Ding, D. J. Griggs, M. Noguer, P. J. van der Linden, and D. Xiaosu (ed.). 2001. Climate change 2001: the scientific basis. Contribution of Working Group I to the third assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom.
- 71.Huq, A. 1984. The role of planktonic copepods in the survival and multiplication of V. cholerae in the aquatic environment. Ph.D. thesis. University of Maryland, College Park.
- 72.Huq, A., M. A. R. Chowdhury, A. Felsenstein, R. R. Colwell, R. Rahman, and K. M. B. Hossain. 1988. Detection of V. cholerae from aquatic environments in Bangladesh, p. 259-264. In M. Yasuno and B. A. Whitton (ed.), Biological monitoring of environment pollution. Tokai University, Tokyo, Japan.
- 73.Huq, A., and R. R. Colwell. 1996. A microbiological paradox: viable but nonculturable bacteria with special reference to Vibrio cholerae. J. Food Prot. 59:96-101. [DOI] [PubMed] [Google Scholar]
- 74.Reference deleted.
- 75.Huq, A., R. R. Colwell, R. Rahman, A. Ali, M. A. R. Chowdhury, S. Parveen, D. A. Sack, and E. Russek-Cohen. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent monoclonal antibody and culture method. Appl. Environ. Microbiol. 56:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huq, A., S. Islam, E. Russek-Cohen, M. Yunus, A. Aziz, A. Mahmud, N. H. Khan, J. Chakrabarty, B. Sack, and R. R. Colwell. 2001. Simple water filtration for cholera prevention, p. 15. Proceedings of the US-Japan Cholera Meeting. U.S.-Japan Cooperative Medical Science Program, Tokyo, Japan.
- 77.Huq, A., I. N. G. Rivera, and R. R. Colwell. 2000. Epidemiological significance of viable but nonculturable microorganisms, p. 301-323. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. American Society for Microbiology Press, Washington, D.C.
- 78.Huq, A., R. Sack, and R. Colwell. 2001. Cholera and global ecosystems, p. 327-347. In J. Aron and J. Patz (ed.), Ecosystem change and public health: a global perspective. Johns Hopkins University Press, Baltimore, Md.
- 79.Huq, A., E. B. Small, P. A. West, M. I. Huq, R. Rahman, and R. R. Colwell. 1983. Ecological relationship between Vibrio cholerae and planktonic copepods. Appl. Environ. Microbiol. 45:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Islam, M. S., B. S. Draser, and D. J. Bradley. 1990. Long-term persistence of toxigenic V. cholerae O1 in the mucilaginous sheath of a blue-green algae, Anabaena variabilis. J. Trop. Med. Hyg. 93:133-139. [PubMed] [Google Scholar]
- 81.Islam, M. S., B. S. Draser, and D. J. Bradley. 1989. Attachment of toxigenic Vibrio cholerae O1 to various freshwater plants and survival with filamentous green algae Rhizoclonium fontanum. J. Trop. Med. Hyg. 92:396-401. [PubMed] [Google Scholar]
- 82.Islam, M. S., Z. Rahim, M. J. Alam, S. Begum, S. M. Moniruzzaman, A. Umeda, K. Amakao, M. J. Albert, R. B. Sack, and R. R. Colwell. 1999. Association of Vibrio cholerae O1 with the cyanobacterium, Anabaena sp., elucidated by polymerase chain reaction and transmission electron microscopy. Trans. R. Soc. Trop. Med. Hyg. 93:36-40. [DOI] [PubMed] [Google Scholar]
- 83.Jannasch, H. W. 1969. Estimations of bacterial growth rates in natural waters. J. Bacteriol. 99:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang, S. C., V. Louis, N. Choopun, A. Sharma, A. Huq, and R. R. Colwell. 2000. Genetic diversity of Vibrio cholerae in Chesapeake Bay determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang, S. C., M. Matte, G. Matte, A. Huq, and R. R. Colwell. 2000. Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang, S. C., and J. H. Paul. 1998. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 64:2780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaneko, T., and R. R. Colwell. 1975. Adsorption of Vibrio parahaemolyticus onto chitin and zooplanktonic copepods. Appl. Microbiol. 29:269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaneko, T., and R. R. Colwell. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaneko, T., and R. R. Colwell. 1975. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl. Microbiol. 30:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaper, J. B., and J. Hacker (ed.). 1999. Pathogenicity islands and other mobile virulence elements of Vibrio cholerae. American Society for Microbiology, Washington, D. C.
- 92.Karaolis, D. K., R. Lan, and P. R. Reeves. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keyhani, N. O., and S. Roseman. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108-122. [DOI] [PubMed] [Google Scholar]
- 94.Khan, M. U., M. Shahidullah, M. S. Haque, and W. U. Ahmed. 1984. Presence of vibrios in surface water and their relation with cholera in a community. Trop. Geogr. Med. 36:335-340. [PubMed] [Google Scholar]
- 95.Kiorboe, T., and T. J. Neilson. 1994. Regulation of zooplankton biomass and production and production in a temperate coastal ecosystem. 1. Copepods. Limnol. Oceanogr. 39:493-507. [Google Scholar]
- 96.Kita-Tsukamoto, K., H. Oyaizu, K. Namba, and U. Simidu. 1993. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int. J. Syst. Bacteriol. 43:8-19. [DOI] [PubMed] [Google Scholar]
- 97.Kurath, G., and Y. Morita. 1983. Starvation-survival physiological studies of a marine Pseudomonas sp. Appl. Environ. Microbiol. 45:1206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Langford, I. H., and G. Bentham. 1995. The potential effects of climate change on winter mortality in England and Wales. Int. J. Biometeorol. 38:141-147. [DOI] [PubMed] [Google Scholar]
- 99.Levitus, S., J. I. Antonov, J. Wang, T. L. Delworth, K. W. Dixon, and A. L. Broccoli. 2001. Anthropogenic warming of the earth's climate system. Science 292:267-270. [DOI] [PubMed] [Google Scholar]
- 100.Linthicum, K. J., A. Anyamba, C. J. Tucker, P. W. Kelley, M. F. Myers, and C. J. Peters. 1999. Climate and satellite indicators to forecast rift valley fever epidemics in Kenya. Science 285:397-400. [DOI] [PubMed] [Google Scholar]
- 101.Lipp, E. K., R. Kurz, R. Vincent, C. Rodriguez-Palacios, S. R. Farrah, and J. B. Rose. 2001. Seasonal variability and weather effects on microbial fecal pollution and enteric pathogens in a subtropical estuary. Estuaries 24:266-276. [Google Scholar]
- 102.Lipp, E. K., C. Rodriguez-Palacios, and J. B. Rose. 2001. Occurrence and distribution of the human pathogen Vibrio vulnificus in a subtropical Gulf of Mexico estuary. Hydrobiologia 460:165-173. [Google Scholar]
- 103.Lipp, E. K., N. Schmidt, M. E. Luther, and J. B. Rose. 2001. Determining the effects of El Niño-southern oscillation events on coastal water quality. Estuaries 24:491-497. [Google Scholar]
- 104.Lobitz, B., L. Beck, A. Huq, B. Wood, G. Fuchs, A. S. G. Faruque, and R. R. Colwell. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. USA 97:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reference deleted.
- 106.Madico, G., W. Checkley, R. H. Gilman, N. Bravo, L. Cabrera, M. Calderon, and A. Ceballos. 1996. Active surveillance for Vibrio cholerae O1 and vibriophages in sewage water as a potential tool to predict cholera outbreaks. J. Clin. Microbiol. 34:2968-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reference deleted.
- 108.Martins, M. T., G. V. A. Pessoa, P. S. Sanchez, M. I. Z. Sato, C. A. Coimbrao, C. K. Monteiro, and E. Marques. 1991. Occurrence of V. cholerae O1 non-toxigenic in wastewaters from Sao Paulo, Brazil. Water Sci. Technol. 24:363-366. [Google Scholar]
- 109.Mezrioui, N., K. Oufdou, and B. Baleux. 1995. Dynamics of non-O1 Vibrio cholerae and fecal coliforms in experimental stabilization ponds in the arid region of Marrakesh, Morocco, and the effect of sunlight on their experimental survival. Can. J. Microbiol. 41:489-498. [DOI] [PubMed] [Google Scholar]
- 110.Miller, C. J., B. S. Draser, and R. J. Heyes. 1984. Response to toxigenic V. cholerae O1 to physicochemical stresses in the aquatic environment. J. Hyg. 93:475-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Montilla, R., M. A. Chowdhury, A. Huq, B. Xu, and R. R. Colwell. 1996. Serogroup conversion of Vibrio cholerae non-O1 to Vibrio cholerae O1: effect of growth state of cells, temperature, and salinity. Can. J. Microbiol. 42:87-93. [DOI] [PubMed] [Google Scholar]
- 112.Morris, J. G. 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179-191. [DOI] [PubMed] [Google Scholar]
- 113.Motes, M., A. DePaola, S. Zywno-Van Ginkel, and M. McPhearson. 1994. Occurrence of toxigenic Vibrio cholerae O1 in oysters in Mobile Bay, Alabama: an ecological investigation. J. Food Prot. 57:975-980. [DOI] [PubMed] [Google Scholar]
- 114.Mourina-Perez, R. R. 1998. Oceanography and the 7th cholera pandemic. Epidemiology 9:355-357. [DOI] [PubMed] [Google Scholar]
- 115.Munro, P. M., and R. R. Colwell. 1996. Fate of Vibrio cholerae O1 in seawater microcosms. Water Res. 30:47-50. [Google Scholar]
- 116.Murphree, R. I., and M. L. Tamplin. 1991. Uptake and retention of Vibrio cholerae O1 in the Eastern oyster, Crassostrea virginica. Appl. Environ. Microbiol. 61:3656-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nalin, D. R., V. Daya, A. Reid, M. M. Levine, and L. Cisneros. 1979. Adsorption and growth of Vibrio cholerae on chitin. Infect. Immun. 25:768-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 119.Ogg, J. E., M. B. Shrestha, and L. Ponadyl. 1978. Phage-induced changes in Vibrio cholerae: serotype and biotype conversions. Infect. Immun. 19:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oppenheimer, J. R., M. G. Ahmad, A. Huq, K. A. Haque, A. K. M. A. Alam, K. M. S. Aziz, S. Ali, and A. S. M. Haque. 1978. Limnological studies in three ponds in Dhaka, Bangladesh. Bangladesh J. Fisher. 1:1-28. [Google Scholar]
- 121.O'Shea, M. L., and R. Field. 1992. Detection and disinfection of pathogens in storm-generated flows. Can. J. Microbiol. 38:267-276. [DOI] [PubMed] [Google Scholar]
- 122.Pacini, F. 1854. Observazioni microscopiche e deduzioni patologiche sul cholera asiatico. Gaz. Med. Ital. 6:405-412. [Google Scholar]
- 123.Reference deleted.
- 124.Pascual, M., X. Rodo, S. P. Ellner, R. R. Colwell, and M. J. Bouma. 2000. Cholera dynamics and El Niño-southern oscillation. Science 289:1766-1769. [DOI] [PubMed] [Google Scholar]
- 125.Patel, M., and M. Isaacson. 1999. The effect of iron on the toxigenicity of Vibrio cholerae. Am. J. Trop. Med. Hyg 60:392-396. [DOI] [PubMed] [Google Scholar]
- 126.Patel, M., M. Isaacson, and E. Gouws. 1995. Effect of iron and pH on the survival of Vibrio cholerae in water. Trans. R. Soc. Trop. Med. Hyg. 89:175-177. [DOI] [PubMed] [Google Scholar]
- 127.Politzer, R. 1959. Cholera. World Health Organization, Geneva, Switzerland.
- 128.Postgate, J. R. 1969. Viable counts and viability, p. 611-628. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology. Academic Press, Inc., London, United Kingdom.
- 129.Reference deleted.
- 130.Rees, R. 1996. Under the weather: climate and disease, 1700-1900. History Today 46:35-42. [Google Scholar]
- 131.Rhine, J. A., and R. K. Taylor. 1994. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol. Microbiol. 13:1013-1020. [DOI] [PubMed] [Google Scholar]
- 132.Rivera, I. G., M. A. Chowdhury, A. Huq, D. Jacobs, M. T. Martins, and R. R. Colwell. 1995. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl. Environ. Microbiol. 61:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodrigues, D. P., and E. Hofer. 1986. Vibrio species from the water-oyster ecosystem of Sepetitiba Bay in Rio de Janeiro State, Brazil. Rev. Microbiol. 17:332-338. [Google Scholar]
- 134.Romalde, J., J. L. Barja, and A. E. Toranzo. 1990. Vibrios associated with red tides caused by Mesodinium rubrum. Appl. Environ. Microbiol. 56:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rose, J. B., A. Huq, and E. K. Lipp. 2001. Health, climate and infectious disease: a global perspective. American Academy of Microbiology, Washington, D.C. [PubMed]
- 136.Rose, J. B., S. Daeschner, D. R. Easterling, F. C. Curriero, S. Lele, and J. A. Patz. 2000. Climate and waterborne disease outbreaks. J. Am. Water Works Assoc. 92:77-87. [Google Scholar]
- 137.Rose, J. B., P. R. Epstein, E. K. Lipp, B. H. Sherman, S. M. Bernard, and J. A. Patz. 2001. Climate variability and change in the United States: potential impacts on water- and foodborne diseases caused by microbiological agents. Environ. Health Perspect. 109:211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rosenberg, C. E. 1962. The cholera years. The University of Chicago Press, Chicago, Ill.
- 139.Schneider, D. R., and C. D. Parker. 1982. Purification and characterization of the mucinase of Vibrio cholerae. J. Infect. Dis. 145:474-482. [DOI] [PubMed] [Google Scholar]
- 140.Schulman, J. L., and E. D. Kilbourne. 1963. Experimental transmission of influenza virus infection in mice: some factors affecting the incidence of transmitted infection. J. Exp. Med. 118:267-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seas, C., J. Miranda, A. I. Gil, R. Leon-Baru, J. A. Patz, A. Huq, R. R. Colwell, and R. B. Sack. 2000. New insights on the emergence of cholera in Latin America during 1991: the Peruvian experience. Am. J. Trop. Med. Hyg. 62: 513-517. [DOI] [PubMed] [Google Scholar]
- 142.Shilo, M. 1975. Factors involved in dynamics of algal blooms in nature, p. 127-132. In W. H. van Dobben and R. H. Lowe-McConnell (ed.), Unifying concepts in ecology. Dr. W. Junk B.V. Publishers, The Hague, The Netherlands.
- 143.Shukla, B. N., D. V. Singh, and S. C. Sanyal. 1995. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol. Med. Microbiol. 12:113-120. [DOI] [PubMed] [Google Scholar]
- 144.Silvery, J. K. G., and A. W. Roach. 1964. Studies on microbiotic cycles in surface waters. J. Am. Water Works Assoc. 56:60-72. [Google Scholar]
- 145.Simidu, U., K. Ashino, and E. Kaneko. 1971. Bacterial flora of phyto- and zoo-plankton in the inshore water of Japan. Can. J. Microbiol. 17:1157-1160. [DOI] [PubMed] [Google Scholar]
- 146.Singh, D. V., M. H. Matte, G. R. Matte, S. Jiang, F. Sabeena, B. N. Shukla, S. C. Sanyal, A. Huq, and R. R. Colwell. 2001. Molecular analysis of Vibrio cholerae O1, O139, and non-O1 and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Singleton, F. L., R. W. Attwell, M. S. Jangi, and R. R. Colwell. 1982. Influence of salinity and organic nutrient concentration on survival and growth of Vibrio cholerae in aquatic microcosms. Appl. Environ. Microbiol. 43:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Singleton, F. L., R. W. Attwell, M. S. Jangi, and R. R. Colwell. 1982. Effects of temperature and salinity on Vibrio cholerae growth. Appl. Environ. Microbiol. 44:1047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Spira, W. M., A. Huq, O. S. Ahmed, and Y. A. Saeed. 1981. Uptake of Vibrio cholerae biotype eltor from contaminated water hyacinth (Eichornia crassipes). Appl. Environ. Microbiol. 42:550-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stevenson, L. H. 1978. A case for bacterial dormancy in aquatic systems. Microb. Ecol. 4:127-133. [DOI] [PubMed] [Google Scholar]
- 151.Tamplin, M. L., and R. R. Colwell. 1986. Effects of microcosm salinity and organic substrate concentration on production of Vibrio cholerae enterotoxin. Appl. Environ. Microbiol. 52:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tarsi, R., and C. Pruzzo. 1999. Role of surface proteins in Vibrio cholerae attachment to chitin. Appl. Environ. Microbiol. 65:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Torrence, C., and P. J. Webster. 1999. Interdecadal changes in the ENSO-monsoon system. J. Climate 12:2679-2690. [Google Scholar]
- 154.Torres Codeço, C. 2February2001. Endemic and epidemic dynamics of cholera: the role of the aquatic reservoir. BMC Infect. Dis. 1:1. [Online.] http://www.biomedcentral.com/1471-2334/1/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valentine, R. C., and J. R. G. Bradfield. 1954. The urea method for bacterial viability counts with electron microscope and its relation to other viability counting method. J. Gen. Microbiol. 11:349-357. [DOI] [PubMed] [Google Scholar]
- 156.Venkateswaran, K., T. Takai, I. M. Navarro, H. Nakano, H. Hasimoto, and R. J. Siebeling. 1989. Ecology of Vibrio cholerae non-O1 and Salmonella spp. and role of zooplankton in their seasonal distribution in Fukuyama coastal waters, Japan. Appl. Environ. Microbiol. 55:1591-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 158.Watkins, J. D., and A. Huq. 2001. Relationship between oceans and human health, p. 356-363. In C. E. Koop, C. E. Pearson, and M. R. Schwarz (ed.), Critical issues in global health. Jossey-Bass, San Francisco, Calif.
- 159.Wittman, R. J., and G. J. Flick. 1995. Microbial contamination of shellfish: prevalence, risk to human health and control strategies. Annu. Rev. Public Health 16:123-140. [DOI] [PubMed] [Google Scholar]
- 160.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.World Health Organization. 2000. Cholera. Wkly. Epidemiol. Rec. 31:249-256. [Google Scholar]
- 162.World Health Organization. 1999. Climate and health (working group meeting) W.H.O./SDE/OEH/99.6. World Health Organization, Geneva, Switzerland.
- 163.World Health Organization. 1999. El Niño and health. W.H.O./SDE/99.4. World Health Organization, Geneva, Switzerland.
- 164.Xu, H. S., N. C. Roberts, F. L. Singleton, R. W. Attwell, D. J. Grimes, and R. R. Colwell. 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 8:313-323. [DOI] [PubMed] [Google Scholar]
- 165.Yu, C., A. M. Lee, B. L. Bassler, and S. Roseman. 1991. Chitin utilization by marine bacteria: a physiological function for bacterial adhesion to immobilized carbohydrates. J. Biol. Chem. 266:24260-24267. [PubMed] [Google Scholar]