Abstract
Pseudomonas aeruginosa isolates from an outbreak in Canada were highly resistant to carbapenems and ceftazidime but not piperacillin. They produced a novel integron-associated metallo-β-lactamase, designated IMP-7, with 91% identity to IMP-1. blaIMP-7 was not detected with standard blaIMP-specific primers, owing to mismatches in the forward primer.
Multidrug resistance in Pseudomonas aeruginosa arises if a strain undergoes multiple mutations, derepressing chromosomal AmpC β-lactamases, up-regulating efflux, and decreasing permeability. Alternatively, P. aeruginosa can develop multidrug resistance by acquiring plasmids, transposons, or integrons that encode potent β-lactamases and aminoglycoside-modifying enzymes. Recent interest has centered on the emergence of strains with IMP and VIM metallo-β-lactamases (9). These enzymes hydrolyze most β-lactams, including carbapenems, and are encoded by integrons that often also specify aminoglycoside 6′-N-acetyltransferases (6, 11, 12).
Outbreaks of P. aeruginosa infection.
P. aeruginosa isolates resistant to carbapenems, aminoglycosides, ciprofloxacin, and ceftazidime but apparently susceptible to piperacillin were recovered during 1995 and 1996 from nine patients in two rehabilitation wards at the Bow Valley Center at Calgary General Hospital (CGH) in Calgary, Alberta, Canada. Most of the isolates were from cultures of urine from patients with spinal cord injuries. At the end of 1996, the rehabilitation program was moved to the Foothills Hospital (FHH), also in Calgary. From January to October 1997, 15 patients in FHH were identified with nosocomial acquisition of P. aeruginosa with a multidrug resistance phenotype similar to that seen at CGH in 1995 and 1999. Most, but not all, of the patients in FHH had direct contact with the rehabilitation or intensive care units. No such P. aeruginosa isolates had been encountered at FHH in 1996. Most were again from urine, but there were also isolates from burn wounds, surgical wounds, sacral and perineal ulcers, lower respiratory specimens, and external ear swabs. No isolates were from cultures of blood or specimens from other normally sterile sites. Cultures of environmental specimens did not reveal a source of the organism, and carbapenems were rarely used at either hospital. Isolation and infection control practices were reinforced on several occasions, and the outbreak ceased late in 1997.
Characterization of isolates.
Susceptibility was determined by various means (see below): with a Vitek system (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, Mo.), by broth microdilution with Pasco panels (Becton-Dickinson, Wheatridge, Colo.), by the Kirby-Bauer disk methodology, by agar dilution (10), and with the Etest (AB Biodisk Solna, Sweden). Isolates were tested by disk diffusion with ceftazidime in the presence of 2-mercaptopropionic acid to detect metallo-β-lactamases (1). Total DNA was fingerprinted by pulsed-field gel electrophoresis (PFGE) after digestion with SpeI (New England Biologicals, Beverly, Mass.) (15). Crude cell extracts were prepared by freeze-thawing and were assayed against 0.1 mM imipenem by spectrophotometry at 297 nm, as described previously (18). Conjugative transfer of carbapenem resistance to P. aeruginosa PU21 (5) was attempted by plate mating with selection on agar with rifampin (100 μg/ml) and meropenem (10 μg/ml).
Detection and analysis of metallo-β-lactamase genes.
PCR assays for blaIMP and blaVIM were performed by use of previously published primers and amplification conditions (13, 17). Genomic DNA fragments from a representative isolate (isolate 98/P/6327) were cloned into pBC SK(+) (Stratagene, Cambridge, United Kingdom). Recombinant plasmids were electroporated into Escherichia coli XL-1 Blue MRF′ (Stratagene), and transformants likely to contain β-lactamase genes were selected with ampicillin (10 μg/ml), as described previously (18). One recombinant plasmid was used for initial sequencing with primers specific for conserved class I integron sequences (7). The sequence obtained was compared with published sequences by using CLUSTAL W software (16).
Resistance phenotypes.
Multiple isolates of P. aeruginosa retrieved from CGH in 1995 and 1996 and FHH in 1997 had unusual antibiograms, with resistance to gentamicin, tobramycin, ciprofloxacin, and ceftazidime but with susceptibility to piperacillin, as determined by the criteria of NCCLS (10). Carbapenems were not routinely tested with the Vitek and Pasco panels used in the study. The isolates gave no zones around 10-μg imipenem or meropenem disks, nor did they give zones around those containing cefepime at 30 μg. Meropenem and ceftazidime MICs were both subsequently found to be >512 μg/ml by agar dilution. The MICs of all relevant drugs were determined in parallel for a representative group of strains with the Etest, confirming the resistance of the isolates to all drugs except piperacillin and piperacillin in combination with tazobactam (Table 1).
TABLE 1.
MICs determined with the Etest for the outbreak strain (three representative isolates), E. coli XL-1 Blue MRF′ containing vector pBC SK(+), and E. coli XL-1 Blue MRF′ containing vector pBC SK(+) with recombinant phagemid pARL98-3
| Antibiotic | MIC (μg/ml)
|
||
|---|---|---|---|
| Outbreak strain | pBC SK(+) | pARL98-3 (blaIMP-7) | |
| Imipenem | >32 | 0.25 | 0.25 |
| Meropenem | >32 | 0.012 | 0.125 |
| Ampicillin | —a | 4 | 32 |
| Aztreonam | 16 | 0.25 | 0.125 |
| Cefoxitin | — | 16 | >256 |
| Cefpirome | — | 0.125 | 1.0 |
| Ceftazidime | >256 | 0.25 | >256 |
| Piperacillin | 8–16 | 1 | 1 |
| Cefepime | >32 | — | — |
| Piperacillin-tazobactam | 8–16 | — | — |
| Tobramycin | 64–128 | — | — |
| Amikacin | 32 | — | — |
| Ciprofloxacin | >32 | — | — |
—, not done.
Isolates with this antibiogram from both CGH and FHH showed consistent PFGE banding profiles, indicating clonality; other P. aeruginosa isolates from the same period but with different antibiograms exhibited different banding patterns. Attempts to transfer carbapenem resistance to P. aeruginosa PU21 were unsuccessful.
Characterization of β-lactamases.
The level of resistance to ceftazidime was reduced in the presence of 2-mercaptopropionic acid, suggesting the involvement of a metallo-β-lactamase (1). Crude cell extracts from four representative isolates hydrolyzed 0.1 mM imipenem rapidly in spectrophotometric assays (8), confirming carbapenemase production. Nevertheless, PCR assays for blaIMP and blaVIM were repeatedly negative. Although the blaIMP-specific primers were originally designed to amplify blaIMP-1, they also detect blaIMP-2 (12), blaIMP-3 (4), and blaIMP-4 (3); likewise, the blaVIM-specific primers detect both blaVIM-1 (6) and blaVIM-2 (11).
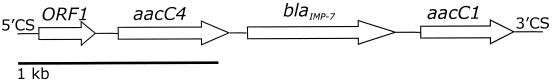
On the basis of this failure to detect alleles of known carbapenemase genes, genomic DNA from a representative isolate was cloned. A cell extract prepared from a selected transformant hydrolyzed imipenem, although phenotypic carbapenem resistance was not apparent (Table 1). The recombinant plasmid in this clone contained an insert of >10 kb, and the plasmid was designated pARL98.3. Sequencing of a 3.0-kb segment between the conserved sequences characteristic of type I integrons revealed four complete gene cassettes (Fig. 1). The first sequence was an unknown open reading frame with all the characteristics of an integron cassette (14): namely, a core site (GTTRRRY), an inverse core site, and a 59-bp element. The second and fourth cassettes contained aacC4 and aacC1, respectively, both of which encode aminoglycoside acetyltransferases. The third cassette, designated blaIMP-7, encoded a class B β-lactamase that shares >86% amino acid identity with known IMP enzymes (Fig. 2). The failure of the original blaIMP-specific primers to amplify blaIMP-7 may be explained by four mismatches among 20 bases in the forward primer (data not shown). The reverse primer showed complete homology. Alternative primers specific for blaIMP (1) also have mismatches with blaIMP-7 and so would probably not detect the new allele.
FIG. 1.
Orientation of class I integron cassettes in clone pARL98.3. 5′CS and 3′CS, conserved sequences of a class I integron; ORF1, unknown open reading frame; aacC4, aminoglycoside resistance gene; blaIMP-7, class B β-lactamase gene; aacC1, aminoglycoside resistance gene.
FIG. 2.
Sequence of IMP-7 β-lactamase in comparison to those of other published IMP variants.
Imipenem resistance was observed in 5.2% of 1,466 P. aeruginosa isolates during a recent survey in Canada (2). Mechanisms of resistance were not defined, but most of these isolates probably lacked the OprD porin; this mechanism, which arises by a simple point mutation, reduces meropenem susceptibility and confers imipenem resistance. Carbapenemase-mediated imipenem resistance in P. aeruginosa is much rarer and has not previously been reported from the Americas. The catalogue of acquired carbapenemases that have been reported is, however, growing sharply (9).
It remains uncertain whether IMP-7 was the sole cause of resistance in the isolates tested in the present study, and permeability lesions may also be needed in order for IMP enzymes to protect bacteria against carbapenems (13). Thus, cloned IMP-7 gave only slight protection against meropenem in E. coli XL-1 Blue and no protection against imipenem or aztreonam (Table 1), whereas resistance to ceftazidime and cefoxitin was evident in the transformants.
Microbiologists should be alert to the emergence of an integron-associated carbapenemase gene in P. aeruginosa in North America and to the risk that this will spread among regions and species. The need for infection control, which ultimately brought these outbreaks to an end, and for the cautious and prudent use of carbapenems should be underscored. The presence of two aminoglycoside resistance genes in the integron evaluated in the present study implies that it might be selected by these drugs as well as by β-lactams, and it is notable that neither CGH nor FHH used carbapenems extensively. Retention of moderate susceptibility to piperacillin by the isolates examined in the present study is interesting but anomalous, since IMP enzymes hydrolyze the compound, but was observed previously in some IMP-1 producers (13). The clinical efficacy of piperacillin against metallo-β-lactamase producers has not been demonstrated.
Imipenem MICs above 8 to 32 μg/ml should raise the possibility of the presence of a carbapenemase in P. aeruginosa, especially if the isolates are highly resistant to ceftazidime but not, perhaps, piperacillin. PCR has been used to screen for IMP and VIM genes, but as shown here, some blaIMP alleles are sufficiently divergent to preclude detection with standard primer pairs. Tests based on inhibition of metallo-β-lactamase activity by thiol compounds are better screens (1) but use compounds with significant toxicities. Because of this constraint and because some carbapenemases (e.g., SME-1, NMC-A, and OXA-23 to OXA-27) are not metalloenzymes, hydrolysis assays with crude cell extracts are probably the best first step in carbapenemase detection.
Nucleotide sequence accession number.
The sequence of the integron obtained in the present study was assigned GenBank accession no. AF318077.
REFERENCES
- 1.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondeau, J. M., R. Laskowski, and S. Borsos. 1999. In-vitro activity of cefepime and seven other antimicrobial agents against 1518 non-fermentative Gram-negative bacilli collected from 48 Canadian health care facilities. Canadian Afermenter Study Group. J. Antimicrob. Chemother. 44:545–548. [DOI] [PubMed] [Google Scholar]
- 3.Chu, Y. W., M. Afzal-Shah, E. T. S. Houang, M. F. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyobe, S., H. Kusadokoro, J. Ozaki, N. Matsumura, S. Minami, S. Haruta, T. Sawai, and K. O’Hara. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:2023–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacoby, G. A. 1974. Properties of an R plasmid in Pseudomonas aeruginosa producing amikacin (BB-K8), butirosin, kanamycin, tobramycin, and sisomicin resistance. Antimicrob. Agents Chemother. 6:807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore, D. M., and J. D. Williams. 1996. β-Lactams: mode of action and mechanisms of resistance, p. 502–578. In V. Lorian (ed.), Antibiotics in laboratory medicine. The Williams & Wilkins Co., Baltimore, Md.
- 9.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489–495. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing; ninth informational supplement. M100–S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669–1683. [DOI] [PubMed] [Google Scholar]
- 15.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial typing. J. Clin. Microbiol. 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsakris, A., S. Pournaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodford, N., M. F. Palepou, G. S. Babini, B. Holmes, and D. M. Livermore. 2000. Carbapenemases of Chryseobacterium (Flavobacterium) meningosepticum: distribution of blaB and characterization of a novel metallo-β-lactamase gene, blaB3, in the type strain, NCTC 10016. Antimicrob. Agents Chemother. 44:1448–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]